Abstract
Genome-wide association (GWA) studies of complex diseases including coronary heart disease (CHD) challenge investigators attempting to identify relevant genetic variants among hundreds of thousands of markers being tested. A selection strategy based purely on statistical significance will result in many false negative findings after adjustment for multiple testing. Thus, an integrated analysis using information from the learned genetic pathways, molecular functions, and biological processes is desirable. In this study, we applied a customized method, variable set enrichment analysis (VSEA), to the Framingham Heart Study data (404 467 variants, n=6421) to evaluate enrichment of genetic association in 1395 gene sets for their contribution to CHD. We identified 25 gene sets with nominal P<0.01; at least four sets are previously known for their roles in CHD: vascular genesis (GO:0001570), fatty-acid biosynthetic process (GO:0006633), fatty-acid metabolic process (GO:0006631), and glycerolipid metabolic process (GO:0046486). Although the four gene sets include 170 genes, only three of the genes contain a variant ranked among the top 100 in single-variant association tests of the 404 467 variants tested. Significant enrichment for novel gene sets less known for their importance to CHD were also identified: Rac 1 cell-motility signaling pathway (h_rac1 Pathway, P<0.001) and sulfur amino-acid metabolic process (GO:0000096, P<0.001). In summary, we showed that the pathway-based VSEA can help prioritize association signals in GWA studies by identifying biologically plausible targets for downstream searches of genetic variants associated with CHD.
Keywords: gene set enrichment, pathway-based analysis, SNP, genome-wide association
Introduction
The introduction of genome-wide association (GWA) represents a revolutionary advance in the genetic investigation of complex diseases. Yet, despite the early promise of GWA studies in cardiovascular and other complex diseases, reported effect sizes of single-nucleotide polymorphisms (SNPs) (both individual and cumulative) explain disappointingly small proportions of the estimated trait heritability.1, 2, 3 Nonetheless, the information gained from GWA studies continues to offer insight regarding the genetic and molecular mechanisms of disease.
The most common approaches to GWA studies focus on the analysis of individual SNPs and their neighboring genes; only the strongest evidence of association for top-ranked SNPs is typically reported. This approach is hampered by the consideration of large numbers of variables (ie, genotypes), the vast majority of which will not meet criteria for genome-wide significance and fewer still that will ultimately be functionally important. Thus, even in studies of large cohorts, true signals remain difficult to identify.
To increase the yield from GWA studies and to ultimately explain a great proportion of the trait heritability, analysis approaches should be adapted to capitalize on available complementary data that allow testing for association on the basis of functional units such as genes, gene sets, and pathways.4 These approaches would decrease the number of statistical tests while taking advantage of known biology. The pathway-based gene-expression analysis approach called gene-set enrichment analysis (GSEA) was adapted for use in GWA studies.5, 6, 7 This adaptation uses the maximum single-SNP test statistic from a gene to score the strength of association between the gene and the trait of interest. It then applied the GSEA procedure to test whether a certain gene set is significantly ‘enriched' with high-scored genes.
We recently presented a further extension of the GSEA method, variable set enrichment analysis (VSEA), which normalizes the maximum SNP statistics based on permutation results so that signals are comparable for genes that have different number of SNPs and different linkage disequilibrium structure.8 In this report, we hypothesize that by applying VSEA to GWA data from the Framingham Heart Study, we will identify gene sets associated with coronary heart disease (CHD) that would be otherwise missed by conventional single-SNP analyses.
Materials and methods
Framingham Heart Study genome-wide data
The Framingham Heart Study is a large-scale population-based cardiovascular study based in Framingham, Massachusetts, USA, which started in 1948 and currently consists of three generations of cohorts.9 To find the pathways related to the CHD, we used the genome-wide data (Affymetrix 5.0 GeneChip array with a 500K SNPs, Santa Clara, CA, USA) from Caucasian individuals representing the three cohorts from the Framingham SHARe project (SNP Health Association Resource) downloaded from the National Center for Biotechnology Information database of Genotypes and Phenotypes website (http://www.ncbi.nlm.nih.gov/gap). The analysis was performed on the Framingham Cohort data, version 4 (embargo release date 4 December 2009). The primary phenotype, prevalent CHD, was defined by the Framingham Heart Study as a composite of recognized and unrecognized myocardial infarction, coronary insufficiency, and CHD death.10
Quality control
The original dataset included 6476 Caucasian subjects (2959 men and 3517 women) and 498 014 SNPs. Mendelian genotype errors were checked and those with errors were set to missing. The quality of the subjects' dataset was then verified: subjects with missing rate >5% and either low (≤25%) or high (≥30%) mean heterozygosity were removed from the dataset, resulting in 6438 individuals. Next, the qualities of individual SNPs were checked: monomorphic SNPs, SNPs with missing rate >5% or a missing rate >1% combined with a minor allele frequency <0.05, those with a Hardy–Weinberg equilibrium test P-value <10−6, and those without an annotated geneID were removed, resulting in 404 467 SNPs for the association analysis. Finally, population substructure was examined by multidimensional scaling using information from the HapMap samples of European (CEU), East Asian (CHB and JPT), and African (YRI) origins. A total of 17 subjects were removed because of poor clustering with CEU subjects. The final analyzable dataset included 6421 subjects (2935 males and 3486 females).
