Abstract
In species with large geographic ranges, genetic diversity of different populations may be well studied, but differences in loci and sample sizes can make the results of different studies difficult to compare. Yet, such comparisons are important for assessing the status of populations of conservation concern. We propose a simple approach of using a single well-studied reference population as a ‘yardstick' to calibrate results of different studies to the same scale, enabling comparisons. We use a well-studied large carnivore, the brown bear (Ursus arctos), as a case study to demonstrate the approach. As a reference population, we genotyped 513 brown bears from Slovenia using 20 polymorphic microsatellite loci. We used this data set to calibrate and compare heterozygosity and allelic richness for 30 brown bear populations from 10 different studies across the global distribution of the species. The simplicity of the reference population approach makes it useful for other species, enabling comparisons of genetic diversity estimates between previously incompatible studies and improving our understanding of how genetic diversity is distributed throughout a species range.
Keywords: genetic diversity, conservation, population comparison, genetics, inbreeding, Ursus arctos
Introduction
Loss of biodiversity is one of the critical challenges faced both by our planet and our species, as many plants and animals have been eradicated from human-dominated landscapes or remain in small populations that face a serious threat of extinction (UNEP, 1992). Conservation of these remaining populations may, in the long run, critically depend on genetic factors (Allendorf and Luikart, 2007; Frankham, 2009). Genetic diversity indicates a population's fitness and evolutionary potential, and consequently its adaptive potential and resilience to environmental change (Reed and Frankham, 2003; Allendorf and Luikart, 2007), which makes it a critical issue for conservation. Increased accessibility and decreasing costs are making the use of genetics in biodiversity conservation more attractive than ever, and increasingly large amounts of genetic data are available for species of conservation concern. Comparing these data between different populations along the range of a species would be useful for understanding and evaluating their genetic health and assessing the risk of inbreeding depression. However, genetic diversity of different populations is often evaluated using different methods and markers, making such comparisons difficult (see Swenson et al., 2011).
We propose a simple approach for calibrating genetic diversity of different populations, reported by different studies, to the same scale relative to a reference population. By using this one well-studied population as a ‘yardstick', we can perform large-scale comparisons of genetic diversity across a species range using the existing data. We demonstrate the utility of this concept using the brown bear (Ursus arctos), a widely distributed carnivore species that has been extensively studied using genetic methods.
Throughout most of its global range, the brown bear is suffering from habitat loss and overharvest, and more than 50% of its range and numbers have been lost since the mid-1800s (Servheen et al., 1999). Large populations remain in Northeastern and Northwestern Russia, Alaska and Canada, but only smaller isolated populations remain in the rest of the bear's former range in Europe, the contiguous United States and the southern portions of the range in Asia. Although genetic diversity of different brown bear populations has been well documented, different studies typically use different types or panels of markers, making the results difficult to compare (Swenson et al., 2011).
Centuries of persecution wiped out the bears from most of the Western Europe, and by the mid 20th century only a few isolated remnant populations remained in the Apennine Mountains, Italian Alps, Cantabrian Mountains and Pyrenees (Zedrosser et al., 2001). Bears in Central, Eastern and Northern Europe fared somewhat better, with indigenous populations remaining in the Dinaric Mountains, the Carpathians and Northern Europe, but most of these populations were much smaller than today (Zedrosser et al., 2001). This situation started to change in the second half of last century, when many remaining populations recovered and expanded as bears started making a comeback due to conservation and management efforts (Zedrosser et al., 2001). The last decade of that century also marked the beginning of reintroductions of this species to Western Europe (Clark et al., 2002). Although the overall situation is improving, many populations are still critically small (Linnell et al., 2005). This makes understanding of genetic diversity both within and between European brown bear populations particularly important, as it can facilitate selection of the most appropriate source for reintroductions or population augmentations, as well as help identify the populations that need assistance.
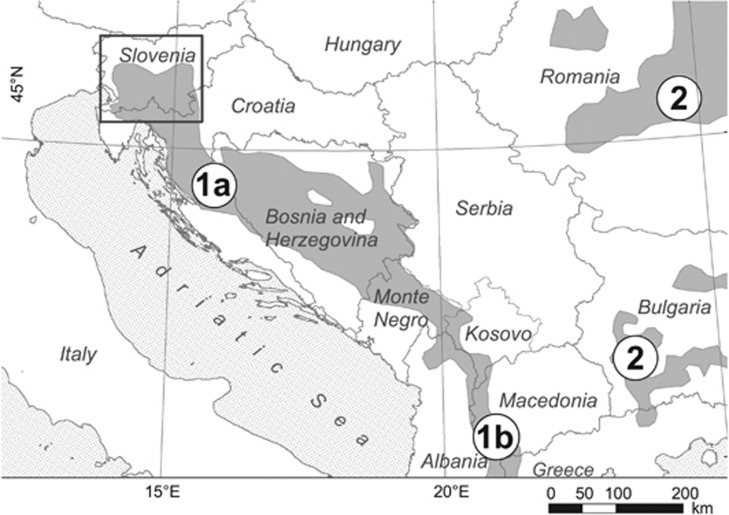
We studied the Northern Dinaric bear population and used it as a reference population in this case study example. This population stretches from Slovenia through Croatia and Bosnia, and Herzegovina into Western Serbia and Montenegro (Zedrosser et al., 2001), and has effective population size of approximately 280 bears (Skrbinšek et al., 2012). It is a part of the larger Alps-Dinara-Pindos population, which spans over 11 countries, is thought to have approximately 2100–2500 individuals, and is considered stable over most of its range (Zedrosser et al., 2001).
