Abstract
Ligands of the thiazolidinedione (TZD) class of compounds, pioglitazone (Actos™) and rosiglitazone (Avandia™) are currently approved for treatment of type 2 diabetes and are known to bind to the PPAR-γ nuclear receptor subtype. Recent evidence suggesting PPAR-γ independent action of the TZDs led to the discovery of a novel integral outer mitochondrial membrane protein, mitoNEET. In spite of the several reported X-ray crystal structures of the unbound form of mitoNEET, the location and nature of the mitoNEET ligand binding sites (LBS) remain unknown. In this study, a molecular blind docking (BD) method was used to discover potential mitoNEET LBS and novel ligands, utilizing the program AutoDock Vina (v 1.0.2). Validation of BD was performed on the PPAR-γ receptor (PDB ID: 1ZGY) with the test compound rosiglitazone, demonstrating that the binding conformation of rosiglitazone determined by AutoDock Vina matches well with that of the cocrystallized ligand (root mean square deviation of the heavy atoms 1.45 Å). The locations and a general ligand binding interaction model for the LBS were determined, leading to the discovery of novel mitoNEET ligands. An in vitro fluorescence binding assay utilizing purified recombinant mitoNEET protein was used to determine the binding affinity of a predicted mitoNEET ligand, and the data obtained is in good agreement with AutoDock Vina results. The discovery of potential mitoNEET ligand binding sites and novel ligands, opens up the possibility for detailed structural studies of mitoNEET–ligand complexes, as well as rational design of novel ligands specifically targeted for mitoNEET.
Keywords: Mitoneet, Autodock, Docking, Fluorescence, Iron–sulfur, Thiazolidinedione
MitoNEET is a recently discovered iron–sulfur (2Fe–2S) outer mitochondrial integral membrane protein that binds the same ligands, specifically pioglitazone [1,2] and rosiglitazone [3], as the peroxisome proliferator-activated receptor gamma (PPAR-γ) nuclear receptor, and with similar binding affinity. Pioglitazone (1) and rosiglitazone (2) are members of the thiazolidinedione (TZD) class of compounds (Fig. 1), currently approved for treatment of type 2 diabetes. Recent evidence suggesting PPAR-γ independent action of the TZDs led to the discovery of mitoNEET through a cross-linking study with a photoactive form of pioglitazone [1]. Moreover, accumulating evidence indicates that many of the clinical effects of TZDs may originate by binding to mitoNEET. A recent report [4] suggests that mitoNEET plays a key role in regulating electron transport and oxidative phosphorylation, as there is a decrease in the complex I-dependent oxygen consumption in the mitochondria isolated from mitoNEET-deficient mice heart.
Fig. 1.
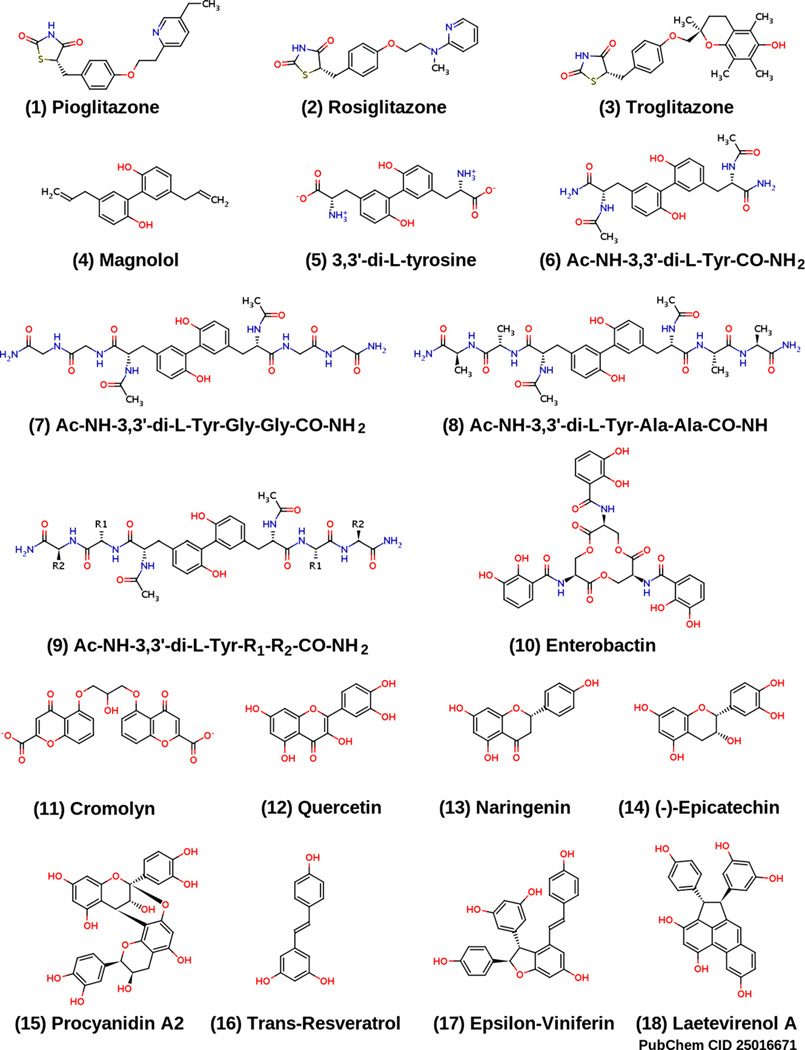
Structures of 15 compounds predicted to bind to mitoNEET, as well as the structures of pioglitazone (1) and rosiglitazone (2).
Pioglitazone was also reported to alter the oxidative capacity of mitochondria and exhibit negative regulatory effect on complex I activity in liver, muscle, and astroglioma cells [5,6], with positive regulatory effect in neuronal cells [7]. These correlations together with the mitochondrial localization of mitoNEET, strongly suggest that pioglitazone (and other TZDs) may exert their actions on mitochondria by regulating activities of the respiratory complexes via interaction with mitoNEET through yet unknown mechanism(s).
In other studies, pioglitazone has been shown as a potential treatment for several neurodegenerative diseases, particularly Alzheimer disease (AD) [8], amyotrophic lateral sclerosis (ALS) [9,10], multiple sclerosis (MS) [11–13], and Parkinson’s disease (PD) [14]. Moreover, pioglitazone was shown to prevent LPS-induced activation of microglia, increased oxidative stress, and impaired mitochondrial function, all of which are specific in dopaminergic neurodegeneration [15,16], implicated in PD [14]. Published evidence indicates that increased oxidative stress [17] is implicated in PD, suggesting that pioglitazone may have anti-inflammatory and anti-oxidative properties.
In addition, mitoNEET is likely to play a vital role in mitochondrial energy homeostasis and metabolism [4], transporting and sequestering iron, electron transfer, and oxidation of fatty acids [1]. Pioglitazone binding to mitoNEET was also found to stabilize the mitoNEET dimer formation, which correlates with decreased oxidative stress [18]. The fact that PPAR-γ ligands bind to mitoNEET suggests an alternate mode of action for these ligands, with the possibility of rationally designing novel targeted agents that bind to mitoNEET, and are structurally distinct from the TZD class of compounds, that have undesirable side effects and toxicity [19].
Currently, a major challenge is to understand the nature of TZD-mitoNEET binding at a much higher level of detail [3]. However, neither quantitative determination of association constants for TZD-mitoNEET binding, nor the precise location of the mitoNEET ligand binding site(s) (LBS) have yet been determined, in part because of the high insolubility of the TZD class of molecules in aqueous solutions, requiring drug solubilization with relatively high concentrations of DMSO, which denatures the structure of mitoNEET [3]. The present lack of structurally diverse and water-soluble ligands known to bind to mitoNEET (besides the TZDs) may likely be a key contributing factor for the absence of any X-ray and/or NMR structures reported to date that provide high resolution structure of mitoNEET complexed with a ligand, despite several reported X-ray crystal structures of the unbound mitoNEET (apo) structure [2,20,21]. Thus, the nature of the mitoNEET LBS remains unknown. Our discovery of potential mitoNEET ligands, with good water solubility and high binding affinity (e.g., molecules 5 and 11, Fig. 1) holds great promise for future high-resolution X-ray and NMR structural studies of the mitoNEET.