VSEA
VSEA is a novel GWA analysis method that tests for aggregated effect of many genes linked by biological functions or statistical gene–gene interaction.8 It is based on the method called GSEA, originally developed for differential gene expression analysis. GSEA derives an enrichment score to detect gene sets significantly enriched with differentially expressed genes.5 To facilitate analysis of SNP data in GWA studies, VSEA employs a permutation-based normalized gene score to aggregate effects of multiple individual SNPs in each gene of a gene set. Permutation was done 1000 times first by calculating the enrichment scores from the datasets where the disease status was randomly shuffled. The P-values were then calculated from the frequency of seeing a larger enrichment score in the observed than in the shuffled dataset.8
For the VSEA analyses described in this paper, we used a library of 1395 gene sets compiled from the collections of the genetic pathways, molecular functions, and/or biological processes in the Kyoto Encyclopedia of Genes and Genomes (www.genome.ad.jp/kegg/pathway.html), BioCarta (http://www.biocarta.com/Default.aspx), and Gene Ontology (http://geneontology.org/) databases.8 Based on the manufacturer's annotation of the Affymetrix 500K GeneChip array, we refined the gene-set library by removing genes that had no SNP included on the genotyping array. To reduce the impact of multiple testing and to avoid testing overly narrow or broad functional categories, our analysis only considered gene sets and the pathways that contained at least 3 and at most 200 genes. The final panel included 1395 gene sets representing 404 467 SNPs, which were attributed to 15 474 genes.
Single-SNP and pairwise SNP–SNP interactions
The aggregated effect of multiple genes in a gene set may reflect the sum of individual gene effects, interactions of pairs or more genes, or both. To allow comparison of VSEA with conventional analytical approaches, genome-wide single-SNP association was determined using the allelic χ2 test by PLINK for prevalent CHD.11 Familial relationships in the sample were ignored based on a previous study that found very similar association test P-values in the Framingham GWAS data, whether familial relationships were considered or omitted.12 This enabled us to perform the large number of permutation tests in a practical time. An earlier simulation study evaluating the effect of such practice (ignoring familial relationships) in association analysis found that the effect-size estimates and power are not significantly affected, although Type I error rates increase as the disease heritability increases.13
The contribution of pairwise SNP–SNP interactions to the aggregated effect detected by VSEA was assessed by analyzing pairwise interactions between SNPs from genes in the highest-ranked gene sets after the VSEA test was performed. Pairwise SNP–SNP interactions were detected by significant difference between genotype correlation in cases and that in controls using Fisher's Z transformation.14, 15, 16, 17 The significant difference of correlations between cases and controls reflects altered pairing preferences of alleles at the two loci and may be the result of some underlying molecular mechanism that was active in CHD. To entertain the idea of such underlying mechanisms, we performed interaction analysis of pairs of SNPs in top-ranked gene sets and organized all the significant pairwise interactions detected in a gene set into clusters (networks) of genes linked by SNP interactions.
Multiple testing
There is no generally accepted method of adjustment for testing the large number of the distinct pathways considered in this study. The VSEA procedure corrects for multiple testing due to genes shared by the different pathways/gene sets. However, adjustment of P-values for testing many distinct gene sets may lead to overly conservative results, especially when using gene sets derived from general-purpose databases (as was done in this study), because gene sets often contain many genes that are irrelevant to the disease trait of interest. Therefore, we used the unadjusted nominal P-values for the VSEA analyses. To prevent potential false positives in our validation analysis of pairwise SNP–SNP interactions among the genes in top-ranked gene sets, we imposed the stringent Bonferroni correction for multiple testing.
Results
After performing data quality control, the analysis sample consisted of 6421 subjects (2935 males and 3486 females) with 404 467 SNPs, of which 326 750 SNPs are associated to 15 474 genes. After removing gene sets that are too small or too large, the final panel includes 1395 gene sets, representing 207 120 SNPs, which were attributed to 8161 genes. Sample characteristics of several known risk factors of CHD are shown in Table 1. CHD events were identified in 221 individuals. Compared with subjects without CHD events (non-CHD controls), CHD cases were more likely to have higher systolic and diastolic blood pressure, total cholesterol and triglyceride, and lower high-density lipoprotein cholesterol.