In this paper, we (1) introduce the reference population approach for calibrating and comparing genetic diversity reported by different studies of different populations, (2) survey the baseline genetic diversity data of the bears in Northern Dinaric Mountains and (3) use the reference population approach with the bears in Northern Dinaric Mountains as a reference population to calibrate and compare genetic diversity reported by different studies of bear populations across the range of the species.
Materials and methods
Comparing genetic diversity using the reference population approach
Different studies of genetic diversity typically vary in the number of samples and the sets of genetic markers they apply. Although this limits the degree to which the reported diversity indices are directly comparable, we can calculate the genetic diversity indices relative to the diversity indices of a single well-studied population (large sample size, a large number of loci) that we use as a ‘yardstick' (the reference population).
For each pairwise comparison of a population with the reference, the genetic marker set of both the reference and the compared population is reduced to the loci they have in common. To correct for differences in sample size, individual genotypes from the larger sample size (typically the reference population) are randomly resampled with replacement many times (∼1000) to the sample size of the smaller data set (Leberg, 2002). Average allelic richness, expected heterozygosity and their standard errors are then calculated over all random subsamples, thus correcting for differences in sample size. The standard errors are calculated as a mean of standard errors of each subsample. Finally, a heterozygosity ratio (Her) and allelic diversity ratio (Art) indices are calculated for the compared population as Her=Hex/HesR and Art=Ax/AsR, where Hex and Ax are expected heterozygosity and allelic diversity for the compared population, and HesR and AsR the subsampling-corrected values of these indices in the reference population (assuming that the reference population had more samples). Standard errors of the Her and Art indices are calculated as the standard error (s.e.) of division,
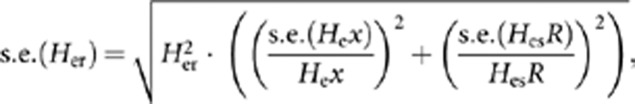 |
and
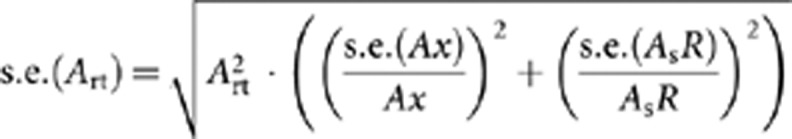 |
Genetic diversity of brown bears in Northern Dinaric Mountains—the reference population
Tissue and blood samples were collected from 2003 to 2008 from 505 dead bears and 8 bears captured for translocation (to France in 2006) or telemetry in the northernmost part of the Northern Dinaric population, in Slovenia (Figure 1). We analyzed 22 microsatellite loci for these 513 bears in three multiplex PCRs. Locus names, primer sequences, dyes, primer concentrations, analytic and quality assurance protocols used are detailed in Appendix 1. Further analytic protocols used for these loci are described in Skrbinšek et al. (2010). We randomly selected 10% of samples and repeated the genotyping to estimate error rates, as suggested by Pompanon et al. (2005). The actual number of repeats was considerably higher as the entire multiplex was repeated if the genotype at any locus was unclear. We used the methods recommended by Broquet and Petit (2004) to estimate the frequency of allelic dropouts and false alleles, and program Micro-Checker (Van Oosterhout et al., 2004) to check the data for the presence of null alleles, and scoring errors due to stuttering and dropout of large alleles.
Figure 1.
Alps-Dinara-Pindos bear population and sampling area. Shaded areas show brown bear range. (a) Alps-Dinara-Pindos population NW, NW Dinaric Mountains; (b) Alps-Dinara-Pindos population SE; (2) Carpathian population (after Zedrosser et al., 2001). Rectangle—sampling area.