The LBS reported in our study is roughly comparable to the major LBS reported in two previous publications [20,22]. In the more recent report [22], molecular docking software employing the SiteFinder module in MOE 2008.10 (Chemical Computing Group) found five potential binding pockets on mitoNEET, by probing the surface of the protein with alpha-spheres [22]. In contrast, AutoDock Vina [23] docks a flexible ligand against the protein using a sophisticated gradient optimization method [23]. The study also used a different crystal structure (PDB ID: 2QH7, personal communications), compared to the one we used (2QD0). The residues in one of the major binding sites identified [22] seem to agree with the rough location of a binding pocket that was first suggested in an earlier study [20], where the authors used AutoDock 3 (personal communications), but the same crystal structure [20] (2QD0) used in our study. 2QD0 has five additional residues on the N-terminus of chain A, while 2QH7 possesses additional lysine and glutamic acid residues on the N-terminus and C-terminus of chains A and B, respectively.
Molecular docking is a powerful computational technique used in structure-based drug design. The availability of reliable structures of the target receptor is crucial for docking. Multiple receptor conformations can be used to incorporate receptor flexibility into the docking algorithm, to improve the accuracy of binding pose prediction. A recent computational study employing “ensemble docking” into multiple crystallographically derived protein structures [24], provides a good example of this approach. We are planning to include receptor flexibility in future studies. As there are no published structures of mitoNEET complexed with a ligand, we have used a technique called blind docking (BD) [25,26] to determine the potential binding site(s). BD is used for detecting possible binding sites and ligand conformations by scanning the entire surface of a protein target, and finding a location with the highest binding affinity on the protein. Based on previous studies [25,26], BD has been shown to reliably predict the binding location and mode of numerous known protein–ligand complexes, and can be used in the case where the binding site(s) on a protein is unknown.
In this study, the program AutoDock Vina (v 1.0.2) [23] was used to perform the blind docking; this program was shown to be more accurate compared to previous versions of AutoDock used in BD studies [25,26]. Known mitoNEET ligands (pioglitazone and rosiglitazone) were screened against the protein surface, and a binding site was found on the surfaces of both mitoNEET protomers. Using the binding site information, a number of potential, structurally diverse novel ligands, unrelated to the TZDs, were discovered. Of these ligands, laetevirenol A (18) and troglitazone (3) were found to have the highest predicted binding affinity, and using an in vitro fluorescence binding assay, determined the binding affinity of a predicted mitoNEET ligand. The discovery of potential mitoNEET binding sites, and novel ligands, opens up the possibility for detailed structural investigation of mitoNEET–ligand complexes, as well as rational design of structurally diverse novel ligands specifically targeted for mitoNEET, and with fewer side effects and toxicity compared to the TZDs [19].
1. Materials and methods
For the molecular docking study, protein structures were obtained from the RCSB Protein Data Bank (http://www.rcsb.org); the mitoNEET structure PDB ID was 2QD0, and the PPAR-γ PDB ID was 1ZGY. Ligand structures were obtained from the PubChem database (http://pubchem.ncbi.nlm.nih.gov). If ligand structures were not available from PubChem, they were drawn using MarvinSketch 5.3.1 (ChemAxon). The UCSF Chimera program (http://www.cgl.ucsf.edu/chimera) was used to prepare the structures for input to AutoDockTools (ADT) version 1.5.4 (http://mgltools.scripps.edu) by adding Gasteiger charges (computed using ANTECHAMBER [27]) and running 10,000 steps of energy minimization. UCSF Chimera uses the MMTK software module [28] to perform steepest descent energy minimization; the generalized amber force field was used [29]. The cocrystallized ligand in the PPAR-γ PDB structure was removed. For both protein structures, lower occupancy residue structures were deleted, and any incomplete side chains were replaced using the Dunbrack rotamer library [30]. Since the mitoNEET protein is a homodimer, UCSF Chimera was also used to delete residues 38–42 on chain B of the mitoNEET protein (PDB ID: 2QD0), thus making the individual protomer sequences identical. ADT was used to remove crystal waters, and add polar hydrogens to both protein structures. The distance between donor and acceptor atoms that form a hydrogen bond was defined as 1.9 Å with a tolerance of 0.5 Å, and the acceptor–hydrogen–donor angle was not less than 120° [31]. The structures were then saved in PDBQT file format, for input into AutoDock Vina version 1.0.2 (http://vina.scripps.edu). For the mitoNEET protein, the search space was defined as a cube 58 Å on each side, centered on the protein (X, Y, Z coordinates: 0.463, −0.322, −16.163). For PPAR-γ, the search space was a box with XYZ dimensions 50 Å × 56 Å × 72 Å respectively, and was centered on 28.744, −1.359, 14.898. Both search space dimensions encompass the entirety of the proteins, as is required for blind docking. The AutoDock Vina parameter “Exhaustiveness”, which determines how comprehensively the program searches for the lowest energy conformation, was set to the default value, eight, for both docking setups. Once the blind docking technique was validated, the search spaces were decreased to encompass only the ligand binding sites. All computations were carried out on a Compaq Evo Desktop D510 personal computer (2.4 GHz Pentium 4 processor, Intel, Santa Clara, CA) running Red Hat Enterprise Linux Client release 5.5 (Tikanga), as well as the Massachusetts Institute of Technology Athena workstations (2.66 GHz Intel Core 2 Duo processor) running Ubuntu Linux. The time required for each simulation run was on the order of 300 s on the Pentium machine, and approximately 120 s on the Core 2 Duo machines.
MitoNEET was prepared and purified as outlined [4], and was generously provided by Dr. Sandra Wiley, from the Department of Pharmacology at the University of California, San Diego. Magnolol was purchased from Sigma–Aldrich Co. (St. Louis, MO; product #: M3445). The Tris buffer used in this study was of analytical grade. The protein and magnolol samples were dissolved in the Tris–HCl buffer solution (50 mM Tris; 150 mM NaCl; pH 7.5 ± 0.1), at a temperature of 298 K. Fluorescence binding studies were performed on a QM-4-SE fluorimeter from Photon Technology International (Montreal, Quebec) using FELIX software, and 2 nm excitation and 6 nm emission bandwidth slits. Measurements were performed at 298 K in a volume of 400 µL Tris–HCl buffer solution, in 500 µL microcuvettes. The excitation wavelength was 295 nm, and the emission spectra were obtained by scanning from 310 to 500 nm at a rate of 1 nm per second. A similar scan of the buffer alone was subtracted from all other scans. Due to the presence of two tryptophan residues in mitoNEET, the optimal excitation wavelength is 295 nm, in order to minimize contribution from tyrosine and phenylalanine residues.