Table 1. Population characteristics of the Framingham Heart Study population used in the current study.
| CHD cases | Non-CHD controls | P-valuea | |
|---|---|---|---|
| n=221 | n=6200 | ||
| Female, n (%) | 74 (33%) | 3412 (55%) | |
| Age, years | 39±8 | 38±9 | |
| Body mass index, kg/m2 | 26±4 | 26±5 | 0.71 |
| Systolic blood pressure, mmHg | 125±14 | 118±14 | 5.3 × 10−13 |
| Diastolic blood pressure, mmHg | 80±10 | 76±10 | 2.7 × 10−9 |
| Total cholesterol, mmol/dl | 5.3±1.1 | 3.1±0.9 | 2.9 × 10−7 |
| High-density lipoprotein cholesterol, mmol/dl | 1.2±0.3 | 1.4±0.4 | 2.0 × 10−9 |
| Triglyceride, mmol/dl | 1.5±1.3 | 1.2±0.9 | 7.7 × 10−4 |
P-values after age- and sex-adjustment.
Among the 1395 gene sets tested, we identified 25 sets with a permutated P-value <0.01 (Table 2; top 100 gene sets available in Supplementary Table S1). Among the 25 gene sets, four (shown in bold) have been previously implicated in CHD by their participation in lipid metabolism and vascular genesis: fatty-acid biosynthetic process (GO:0006633), fatty-acid metabolic process (GO:0006631), glycerolipid metabolic process (GO:0046486), and vascular genesis (GO:0001570). The identification of these gene sets is supported by the existing body of literature linking these biological processes with atherosclerosis. Among the 170 genes represented by these four gene sets (Supplementary Table S2), only three contained any SNP ranked among top 100 in the single-SNP scan (Supplementary Table S3).
Table 2. Top 25 gene sets from the pathway-based VSEA test.
| Gene set | Pathway ID | P-value | Number of genes | Average single-SNP rank |
|---|---|---|---|---|
| Rac 1 cell-motility signaling pathway | h_rac1Pathway | <0.001 | 18 | 56 573 |
| Actinin binding | GO:0042805 | <0.001 | 4 | 1821 |
| Sulfur amino-acid metabolic process | GO:0000096 | <0.001 | 17 | 72 336 |
| Vasculogenesis | GO:0001570 | <0.001 | 17 | 69 199 |
| Fatty-acid biosynthetic process | GO:0006633 | 0.001 | 51 | 66 381 |
| Neuron differentiation | GO:0030182 | 0.001 | 106 | 47 424 |
| Gene silencing | GO:0016458 | 0.002 | 17 | 79 722 |
| Estrogen-responsive protein Efp controls cell cycle and breast tumors growth | h_EfpPathway | 0.003 | 6 | 37 582 |
| Calpain and friends in cell spread | h_ucalpainPathway | 0.003 | 11 | 41 681 |
| Rho cell-motility signaling pathway | h_rhoPathway | 0.004 | 17 | 78 688 |
| Phosphoinositide binding | GO:0035091 | 0.004 | 163 | 62 020 |
| Limonene and pinene degradation | hsa00903 | 0.005 | 28 | 54 080 |
| rRNA binding | GO:0019843 | 0.005 | 14 | 58 365 |
| Fatty-acid metabolic process | GO:0006631 | 0.006 | 120 | 67 524 |
| Mitosis | GO:0007067 | 0.006 | 152 | 86 502 |
| Drug transport | GO:0015893 | 0.006 | 13 | 36 267 |
| Hydrolyase activity | GO:0016836 | 0.007 | 47 | 72 672 |
| Two-component signal transduction system (phosphorelay) | GO:0000160 | 0.007 | 8 | 43 192 |
| Response to bacterium | GO:0009617 | 0.007 | 12 | 27 606 |
| Ubiquitin-mediated proteolysis | hsa04120 | 0.008 | 39 | 67 463 |
| Histone deacetylase activity | GO:0004407 | 0.008 | 12 | 81 299 |
| Semaphorin receptor activity | GO:0017154 | 0.008 | 5 | 5989 |
| DNA repair | GO:0006281 | 0.009 | 200 | 79 037 |
| Glycerolipid metabolic process | GO:0046486 | 0.01 | 21 | 39 609 |
| Myelination | GO:0042552 | 0.01 | 19 | 53 658 |
Bolded gene sets previously implicated in CHD.
The pathways shown underlined in Table 2 are examples of novel gene sets. Although these gene sets are less known for their association with CHD, a pathophysiological role in cardiovascular diseases is plausible. For example, many of the genes in the Rac 1 cell-motility signaling pathway (h_rac1 Pathway) are myosin-/actin-associated genes that have been shown to have roles in left ventricular hypertrophy and hypertrophic cardiomyopathy (RAC 1, MYL2, TRIO, and PPP1R12B). Other genes in this pathway have been shown to modulate cardiovascular risk traits including insulin sensitivity, glucose tolerance, and obesity (PIK3CB and RPS6KB1). Similarly, genes involved in the sulfur amino-acid metabolic process (GO:0000096) are related to cardiovascular diseases, through roles in oxidative stress (GCLC, GCLM, and MSRA) and/or metabolism of homocysteine (GCLC, BHMT, MTHFR, MTR, MRTT, and CBS), a well-known risk factor of CHD. There are also a few genes related to oxidative stress (CDO1, ADI1, and SOUX), although their roles in cardiovascular disease are not well studied.