2. Results and discussion
2.1. Validation of AutoDock Vina blind docking method
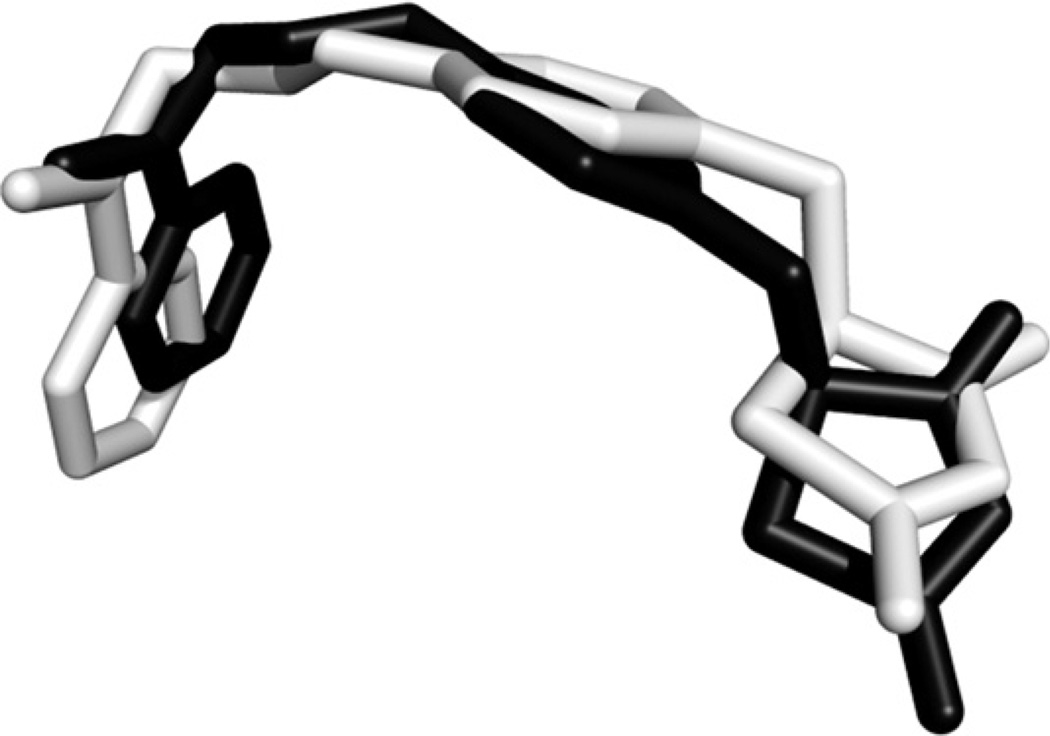
Blind docking (BD)was performed on the PPAR-γ receptor (PDB ID: 1ZGY) to validate its accuracy at reproducing the experimental binding mode of rosiglitazone. The results of BD indicate that the binding conformation of rosiglitazone determined by AutoDock Vina matches well with that of the cocrystallized ligand (Fig. 2). The root mean square deviation (RMSD) of the heavy atoms is only 1.45 Å (Fig. 2). This result, in addition to previous extensive validation [23,25,26], demonstrates the ability of AutoDock Vina to accurately predict the binding conformations of ligand–protein complexes.
Fig. 2.
Superposition of the computed rosiglitazone binding conformation (black) docked in the PPAR-γ receptor ligand binding pocket, and the cocrystallized ligand (light gray). RMSD calculation was based on the heavy atoms, and was performed using the “rmsd” command of UCSF Chimera (http://www.cgl.ucsf.edu/chimera). PPAR-γ receptor PDB ID: 1ZGY.
2.2. MitoNEET binding site
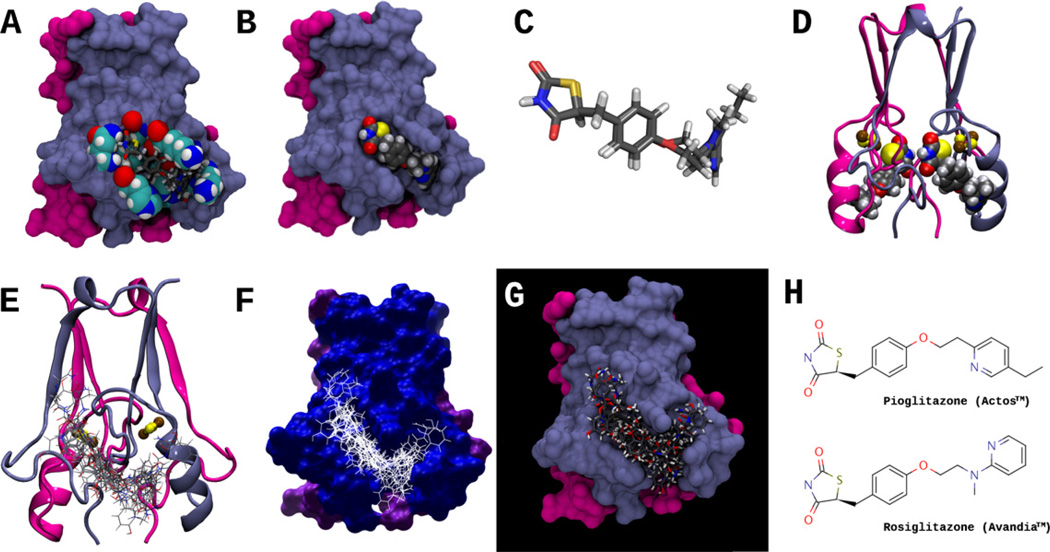
Having validated the BD method on PPAR-γ, we screened pioglitazone and rosiglitazone against the mitoNEET protein surface, in search of binding site(s). The reported crystal structure of the soluble domain [2,20,21] of mitoNEET indicates that it forms a globular homodimer domain, where the two monomer subunits are almost identical (RMS deviation of 0.091 Å over 64 Cα pairs [20]). This information was used to generate the structural representations of the mitoNEET homodimer in Fig. 3. From the docking studies, we identified one potential binding site on the surface of each protomer of the mitoNEET homodimer (Fig. 3). Besides pioglitazone and rosiglitazone, we also docked 15 other small molecules (Fig. 1) against the mitoNEET protein, and the output ensemble is depicted in Fig. 3(E–G), thus defining the entire mitoNEET binding pocket on each protomer (Table 1). We chose this set of compounds as they are structurally related to known PPAR-γ ligands; this will be discussed in further detail.
Fig. 3.
(A) Molecular surface representation of the MitoNEET homodimer; residues that interact with both pioglitazone and rosiglitazone are shown in space-filling representation, and include His48, Ile49, Gln50, Arg76, Lys78, Ala86, Lys89, and His90. Both pioglitazone and rosiglitazone are shown as licorice models, docked in the predicted binding conformation found using AutoDock Vina. Protomer I of the mitoNEET homodimer, is colored light blue, and protomer II is colored magenta. (B) Surface representation of MitoNEET homodimer, showing rosiglitazone docked (space-filling representation) in the MitoNEET binding pocket. (C) Superposition of pioglitazone and rosiglitazone in their docked conformations, as in (A); note that conformations of the TZD ring is nearly identical for both molecules. (D) Ribbon representation of the MitoNEET-rosiglitazone complex, showing rosiglitazone docked to both MitoNEET protomers; the 2Fe–2S clusters are shown in balls and sticks, where iron and sulfur atoms are colored brown and yellow, respectively. (E–G) Ribbon and surface representations of MitoNEET and an ensemble of 17 docked ligands (interactions shown in Fig. 5). (H) The structures of Pioglitazone (Actos™) and Rosiglitazone (Avandia™). Pioglitazone was the first ligand shown to bind to MitoNEET [1].
Table 1.
List of compounds with their corresponding binding affinities and interacting residues in the binding site of the mitoNEET protein. Residues in bold form hydrogen bonds in addition to van der Waals interactions (vdW), while the other residues primarily form vdW interactions with the ligand. Underlined residues have aromatic ring interactions.