Performance of genes in single-SNP analysis
Genome-wide single-SNP association was determined to allow comparison of VSEA with conventional analytical approaches. As an example of the effectiveness of VSEA to identify gene sets potentially relevant to CHD, the best-ranked single-SNP by the χ2 test from genes in the vasculogenesis pathway are shown in Table 3. Except for genes QKI, HEY2, and WARS2, the other genes in this pathway are not ranked highly, thus, these genes would likely be excluded from further follow-up studies if selection was based solely on the significance of the single-SNP test. However, when the VSEA test considered the 17 genes as a unit, their small marginal effects were combined, thus allowing this gene set to be identified as one significantly ‘enriched' for genetic association with CHD. Upon further examination, other top-ranked gene sets showed similar patterns of predominantly weak single-SNP rankings.
Table 3. Ranks of SNPs in the vasculogenesis pathway (GO:0001570).
| Gene | Best SNP rank a | Number of SNPs |
|---|---|---|
| AGGF1 | 41 202 | 6 |
| AMOT | 34 660 | 10 |
| CCM2 | 55 707 | 9 |
| CITED1 | 101 443 | 3 |
| CUL7 | 177 116 | 2 |
| EGFL7 | 139 662 | 3 |
| FOXF2 | 20 621 | 40 |
| GLMN | 215 819 | 1 |
| HEY2 | 55 | 80 |
| KDR | 88 447 | 20 |
| QKI | 3 | 99 |
| RASA1 | 24 703 | 9 |
| SHH | 7393 | 22 |
| SMO | 183 604 | 2 |
| VEGFA | 73 822 | 12 |
| WARS2 | 354 | 24 |
| WT1 | 11 776 | 73 |
Out of 404 467 SNPs ranked.
Pairwise interactions among genes from enriched gene sets
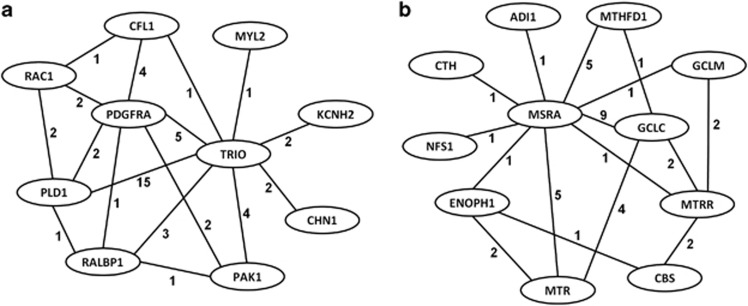
Using the VSEA test, 1005 distinct genes were identified from among the top 25 gene sets. This constitutes a total of 15 960 SNPs after removing those in high LD (r2>0.8), which resulted in 119 209 805 pairwise interaction tests. Using a stringent Bonferroni-adjusted significance level (P<4.2 × 10−10), 439 of the 1005 genes were linked by cross-gene SNP–SNP interactions. When these cross-gene SNP–SNP interactions were superimposed over the top-ranked gene sets, we obtained clusters (subnetworks) of genes within these pathways that reflected concerted action of multiple genes that differentiated the CHD group from the non-CHD group (Figure 1 for interaction subnetworks from the Rac 1 cell-motility signaling and sulfur amino-acid metabolic process pathways).
Figure 1.
Subnetworks of genes from the Rac 1 cell-motility signaling (a) and sulfur amino-acid metabolic process (b) pathways that differentiated the CHD from the non-CHD group. Subnetworks are composed of pairwise SNP–SNP interactions where the number on each edge (line) represents the number of interactions between each pair of genes.
Genes participating in pairwise interactions were then ranked by the number of other gene interaction partners. Genes interacting with ≥30 partners are listed in Table 4. Once again, VSEA has identified many genes with important roles in cardiovascular diseases including two genes (CDH13 and PARD3), which have been recently associated with CHD risk traits.
Table 4. Genes from enriched gene sets with highest number of interaction partners.