| Compound: | Affinity (kcal/mol) | Residues | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Troglitazone (3) | −7.9 | His48 | Ile49 | Gln50 | Arg76 | Lys78 | Ala86 | Lys89 | His90 | Glu93 | ||||||||
| Laetevirenol A (18) | −7.8 | Leu47 | His48 | Trp75 | Ser77 | Lys78 | Ala86 | Lys89 | Glu93 | |||||||||
| Cromolyn (11) | −7.5 | His48 | Arg76 | Lys78 | His90 | |||||||||||||
| 7 | −7.5 | His48 | His58 | Ile49 | Gln50 | Arg76 | Lys7B | Phe80 | His90 | Glu93 | ||||||||
| ProcyanidinA2 (15) | −7.4 | His48 | Ile49 | Gln50 | Arg76 | Lys78 | Phe80 | |||||||||||
| ε-Viniferin (17) | −7.1 | Asn46 | Leu47 | His48 | Trp75 | Arg76 | Lys78 | |||||||||||
| 8 | −7.1 | His48 | Arg76 | Lys78 | Lys79 | Asp84 | ||||||||||||
| Enterobactin (10) | −7.0 | His48 | Ile49 | Arg76 | Asp84 | Lys89 | ||||||||||||
| 6 | −6.9 | Arg76 | Ser77 | Lys78 | Lys89 | His90 | Glu93 | |||||||||||
| Dityrosine (5) | −6.7 | His48 | Arg76 | Ser77 | Lys78 | Lys89 | His90 | Glu93 | ||||||||||
| Quercetin (12) | −6.7 | His48 | Ile49 | Gln50 | Arg76 | Lys78 | Phe80 | Ala86 | ||||||||||
| Naringenin (13) | −6.4 | His48 | Ile49 | Gln50 | Arg76 | Lys78 | Phe80 | |||||||||||
| (−)-Epicatechin (14) | −6.3 | His48 | Ile49 | Gln50 | Arg76 | Lys78 | Phe80 | |||||||||||
| Pioglitazone (1) | −6.3 | His48 | Ile49 | Gln50 | Arg76 | Lys78 | Ala86 | Lys89 | His90 | |||||||||
| Rosiglitazone (2) | −6.2 | His48 | Ile49 | Gln50 | Arg76 | Lys78 | Ala86 | Lys89 | His90 | |||||||||
| Maguolol (4) | −6.0 | His48 | Ile49 | Arg76 | Lys78 | Lys89 | ||||||||||||
| Resveratrol (16) | −5.8 | His48 | Ile49 | Gln50 | Arg76 | Lys78 |
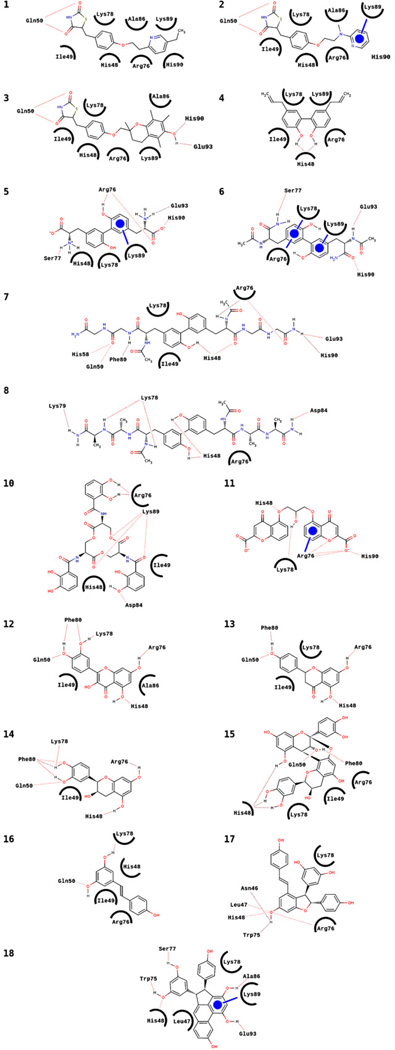
Analysis of the ligand–protein binding interactions for pioglitazone (1) and rosiglitazone (2) indicate very similar interactions with the binding site on mitoNEET (Figs. 3 and 4; Table 1). The main interactions involve the residues His48, Ile49, Arg76, Lys78, Ala86, Lys89, and His90 forming van der Waals (vdW) contacts with the ligands, and Gln50 forming two hydrogen bonds with the TZD rings (Fig. 4, 1–2; Table 1). Rosiglitazone (2) also forms an additional aromatic ring interaction between its pyridine ring and Lys89. The structural similarity of 1 and 2 is reflected in the similar interactions with the mitoNEET binding site residues, as well as the nearly identical predicted binding affinities (−6.3 kcal/mol and −6.2 kcal/mol, respectively) and position of the TZD rings (Fig. 3A and C). In comparison, the much higher binding energy predicted for another TZD, troglitazone (3, −7.9 kcal/mol), can be explained by formation of additional hydrogen bonding interactions between the phenolic hydroxyl of 3 and the hydrogen bond donor and acceptor groups on His90 and Glu93, respectively (Fig. 4, 3).
Fig. 4.
Schematic representations of hydrogen bonding, van der Waals (vdW), and aromatic ring interactions for the ligands listed in Fig. 1, arranged in the same numbering order. Dotted lines are hydrogen bonds, circled residues indicate van der Waals contacts with the ligands, and circles represent aromatic ring interactions (the line indicates the residue interacting with the ligand).
The result that troglitazone has a high predicted binding affinity to mitoNEET is consistent with data obtained from a preliminary experiment we performed, in which liver C3A cells were treated with 25 µM troglitazone for 30 min, resulting in a decrease of the mitochondrial membrane potential (Δψ) compared to control. Indeed, such a decrease in Δψ has been reported in a different study [32], in which troglitazone induced a rapid and dose-dependent drop of Δψ in liver HepG2 cells. Since mitoNEET was found associated with components of the pyruvate dehydrogenase complex [11], TZD binding could block pyruvate driven respiration [11], thereby lowering Δψ, and explaining these experimental results.
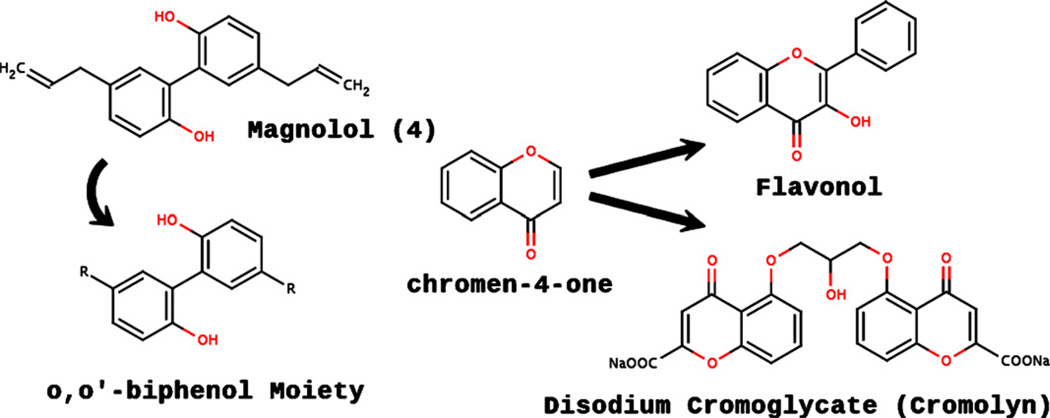
Our discovery of molecules that do not contain the TZD ring moiety and yet are predicted to bind to mitoNEET has not been reported before. In a very recent report [33], pharmacophore-based virtual screening of 3D natural product libraries was used to identify novel neolignan PPAR-γ ligands (e.g., 4, Fig. 1) that bind to the PPAR-γ ligand binding domain, with a binding pattern very similar to that of pioglitazone [33]. Structural analysis of 4 shows that it contains the o,o′-biphenol moiety (Fig. 5), and therefore we screened five molecules (4–8, Fig. 1) that contain the o,o′-biphenol moiety.
Fig. 5.
Graphical illustration depicting the relationship of the o,o′-biphenol and chromen-4-one core structures. “R” represents any functional group.
Molecules 4–8 (Fig. 1) all contain the o,o′-biphenol moiety (Fig. 5) and have predicted binding affinities close to or greater than that of pioglitazone (1) or rosiglitazone (2), (Table 1). 3,3′-di-l-tyrosine (5), forms hydrogen bonds with Arg76, Ser77, His90, Glu93, and an aromatic ring interaction with Lys89 (Fig. 4 and Table 1). Molecule 6 has slightly higher predicted binding affinity than 5, which can be explained by two additional aromatic ring interactions. Molecules 7 and 8 have greater predicted affinities compared to 5 or 6, likely due to their increased surface area and additional hydrogen bonding interactions, analogous to the higher predicted binding affinity of troglitazone (3) relative to pioglitazone (1) and rosiglitazone (2).