| Gene | GeneID | Chromosome | Number of SNPs | Number of interaction partners |
|---|---|---|---|---|
| CDH13 | 1012 | 16 | 575 | 119 |
| DCC | 1630 | 18 | 487 | 80 |
| PARD3 | 56 288 | 10 | 275 | 66 |
| NRP1 | 8829 | 10 | 244 | 65 |
| CD109 | 135 228 | 6 | 228 | 64 |
| GPC5 | 2262 | 13 | 314 | 63 |
| SYT1 | 6857 | 12 | 167 | 61 |
| HDAC9 | 9734 | 7 | 158 | 60 |
| CDH4 | 1002 | 20 | 367 | 60 |
| CA10 | 56 934 | 17 | 371 | 58 |
| SMC2 | 10 592 | 9 | 260 | 57 |
| TNP1 | 7141 | 2 | 214 | 56 |
| PTPRG | 5793 | 3 | 239 | 55 |
| ROBO2 | 6092 | 3 | 215 | 54 |
| CA8 | 767 | 8 | 131 | 53 |
| PPARGC1A | 10 891 | 4 | 264 | 52 |
| TLR4 | 7099 | 9 | 142 | 52 |
| PIK3R1 | 5295 | 5 | 253 | 50 |
| SLC46A2 | 57 864 | 9 | 42 | 49 |
| CNTN4 | 152 330 | 3 | 219 | 47 |
| BRUNOL4 | 56 853 | 18 | 307 | 47 |
| NEGR1 | 257 194 | 1 | 152 | 46 |
| UBE2E2 | 7325 | 3 | 157 | 45 |
| OPCML | 4978 | 11 | 285 | 45 |
| EFNA5 | 1946 | 5 | 183 | 44 |
| CNTN5 | 53 942 | 11 | 257 | 44 |
| ROBO1 | 6091 | 3 | 228 | 40 |
| PTK2 | 5747 | 8 | 46 | 39 |
| CETN3 | 1070 | 5 | 126 | 39 |
| GFRA1 | 2674 | 10 | 90 | 38 |
| CNTN6 | 27 255 | 3 | 165 | 38 |
| MSRA | 4482 | 8 | 127 | 38 |
| GP2 | 2813 | 16 | 51 | 37 |
| ERCC4 | 2072 | 16 | 193 | 37 |
| C20orf23 | 55 614 | 20 | 285 | 36 |
| NRP2 | 8828 | 2 | 120 | 35 |
| SNX16 | 64 089 | 8 | 60 | 35 |
| CD36 | 948 | 7 | 314 | 35 |
| SNAG1 | 112 574 | 5 | 63 | 34 |
| ALOX5AP | 241 | 13 | 132 | 34 |
| IREB2 | 3658 | 15 | 67 | 33 |
| CNTN1 | 1272 | 12 | 23 | 33 |
| CXCR4 | 7852 | 2 | 75 | 32 |
| EPHA7 | 2045 | 6 | 187 | 32 |
| PALLD | 23 022 | 4 | 176 | 31 |
| CCKAR | 886 | 4 | 135 | 31 |
| SLITRK5 | 26 050 | 13 | 28 | 31 |
| DSCAML1 | 57 453 | 11 | 444 | 31 |
| SEMA3A | 10 371 | 7 | 95 | 31 |
| NCAM1 | 4684 | 11 | 140 | 30 |
| THY1 | 7070 | 11 | 86 | 30 |
| GPC6 | 10 082 | 13 | 38 | 30 |
| RAD51L1 | 5890 | 14 | 293 | 30 |
| LIG4 | 3981 | 13 | 159 | 30 |
Discussion
The purpose of this study was to apply a novel method, VSEA, which capitalizes on existing biological data to gain new insight about CHD genetics by testing for association on the basis of functional units such as gene sets and pathways beyond individual SNPs. We identified gene sets enriched with genes that have been previously associated with CHD. We also discovered gene sets with emerging evidence supporting roles in a variety of cardiovascular diseases and related illnesses. Importantly, many CHD genes ranked poorly in single-SNP tests, whereas their member groups were successfully picked up by analyzing pathway-based gene sets. Thus, VSEA identified gene sets that would have been otherwise missed by conventional single-SNP analyses.
There is ample evidence to support the biological plausibility of association with CHD among the identified enriched gene sets. Among the 25 sets with a permutated P-value<0.01, some pathways have been previously linked with CHD and/or CHD risk factors. For example, among genes from the vasculogenesis pathway, published reports have shown that SNPs in VEGFA modulate atherosclerosis severity and the prevalence of myocardial infarction.18, 19 Likewise, WARS2 was recently identified in a meta-analysis of GWA studies for adiposity, a CHD risk factor.20 SNPs in genes from the fatty-acid biosynthetic and metabolic process pathways, particularly those participating in the synthesis of prostaglandins (eg, ALOX5, ALOX5AP, ALOX12, ALOX15, PTGS1, PTGS2, and COX2), have also been identified as risk factors for atherosclerotic plaque burden and CHD events.21, 22, 23, 24, 25, 26 SNPs in other genes from these pathways modulate CHD risk, presumably through their effects on lipids.27, 28, 29, 30, 31 Thus, inclusion of these genes among enriched gene sets is supported by existing scientific literature.