Molecules 4–9 have a number of unique physicochemical and biological properties. For example, all are fluorophores, with characteristic intrinsic fluorescence properties. In particular, 5, which consists of the 3,3′-di-l-tyrosine moiety (dityrosine), is a natural post-translational modification found in peptides and proteins [34]. Furthermore, since formation of the dityrosine cross-link involves oxidative coupling via a phenoxyl radical intermediate, it occurs under oxidative conditions, and therefore dityrosine formation is closely related to the in vivo oxidation state, and was proposed as a useful marker for in vivo protein oxidation [34]. The intrinsic fluorescence properties of molecules 4–9 make them very useful as fluorescent probes [34]. Fluorescence anisotropy measurements of mitoNEET–ligand complex formation for molecules 4–9 are likely to provide invaluable insights as to the nature of conformational changes that take place upon ligand binding to mitoNEET. Systematic studies of mitoNEET–ligand binding for molecules 4–9 should be a powerful tool for probing the structural features of mitoNEET complexed with a large number of ligands, whose structures can be designed and easily synthesized by changing the amino acid residues R1 and R2 in 9 (Fig. 1).
Molecules 4–9 can be formed by dimerization of their respective monomers. For example, magnolol (4) can be formed by dimerization of chavicol, giving rise to the o,o′-biphenol moiety; 5 can be formed by dimerization of l-tyrosine [34]. N-Acetyl-l-tyrosine amide (Ac-NH-l-Tyr-CO-NH2) is a simple model for an oligopeptide of tyrosine, and 6 can be prepared by dimerization of Ac-NH-l-Tyr-CO-NH2. In an analogous fashion, Ac-NH-l-Tyr-Gly-Gly-CO-NH2 and Ac-NH-l-Tyr-Ala-Ala-CO-NH2 tripeptide amides serve as monomer precursors for 7 and 8, respectively. 9 represents a generalized structure of a dimer of Ac-NH-l-Tyr-R1-R2-CO-NH2 (containing the dityrosine moiety), where R1 and R2 denote any of the 20 naturally occurring amino acids.
Epsilon-viniferin (17) and laetevirenol A (18) are stilbene dimers, and have much higher predicted binding affinity (Table 1) compared to resveratrol (16), a stilbene monomer. 18 has been reported to have strong antioxidant properties due to its unusual phenanthrene moiety [35]. The high predicted binding affinity of 18 can be rationalized by the large number of interactions it forms with the mitoNEET binding site (Fig. 4, 18); the relatively high predicted affinity of epsilon-viniferin can be explained in a similar fashion as well.
Since flavonoids have been shown to bind to the PPAR-γ receptor [36], we screened compounds 12–14 based on the chromen-4-one core structure (Fig. 5). Molecules 12–14 have comparable predicted binding affinities, as well as similarities in their interactions with the mitoNEET binding site; the three molecules all form hydrogen bonds with His48, Gln50, Arg76, and Phe80, and also have a van der Waals interaction with Ile49. Compound 15 is a dimeric form of 14, and in analogy to the stilbene dimers, has more interactions than the monomeric form, explaining the higher predicted affinity (Fig. 4, Table 1).
From the chromen-4-one moiety (Fig. 5), one can derive the structure of disodium cromoglycate (11, cromolyn). The main interactions of cromolyn with the mitoNEET binding site involve three hydrogen bonds with Arg76, as well as a hydrogen bond with His48, His90, and Lys78. Arg76 also forms an aromatic ring interaction with one of the chromen-4-one rings of cromolyn, and Lys78 forms vdW contacts with the other chromen-4-one ring. The number of hydrogen bonding, vdW, and aromatic ring interactions are consistent with cromolyn being one of the best predicted binders that we discovered.
Enterobactin (10) is one of the strongest siderophores known, binding to ferric iron (Fe3+) with K ≈ 1051 M−1 [37]. Interestingly, since mitoNEET is an iron–sulfur protein involved in iron sequestration/transport, binding of enterobactin to mitoNEET would likely have a significant effect on the overall mitoNEET protein structure, and we will test this hypothesis. The high predicted binding affinity of enterobactin is reflected by the numerous interactions depicted in Fig. 4, and Table 1.
Molecules 4–8 (Fig. 1) have predicted binding affinities similar to or greater than that of pioglitazone or rosiglitazone, and all contain the dityrosine moiety, which, due to its intrinsic fluorescence, can provide structural and functional information on the mitoNEET receptor protein in the bound state [34]. We are in the process of preparing experiments that will utilize molecules 4–8 as probes, in order to investigate the structure of mitoNEET with ligands in their bound conformation. Cromolyn (11) is marketed as a treatment for allergic reactions, however, it has been recently shown [38] to reduce diet-induced obesity and type 2 diabetes. Interestingly enough, our prediction that cromolyn binds to mitoNEET can explain why it has such an effect, and we will test this hypothesis as well. Molecules 12–18 (Fig. 1) are natural products and should be useful as a starting point for the rational design of novel synthetic ligands that selectively target mitoNEET.
From the ensemble of interactions of the 17 ligands examined in this work, (Figs. 3E–G and 4, and Table 1), a general ligand binding interaction model can be described. The ligand binding site on each of the mitoNEET protomers has a rather large “Y-shaped” configuration (Fig. 3E–G), which explains why it is predicted to bind a structurally diverse set of ligands, including large ligands such as 7 and 8, as well as relatively small ligands such as 4, 13, and 16. One example of a receptor protein that has a similarly shaped binding site, and is also capable of accommodating a large number of structurally diverse ligands is the PPAR-γ receptor [39], which can bind compounds ranging from small molecules such as magnolol [33], and flavonoids [36], to larger ligands such as TZDs, and a large variety of fatty acids including docosahexaenoic acid (DHA, PDB ID: 2VV0) [40]. The large pocket of PPAR-γ can also bind a long molecule such as DHA, and similarly, mitoNEET is predicted to bind long peptide derivatives (7 and 8) with high affinity as well.
For ligands that have polar functional groups at both ends (e.g., 3), separated by a less polar “linker” region, the residues His90 and Glu93 generally form hydrogen bonding interactions with one polar end. A good example is illustrated in the case of molecules 1–3, where the orientations of the TZD ring are very similar, as are the interactions around the TZD moiety. A striking feature that emerges from examination of the interactions of molecules 1–3, is that the ligand with a polar functional group at both ends (3), separated by a less polar “linker” region is predicted to have a much higher binding affinity compared to the ligands which contain a polar moiety at only one end (1–2). Of the molecules screened, 3, 5–8, 11, and 16 all have comparable structural features (two polar regions connected by a less polar “linker”). The residues His90 and Glu93 form hydrogen bonding interactions with one of the polar ends of these molecules in several instances (3, 5–7, 11). The three flavonoids all have very similar interaction patterns involving Gln50, Phe80, His48, and Arg76; the conformations of the three rings are very similar as well. The large mitoNEET binding site can accommodate structurally complex ligands such as 10, 15, 17 and 18, by forming numerous hydrogen bonding, van der Waals, and aromatic ring interactions, reflected by the high predicted binding affinities for these ligands. Although mitoNEET has a large binding site, consisting of 17 residues exhibiting significant interactions, the three residues His48, Arg76, and Lys78 interact with virtually every ligand screened, likely due to their central location in the binding site.