Among genes in sets with less-well characterized associations with CHD, a few have recently been linked with CHD and/or CHD risk factors. For example, a functional promoter polymorphism in GCLC, a member of the sulfur amino-acid metabolic process pathway, has been associated with endothelium-dependent dilation of coronary arteries and myocardial infarction.32 GCLM and MSRA, scavengers of reactive oxygen species, were recently found to protect the myocardium from ischemia-reperfusion injury, a critical determinant of survival following myocardial infarction.33, 34, 35 Several other genes in this pathway (eg, BHMT, MTHFR, MTR, MRTT, and CBS) regulate the metabolism of homocysteine, a risk factor of CHD.36, 37, 38, 39, 40, 41 Many genes in this gene set (GCLC, GCLM, MTHFR, MTHFD1, MTR, and MTRR) are also related to methylation processes, a potential, but understudied, mechanism for CVD.42, 43
Several genes in the Rac 1 cell-motility signaling pathway have also been recently implicated in CHD. For example, Rac 1, a subunit of the Nox2 NADPH oxidase enzyme which is responsible for generating damaging reactive oxygen species in the heart, has been mechanistically linked with ischemia-reperfusion injury, adverse remodeling of the left ventricle, and survival in transgenic mice following myocardial infarction.44, 45, 46 Genes from the Rac 1 pathway that encode actin- and myosin-associated proteins, including RAC 1 and MYL2, have also been associated with left ventricular mass, an intermediate traits that is a known risk factor for cardiovascular morbidity and mortality.47, 48 Also notable is that among the genes with the greatest number of cross-gene interactions, two of the top-ranked genes (CDH13 and PARD3) have been recently associated with CHD risk traits, including left ventricular hypertrophy, dyslipidemia, metabolic syndrome, type 2 diabetes, and adiponectin levels.7, 49, 50, 51
We note that similar gene-set enrichment approaches were used by others to evaluate particular pathways52 or to prioritize candidate genes.53 But, there is an inherent difficulty in defining the potential relevance of any pathway to a specific disease process. Incorporating more specific types of biological functions such as protein–protein interactions as done by Jensen et al54 will certainly improve the functional relevance of detected gene sets. In the absence of well-informed disease-based pathway databases, it is difficult to give an unbiased assessment of validity of the results. Although for some gene sets the relatedness to the disease trait may be more certain, for the majority this is less clear or unknown. The four gene sets identified as relevant to CHD were highlighted based on existing literature. Ultimately, functional studies are necessary to confirm the biological relevance of genetic variation in these pathways to CHD.
In summary, the present study shows the use of VSEA as a robust novel extension to existing analysis methods for GWA data. This study confirmed the interplay of multiple loci among the genes in the pathways responsible for CHD. More importantly, it also showed that analysis methods that capitalize on existing knowledge and directly test for gene–gene interactions can allow an improved understanding of the genetic variants and the pathways responsible for CHD.
Acknowledgments
This research is supported in part by the NIH grants HL091028, HL071782, HL094668 and an AHA grant 0855626G. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (contract no. N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ. et al:Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Weder AB, Eskin E, O'Connor DT. Genome-wide case/control studies in hypertension: only the 'tip of the iceberg'. J Hypertens. 2010;28:1115–1123. doi: 10.1097/HJH.0b013e328337f6bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK. et al:Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Li W, Zheng D. et al:Comparative analysis of mRNA isoform expression in cardiac hypertrophy and development reveals multiple post-transcriptional regulatory modules. PLoS One. 2011;6:e22391. doi: 10.1371/journal.pone.0022391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, de las Fuentes L, Davila-Roman VG, Charles GuC. Variable set enrichment analysis in genome-wide association studies. Eur J Hum Genet. 2011;19:893–900. doi: 10.1038/ejhg.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples LA, Arruda HT, Benjamin EJ.et al:The Framingham Heart Study 100 K SNP genome-wide association study resource: overview of 17 phenotype working group reports BMC Med Genet 20078Suppl 1S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, Wolf PA, Garrison RJ. Monograph chapter 34: some risk factor related to the annual incidence of cardiovascular disease and death using pooled repeated biennial measurements: framingham heart study, 30-year follow-up. Springfield, MA National Technical Information Service; 1987. pp. 1–459. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K. et al:PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Abo RP, Wong J, Thomas A, Camp NJ.Pedigree association: assigning individual weights to pedigree members for genetic association analysis BMC Proc 20093Suppl 7 S121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle PF, O'Connell JR, Pollin TI. et al:Accounting for relatedness in family based genetic association studies. Hum Hered. 2007;64:234–242. doi: 10.1159/000103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinbaum DG, Kupper LL. Applied regression analysis and other multivariable methods. North Scituate, MA Duxbury Press; 1978. [Google Scholar]