2.3. Fluorescence emission spectra
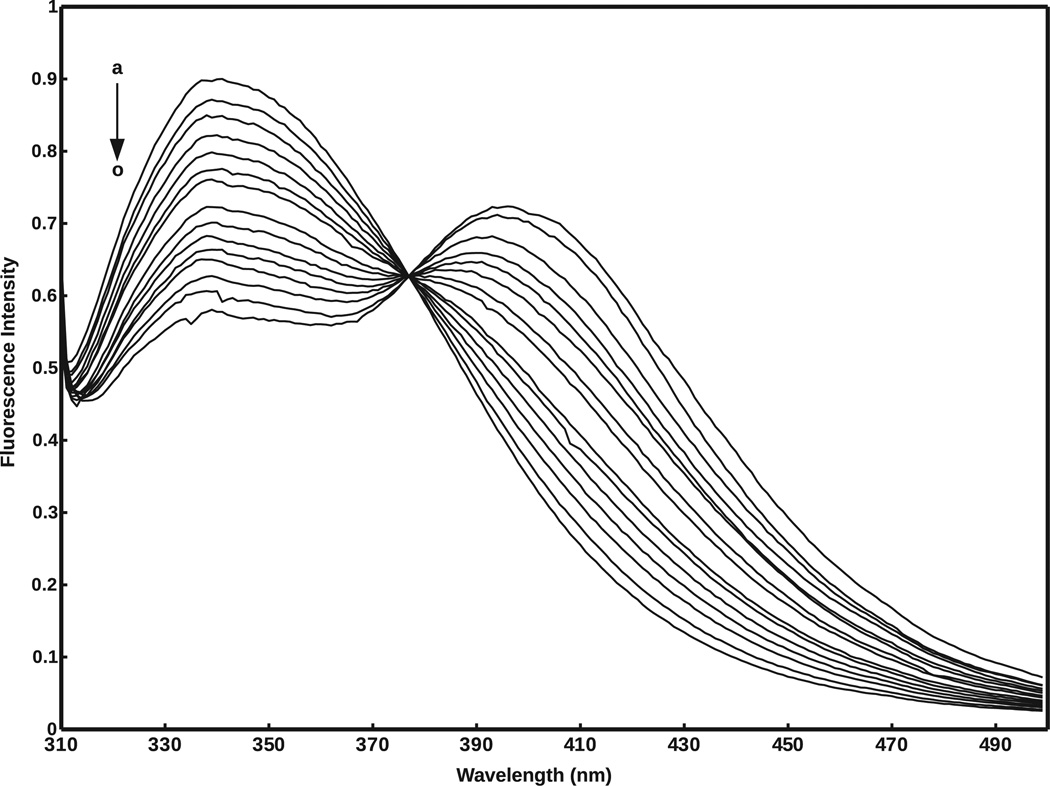
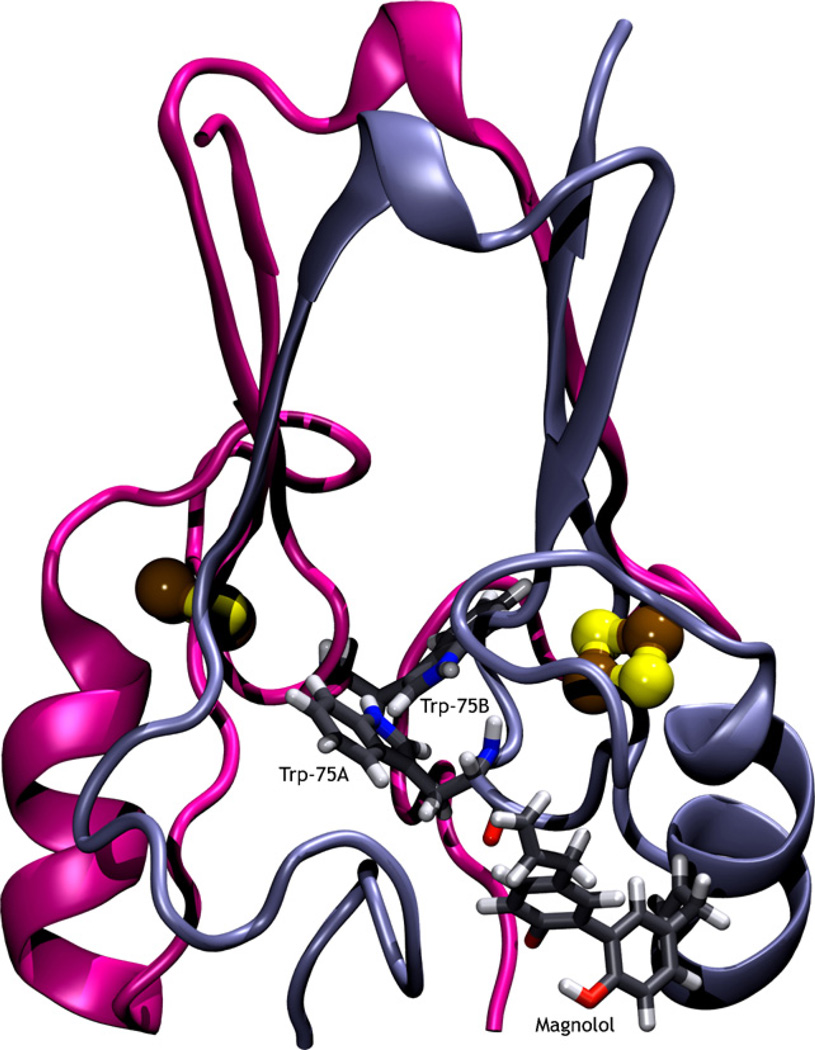
The steady state emission spectra of mitoNEET alone (F0) and in the presence of varying concentrations of magnolol (F), are shown in Fig. 6. In this experiment, the concentrations of mitoNEET were stabilized at 2.4 × 10−5 mol/L, and the concentrations of magnolol varied from 0 to 77.4 × 10−6 mol/L. Fig. 7 shows the predicted binding conformation of magnolol, as well as the location of both tryptophan residues of the mitoNEET homodimer. As shown in Fig. 6, mitoNEET has significant fluorescence emission with a peak at 341 nm, upon excitation at 295 nm, and the fluorescence intensity is quenched (decreases) with increasing concentrations of magnolol. Numerous molecular interactions can cause fluorescence quenching, including resonance energy transfer, ground-state complex formation, and collisional quenching [41]. Since the Trp75 residue in mitoNEET is a fluorophore, the close proximity of this residue to the predicted magnolol binding conformation (Fig. 7) indicates that fluorescence quenching should occur, which agrees with the decreased mitoNEET fluorescence observed experimentally (Fig. 6). The isosbestic point at 377 nm indicates that fluorescence quenching is due to mitoNEET–magnolol complex formation. The addition of magnolol does not shift the maximum wavelength of mitoNEET emission, suggesting that no major conformational changes were induced in the vicinity of the Trp75 residue in mitoNEET.
Fig. 6.
Fluorescence emission spectra of 2.4 × 10−5 mol/L mitoNEET in the presence of various concentrations of magnolol (T = 298 K; λex = 295 nm); a–o: [magnolol] × 10−6 mol/L = 0; 1.9; 3.7; 5.5; 7.4; 10.9; 14.4; 21.2; 27.8; 34.1; 40.2; 46.1; 57.2; 67.6; 77.4.
Fig. 7.
Magnolol bound to the mitoNEET receptor protein. The two iron–sulfur clusters are shown as ball and stick, while the two Trp75 residues and magnolol are shown in licorice. The intrinsic fluorescence of the two Trp75 residues, and their proximity to the binding pocket, enables fluorescence quenching.
2.4. Ligand binding affinity
The fluorescence emission data (Fig. 6) was used to determine the experimental binding affinity of magnolol to mitoNEET, using the modified Stern–Volmer equation [41]:
where F0 − F represents the difference in fluorescence intensity of mitoNEET alone (F0) and in the presence of a quencher (F) at concentration [Q]; fa is the fraction of initial fluorescence that is accessible to the quencher, and Ka is the Stern–Volmer quenching constant for the fraction of the fluorophore (analogous to associative binding constants for the system). Eq. (1) can be used to determine fa and Ka by plotting F0/(F0 − F) versus 1/[Q] yielding fa−1 as the intercept and (faKa)−1 as the slope, from which Ka, and therefore the standard free energy change, ΔG0, can be derived according to:
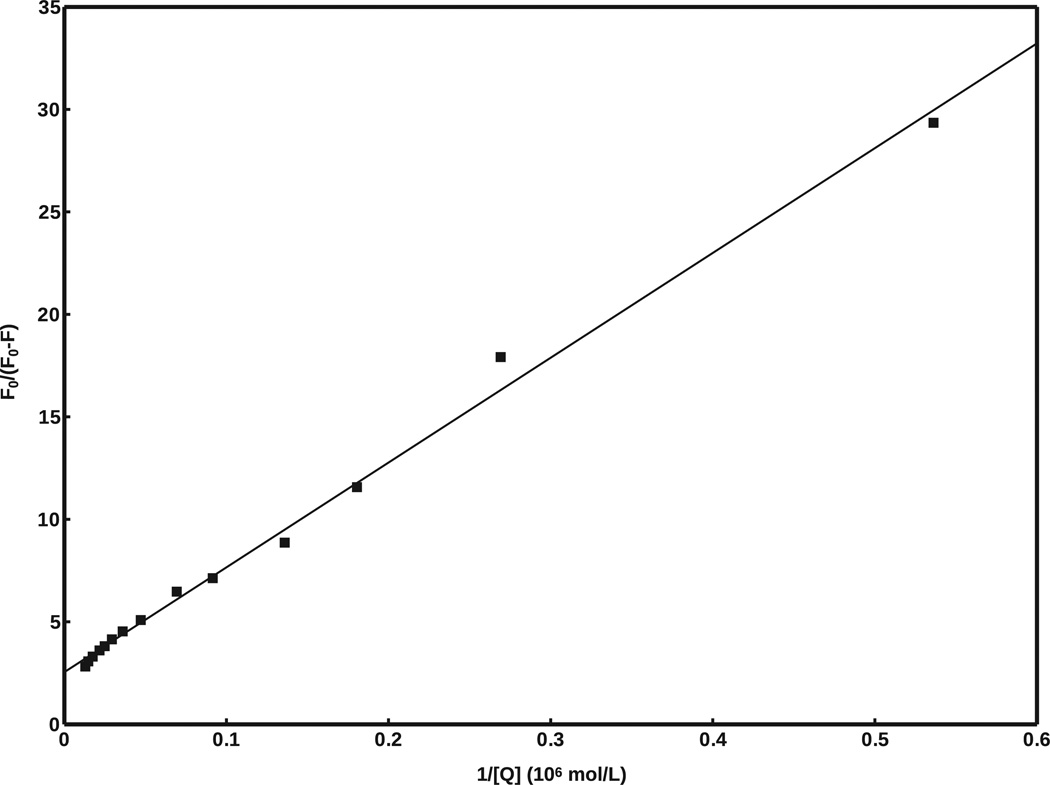
Fig. 8 shows a plot of F0/(F0 − F) versus 1/[Q], where fa = 0.393, and Ka = 4.98 × 105 mol/L. From these values, ΔG0 is calculated to be −6.40 kcal/mol, compared to the value of −6.0 kcal/mol predicted by AutoDock Vina. This result is well within the standard error of AutoDock Vina [23].
Fig. 8.
Modified Stern–Volmer plot of F0/(F0 − F) versus 1/[Q], generated from fluorescence data in Fig. 6. R2 = 0.995.
3. Summary
Even though compounds 4–18 do not contain the TZD ring moiety, our molecular docking and in vitro binding studies predict that they bind with similar or greater affinity to mitoNEET, compared with the known TZD ligands, pioglitazone and rosiglitazone (Table 1). For the first time, these results indicate that it is possible to design novel ligands targeted for mitoNEET that are not structurally related to the TZDs, therefore having less of the undesirable side effects and toxicity associated with TZDs.
In summary, using molecular docking we discovered a novel potential binding site on the surface of each protomer of the mitoNEET homodimer protein. Using this information, new structurally diverse ligands, distinct from the TZDs, and with higher predicted binding affinities, were discovered. Several of these ligands have interesting physicochemical and biological properties. We also discovered a number of natural products that are predicted to bind to mitoNEET with high affinity; they are likely useful as a basis for the rational design of novel ligands targeted for the mitoNEET receptor. An in vitro fluorescence binding assay was used to confirm mitoNEET binding to a novel ligand, demonstrating that the results predicted by AutoDock Vina are consistent with experimentally derived results.
Acknowledgements
The project described was supported by Award Number R01DK059766 from the National Institute Of Diabetes And Digestive And Kidney Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Diabetes And Digestive And Kidney Diseases or the National Institutes of Health. We thank Max R. Bieganski for assistance with creating graphical content and technical support. We also like to acknowledge Dr. Sandra Wiley, from the Department of Pharmacology at the University of California, San Diego, for providing purified recombinant mitoNEET protein samples, and Prof. JoAnne Stubbe, from the Departments of Biology and Chemistry at the Massachusetts Institute of Technology, for providing access to the QM-4-SE fluorimeter used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR. Identification of a novel mitochondrial protein (”mitoNEET“) cross-linked specifically by a thiazolidinedione photoprobe. Am. J. Physiol. Endocrinol. Metab. 2004 Feb 2;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- 2.Paddock ML, Wiley SE, Axelrod HL, Cohen AE, Roy M, Abresch EC, Capraro D, Murphy AN, Nechushtai R, Dixon JE, Jennings PA. MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bak DW, Zuris JA, Paddock ML, Jennings PA, Elliott SJ. Redox characterization of the FeS protein MitoNEET and impact of thiazolidinedione drug binding. Biochemistry. 2009;48:10193–10195. doi: 10.1021/bi9016445. (November 43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. U.S.A. 2007 Mar 13;104:5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunmair B, Staniek K, Gras F, Scharf N, Althaym A, Clara R, Roden M, Gnaiger E, Nohl H, Waldhäusl W, Fürnsinn C. Thiazolidinediones, like metformin, inhibit respiratory complex I: a common mechanism contributing to their antidiabetic actions? Diabetes. 2004 Apr 4;53:1052–1059. doi: 10.2337/diabetes.53.4.1052. [DOI] [PubMed] [Google Scholar]
- 6.Pérez-Ortiz JM, Tranque P, Burgos M, Vaquero CF, Llopis J. Glitazones induce astroglioma cell death by releasing reactive oxygen species from mitochondria: modulation of cytotoxicity by nitric oxide. Mol. Pharmacol. 2007 Aug 2;72:407–417. doi: 10.1124/mol.106.032458. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh S, Patel N, Rahn D, McAllister J, Sadeghi S, Horwitz G, Berry D, Wang KX, Swerdlow RH. The thiazolidinedione pioglitazone alters mitochondrial function in human neuron-like cells. Mol. Pharmacol. 2007 Jun 6;71:1695–1702. doi: 10.1124/mol.106.033845. [DOI] [PubMed] [Google Scholar]
- 8.Heneka MT, Sastre M, Dumitrescu-Ozimek L, Hanke A, Dewachter I, Kuiperi C, O’Banion K, Klockgether T, Van Leuven F, Landreth GE. Acute treatment with the PPARgamma agonist pioglitazone and ibuprofen reduces glial inflammation and Abeta1-42 levels in APPV717I transgenic mice. Brain. 2005 Jun;128(Pt 6):1442–1453. doi: 10.1093/brain/awh452. [DOI] [PubMed] [Google Scholar]
- 9.Schütz B, Reimann J, Dumitrescu-Ozimek L, Kappes-Horn K, Landreth GE, Schürmann B, Zimmer A, Heneka MT. The oral antidiabetic pioglitazone protects from neurodegeneration and amyotrophic lateral sclerosis-like symptoms in superoxide dismutase-G93A transgenic mice. J. Neurosci. 2005;25:7805–7812. doi: 10.1523/JNEUROSCI.2038-05.2005. (August 34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiaei M, Kipiani K, Chen J, Calingasan NY, Beal MF. Peroxisome proliferator-activated receptor-gamma agonist extends survival in transgenic mouse model of amyotrophic lateral sclerosis. Exp. Neurol. 2005 Feb 2;191:331–336. doi: 10.1016/j.expneurol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Feinstein DL, Spagnolo A, Akar C, Weinberg G, Murphy P, Gavrilyuk V, Dello Russo C. Receptor-independent actions of PPAR thiazolidinedione agonists: is mitochondrial function the key? Biochem. Pharmacol. 2005 Jul 2;70:177–188. doi: 10.1016/j.bcp.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Pershadsingh HA, Heneka MT, Saini R, Amin NM, Broeske DJ, Feinstein DL. Effect of pioglitazone treatment in a patient with secondary multiple sclerosis. Neuroinflammation. 2004 Apr 1;1:3. doi: 10.1186/1742-2094-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klotz L, Schmidt M, Giese T, Sastre M, Knolle P, Klockgether T, Heneka MT. Proinflammatory stimulation and pioglitazone treatment regulate peroxisome proliferator-activated receptor gamma levels in peripheral blood mononuclear cells from healthy controls and multiple sclerosis patients. J. Immunol. 2005 Oct 8;175:4948–4955. doi: 10.4049/jimmunol.175.8.4948. [DOI] [PubMed] [Google Scholar]