- Horwitz B. Simulating functional interactions in the brain: a model for examining correlations between regional cerebral metabolic rates. Int J Biomed Comput. 1990;26:149–170. doi: 10.1016/0020-7101(90)90039-w. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jin L, Xiong M. Test for interaction between two unlinked loci. Am J Hum Genet. 2006;79:831–845. doi: 10.1086/508571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell WM, Ali S, Rose-Zerilli MJ, Ye S. VEGF polymorphisms and severity of atherosclerosis. J Med Genet. 2005;42:485–490. doi: 10.1136/jmg.2004.025734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic D, Verhovec R, Globocnik Petrovic M, Osredkar J, Peterlin B. Association of vascular endothelial growth factor gene polymorphism with myocardial infarction in patients with type 2 diabetes. Cardiology. 2007;107:291–295. doi: 10.1159/000099064. [DOI] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC. et al:Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsel-Nitschke P, Gotz A, Medack A. et al:Genetic variation in the arachidonate 5-lipoxygenase-activating protein (ALOX5AP) is associated with myocardial infarction in the German population. Clin Sci (Lond) 2008;115:309–315. doi: 10.1042/CS20070468. [DOI] [PubMed] [Google Scholar]
- Lee CR, North KE, Bray MS, Couper DJ, Heiss G, Zeldin DC. Cyclooxygenase polymorphisms and risk of cardiovascular events: the Atherosclerosis Risk in Communities (ARIC) study. Clin Pharmacol Ther. 2008;83:52–60. doi: 10.1038/sj.clpt.6100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudock ME, Liu Y, Ziegler JT. et al:Association of polymorphisms in cyclooxygenase (COX)-2 with coronary and carotid calcium in the Diabetes Heart Study. Atherosclerosis. 2009;203:459–465. doi: 10.1016/j.atherosclerosis.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Net JB, Versmissen J, Oosterveer DM. et al:Arachidonate 5-lipoxygenase-activating protein (ALOX5AP) gene and coronary heart disease risk in familial hypercholesterolemia. Atherosclerosis. 2009;203:472–478. doi: 10.1016/j.atherosclerosis.2008.07.025. [DOI] [PubMed] [Google Scholar]
- Burdon KP, Rudock ME, Lehtinen AB. et al:Human lipoxygenase pathway gene variation and association with markers of subclinical atherosclerosis in the diabetes heart study. Mediators Inflamm. 2010;2010:170153. doi: 10.1155/2010/170153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegener HH, Diehl KA, Kurth T, Gaziano JM, Ridker PM, Zee RY. Polymorphisms of prostaglandin-endoperoxide synthase 2 gene, and prostaglandin-E receptor 2 gene, C-reactive protein concentrations and risk of atherothrombosis: a nested case-control approach. J Thromb Haemost. 2006;4:1718–1722. doi: 10.1111/j.1538-7836.2006.02054.x. [DOI] [PubMed] [Google Scholar]
- Love-Gregory L, Sherva R, Schappe T. et al:Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Hum Mol Genet. 2011;20:193–201. doi: 10.1093/hmg/ddq449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles JW, Wang H, Itakura H. et al:Association of polymorphisms in platelet and hemostasis system genes with acute myocardial infarction. Am Heart J. 2007;154:1052–1058. doi: 10.1016/j.ahj.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguri M, Kato K, Yokoi K. et al:Association of genetic variants with myocardial infarction in Japanese individuals with metabolic syndrome. Atherosclerosis. 2009;206:486–493. doi: 10.1016/j.atherosclerosis.2009.02.037. [DOI] [PubMed] [Google Scholar]
- Hegener HH, Lee IM, Cook NR, Ridker PM, Zee RY. Association of adiponectin gene variations with risk of incident myocardial infarction and ischemic stroke: a nested case-control study. Clin Chem. 2006;52:2021–2027. doi: 10.1373/clinchem.2006.074476. [DOI] [PubMed] [Google Scholar]
- Anand SS, Xie C, Pare G. et al:Genetic variants associated with myocardial infarction risk factors in over 8000 individuals from five ethnic groups: The INTERHEART Genetics Study. Circ Cardiovasc Genet. 2009;2:16–25. doi: 10.1161/CIRCGENETICS.108.813709. [DOI] [PubMed] [Google Scholar]
- Koide S, Kugiyama K, Sugiyama S. et al:Association of polymorphism in glutamate-cysteine ligase catalytic subunit gene with coronary vasomotor dysfunction and myocardial infarction. J Am Coll Cardiol. 2003;41:539–545. doi: 10.1016/s0735-1097(02)02866-8. [DOI] [PubMed] [Google Scholar]