- 14.Hunter RL, Choi DY, Ross SA, Bing G. Protective properties afforded by pioglitazone against intrastriatal LPS in Sprague–Dawley rats. Neurosci. Lett. 2008 Feb 3;432:198–201. doi: 10.1016/j.neulet.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrera AJ, Castaño A, Venero JL, Cano J, Machado A. The single intranigral injection of LPS as a new model for studying the selective effects of inflammatory reactions on dopaminergic system. Neurobiol. Dis. 2000 Aug 4;7:429–447. doi: 10.1006/nbdi.2000.0289. [DOI] [PubMed] [Google Scholar]
- 16.Hunter RL, Dragicevic N, Seifert K, Choi DY, Liu M, Kim HC, Cass WA, Sullivan PG, Bing G. Inflammation induces mitochondrial dysfunction and dopaminergic neurodegeneration in the nigrostriatal system. J. Neurochem. 2007 Mar 5;100:1375–1386. doi: 10.1111/j.1471-4159.2006.04327.x. [DOI] [PubMed] [Google Scholar]
- 17.Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. (November 5545) [DOI] [PubMed] [Google Scholar]
- 18.Guerin T, Smart EJ. MitoNEET dimerization uncouples mitochondria and reduces oxidative stress. FASEB J. 2009;23:869.2. [Google Scholar]
- 19.Rizos CV, Elisaf MS, Mikhailidis DP, Liberopoulos EN. How safe is the use of thiazolidinediones in clinical practice? Expert Opin. Drug Saf. 2009 Jan 1;8:15–32. doi: 10.1517/14740330802597821. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Zhou T, Ye K, Wang J. Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14640–14645. doi: 10.1073/pnas.0702426104. (September 37) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlan AR, Paddock ML, Axelrod HL, Cohen AE, Abresch EC, Wiley S, Roy M, Nechushtai R, Jennings PA. The novel 2Fe-2S outer mitochondrial protein mitoNEET displays conformational flexibility in its N-terminal cytoplasmic tethering domain. Acta Crystallogr. F. 2009;65:654–659. doi: 10.1107/S1744309109019605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geldenhuys WJ, Funk MO, Barnes KF, Carroll RT. Structure-based design of a thiazolidinedione which targets the mitochondrial protein mitoNEET. Bioorg. Med. Chem. Lett. 2010 Feb 3;20:819–823. doi: 10.1016/j.bmcl.2009.12.088. [DOI] [PubMed] [Google Scholar]
- 23.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010 Jan 2;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craig IR, Essex JW, Spiegel K. Ensemble docking into multiple crystallographically derived protein structures: an evaluation based on the statistical analysis of enrichments. J. Chem. Inf. Model. 2010;50:511–524. doi: 10.1021/ci900407c. [DOI] [PubMed] [Google Scholar]
- 25.Hetényi C, van der Spoel D. Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 2002 Jul 7;11:1729–1737. doi: 10.1110/ps.0202302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hetényi C, van der Spoel D. Blind docking of drug-sized compounds to proteins with up to a thousand residues. FEBS Lett. 2006 Feb 5;580:1447–1450. doi: 10.1016/j.febslet.2006.01.074. Epub 2006 Jan 31. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006 Oct 2;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Hinsen K. The molecular modeling toolkit: a new approach to molecular simulations. J. Comp. Chem. 2000;21:79–85. [Google Scholar]
- 29.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25(9):1157. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 30.Dunbrack RL., Jr Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 31.Desiraju G, Steiner T. Structural Chemistry and Biology. United States: Oxford University Press; 2001. The Weak Hydrogen Bond. [Google Scholar]
- 32.Bova MP, Tam D, McMahon G, Mattson MN. Troglitazone induces a rapid drop of mitochondrial membrane potential in liver HepG2 cells. Toxicol. Lett. 2005 Jan 1;155:41–50. doi: 10.1016/j.toxlet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Fakhrudin N, Ladurner A, Atanasov AG, Heiss EH, Baumgartner L, Markt P, Schuster D, Ellmerer EP, Wolber G, Rollinger JM, Stuppner H, Dirsch VM. Computer-aided discovery, validation, and mechanistic characterization of novel neolignan activators of peroxisome proliferator-activated receptor gamma. Mol. Pharmacol. 2010 Apr 4;77:559–566. doi: 10.1124/mol.109.062141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malencik DA, Anderson SR. Dityrosine as a product of oxidative stress and fluorescent probe. Amino Acids. 2003 Dec 3–4;25:233–247. doi: 10.1007/s00726-003-0014-z. [DOI] [PubMed] [Google Scholar]
- 35.He S, Wu B, Pan Y, Jiang L. Stilbene oligomers from Parthenocissus laetevirens: isolation, biomimetic synthesis, absolute configuration, and implication of antioxidative defense system in the plant. J. Org. Chem. 2008 Jul 14;73:5233–5241. doi: 10.1021/jo8001112. [DOI] [PubMed] [Google Scholar]
- 36.Salam NK, Huang TH, Kota Bp, Kim MS, Li Y, Hibbs DE. Novel PPAR-gamma agonists identified from a natural product library: a virtual screening, induced-fit docking and biological assay study. Chem. Biol. Drug Des. 2008 Jan 1;71:57–70. doi: 10.1111/j.1747-0285.2007.00606.x. [DOI] [PubMed] [Google Scholar]
- 37.Carrano CJ, Raymond KN. Ferric ion sequestering agents. 2. Kinetics and mechanism of iron removal from transferrin by enterobactin and synthetic tricatechols. J. Am. Chem. Soc. 1979;101(18):5401–5404. [Google Scholar]
- 38.Liu J, Divoux A, Sun J, Zhang J, Clément K, Glickman JN, Sukhova GK, Wolters PJ, Du J, Gorgun CZ, Doria A, Libby P, Blumberg RS, Kahn BB, Hotamisligil GS, Shi GP. Genetic deficiency and pharmacological stabilization of mast cells reduce diet-induced obesity and diabetes in mice. Nat. Med. 2009 Aug 8;15:940–945. doi: 10.1038/nm.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markt P, Petersen RK, Flindt EN, Kristiansen K, Kirchmair J, Spitzer G, Distinto S, Schuster D, Wolber G, Laggner C, Langer T. Discovery of novel PPAR ligands by a virtual screening approach based on pharmacophore modeling 3D shape, and electrostatic similarity screening. J. Med. Chem. 2008 Oct 20;51:6303–6317. doi: 10.1021/jm800128k. [DOI] [PubMed] [Google Scholar]
- 40.Itoh T, Fairall L, Amin K, Inaba Y, Szanto A, Balint BL, Nagy L, Yamamoto K, Schwabe JW. Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat. Struct. Mol. Biol. 2008 Sep 9;15:924–931. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. Chapter 8. Springer; 2006. pp. 288–289. [Google Scholar]