- Zhao H, Sun J, Deschamps AM. et al:Myristoylated methionine sulfoxide reductase a protects the heart from ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2011;301:H1513–H1518. doi: 10.1152/ajpheart.00441.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice HM, Moench IA, Rickaway ZT, Dougherty CJ, Webster KA, Weissbach H. MSRA protects cardiac myocytes against hypoxia/reoxygenation induced cell death. Biochem Biophys Res Commun. 2008;366:775–778. doi: 10.1016/j.bbrc.2007.12.043. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Watanabe Y, Saito Y. et al:Mice lacking the glutamate-cysteine ligase modifier subunit are susceptible to myocardial ischaemia-reperfusion injury. Cardiovasc Res. 2010;85:785–795. doi: 10.1093/cvr/cvp342. [DOI] [PubMed] [Google Scholar]
- Rallidis LS, Gialeraki A, Komporozos C. et al:Role of methylenetetrahydrofolate reductase 677C->T polymorphism in the development of premature myocardial infarction. Atherosclerosis. 2008;200:115–120. doi: 10.1016/j.atherosclerosis.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Ilhan N, Kucuksu M, Kaman D, Ozbay Y. The 677 C/T MTHFR polymorphism is associated with essential hypertension, coronary artery disease, and higher homocysteine levels. Arch Med Res. 2008;39:125–130. doi: 10.1016/j.arcmed.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Ma J, Stampfer MJ, Hennekens CH. et al:Methylenetetrahydrofolate reductase polymorphism, plasma folate, homocysteine, and risk of myocardial infarction in US physicians. Circulation. 1996;94:2410–2416. doi: 10.1161/01.cir.94.10.2410. [DOI] [PubMed] [Google Scholar]
- Lewis SJ, Ebrahim S, Davey Smith G. Meta-analysis of MTHFR 677C->T polymorphism and coronary heart disease: does totality of evidence support causal role for homocysteine and preventive potential of folate. BMJ. 2005;331:1053. doi: 10.1136/bmj.38611.658947.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg IS, Park E, Ballman KV. et al:Investigations of a common genetic variant in betaine-homocysteine methyltransferase (BHMT) in coronary artery disease. Atherosclerosis. 2003;167:205–214. doi: 10.1016/s0021-9150(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Klerk M, Lievers KJ, Kluijtmans LA. et al:The 2756A>G variant in the gene encoding methionine synthase: its relation with plasma homocysteine levels and risk of coronary heart disease in a Dutch case-control study. Thromb Res. 2003;110:87–91. doi: 10.1016/s0049-3848(03)00341-4. [DOI] [PubMed] [Google Scholar]
- de Vogel S, Wouters KA, Gottschalk RW. et al:Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3086–3096. doi: 10.1158/1055-9965.EPI-09-0289. [DOI] [PubMed] [Google Scholar]
- Williams KT, Schalinske KL. Tissue-specific alterations of methyl group metabolism and DNA hypermethylation in the Zucker (type 2) diabetic fatty rat. Diabetes Metab Res Rev. 2011;28:123–131. doi: 10.1002/dmrr.1281. [DOI] [PubMed] [Google Scholar]
- Shan L, Li J, Wei M. et al:Disruption of Rac1 signaling reduces ischemia-reperfusion injury in the diabetic heart by inhibiting calpain. Free Radic Biol Med. 2010;49:1804–1814. doi: 10.1016/j.freeradbiomed.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Doerries C, Grote K, Hilfiker-Kleiner D. et al:Critical role of the NAD(P)H oxidase subunit p47phox for left ventricular remodeling/dysfunction and survival after myocardial infarction. Circ Res. 2007;100:894–903. doi: 10.1161/01.RES.0000261657.76299.ff. [DOI] [PubMed] [Google Scholar]
- Looi YH, Grieve DJ, Siva A. et al:Involvement of Nox2 NADPH oxidase in adverse cardiac remodeling after myocardial infarction. Hypertension. 2008;51:319–325. doi: 10.1161/HYPERTENSIONAHA.107.101980. [DOI] [PubMed] [Google Scholar]
- Koren MJ, Devereux RB, Casale PN, Savage DD, Laragh JH. Relation of left ventricular mass and geometry to morbidity and mortality in uncomplicated essential hypertension. Ann Intern Med. 1991;114:345–352. doi: 10.7326/0003-4819-114-5-345. [DOI] [PubMed] [Google Scholar]
- Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- Jee SH, Sull JW, Lee JE. et al:Adiponectin concentrations: a genome-wide association study. Am J Hum Genet. 2010;87:545–552. doi: 10.1016/j.ajhg.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CM. et al:Trait locus of adiponectin on CDH13 that predicts cardiometabolic outcomes. Diabetes. 2011;60:2417–2423. doi: 10.2337/db10-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E, Eyheramendy S, Juhanson P. et al:Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18:2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre AV, Diagram Consortium, MAGIC investigators et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010. p. 6. [DOI] [PMC free article] [PubMed]
- Pers TH, Hansen NT, Lage K. et al:Meta-analysis of heterogeneous data sources for genome-scale identification of risk genes in complex phenotypes. Genet Epidemiol. 2011;35:318–332. doi: 10.1002/gepi.20580. [DOI] [PubMed] [Google Scholar]
- Jensen MK, Pers TH, Dworzynski P, Girman CJ, Brunak S, Rimm EB. Protein interaction-based genome-wide analysis of incident coronary heart disease. Circ Cardiovasc Genet. 2011;4:549–556. doi: 10.1161/CIRCGENETICS.111.960393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.