We used R statistical environment (R Development Core Team, 2011) and ‘adegenet' package (Jombart, 2008) for data handling and calculation of genetic diversity indices—observed heterozygosity (Ho), expected heterozygosity (He) and allelic diversity (A). Probability of identity (PI) and probability of identity of siblings (PIsib) were calculated according to Waits et al. (2001). We used the procedure described in Guo and Thompson (1992) with 1 000 000 steps in Markov chain and 10 000 dememorization steps to detect per-locus significant departures from Hardy–Weinberg equilibrium using the program Arlequin (Excoffier and Lischer, 2010). Holm–Bonferronni multiple test correction with α=0.05 threshold was used to correct for multiple testing.
Using the reference population approach to explore differences in genetic diversity of brown bear populations across species range
We compared genetic diversity of different brown bear populations across the species range using the bears in Northern Dinaric Mountains as the reference. The details of the included studies are presented in the Appendix 2. The marker set we used for the reference population included the majority or all markers used in any other study, allowing for a large panel of loci for most comparisons. As our data set also included several times the number of samples analyzed in any other study, we always used it as the larger data set for resampling. We made 1000 random subsamples for each comparison. Finally, we calculated the Her and Art indices, and used these to compare genetic diversity of bear populations across the species range.
The R code required to run comparisons between populations using the reference population approach (in the form of an R package with user manual and a user-friendly vignette), as well as the genetic data from the Dinaric bear population used for this study, are accessible in the Dryad repository (doi:10.5061/dryad.qt3j5).
Results
Genotyping
No loci showed evidence of long allele dropout or scoring errors due to stuttering. Locus Mu26 had null alleles (estimated frequency using detected null homozygotes=0.117), and was excluded from downstream analyses. Locus G10H did not provide reliable genotyping results and was also excluded. Locus Mu23 had an irregular repeat pattern, as two out of the eight alleles had a single base deletion in the region flanking the (CA)n microsatellite, making their size a single base pair different from the neighbouring alleles. We were able to score the alleles reliably, so we can include this locus in the analyses. However, as the other studies may have missed this, or used primers that did not include the region with this single base polymorphism, using this locus for the reference population could bias the genetic diversity estimates for the reference population high. Considering this, we decided to exclude this locus from the reference population data.
On average 66% of per-locus genotype analyses were repeated more than once (varies between multiplexes: A=69%, B=71%, C=51%). Median allelic dropout rate was 0.19% (0.00–0.70%). We detected false alleles only on locus G10P (0.19%). Taking into account the number of loci, per-locus error rates, the number of samples genotyped and the number of times analyses of each sample were repeated, we can expect that there are still approximately 10 (9.6) single-locus errors in the data set. This makes the estimated remaining per-locus error rate in the entire data set 9.36 × 10−4.
Genetic diversity of bears in Northern Dinaric Mountains (Slovenia)
Average heterozygosities using the 20 remaining loci were 0.731 (He) and 0.738 (Ho). All these loci fit Hardy–Weinberg expectations after Holm–Bonferronni multiple test correction at P=0.05. Average allelic diversity was 6.75 (s.d.=1.77). Per-locus results are summarized in Table 1.
Table 1. Genetic diversity indices for brown bears in Northern Dinaric Mountains.
| Marker | A | Ho | He | PI | PI sib | P(HW) | Size |
|---|---|---|---|---|---|---|---|
| Cxx20 | 5 | 0.77 | 0.76 | 0.10 | 0.40 | 121–141 | |
| G10B | 8 | 0.73 | 0.71 | 0.11 | 0.42 | 0.010 | 130–154 |
| G10C | 9 | 0.76 | 0.74 | 0.11 | 0.41 | 93–115 | |
| G10D | 7 | 0.80 | 0.79 | 0.08 | 0.37 | 168–182 | |
| G10J | 5 | 0.65 | 0.69 | 0.16 | 0.45 | 78–92 | |
| G10L | 5 | 0.63 | 0.64 | 0.20 | 0.48 | 0.054 | 153–163 |
| G10M | 6 | 0.76 | 0.76 | 0.09 | 0.39 | 204–218 | |
| G10P | 8 | 0.78 | 0.78 | 0.07 | 0.38 | 147–175 | |
| G10X | 10 | 0.82 | 0.84 | 0.04 | 0.34 | 0.007 | 132–154 |
| G1A | 4 | 0.66 | 0.65 | 0.19 | 0.47 | 180–190 | |
| Mu05 | 7 | 0.62 | 0.66 | 0.16 | 0.46 | 127–141 | |
| Mu09 | 9 | 0.69 | 0.72 | 0.11 | 0.42 | 174–206 | |
| Mu10 | 4 | 0.69 | 0.68 | 0.17 | 0.45 | 112–126 | |
| Mu11 | 7 | 0.72 | 0.74 | 0.11 | 0.41 | 80–94 | |
| Mu15 | 6 | 0.78 | 0.77 | 0.09 | 0.39 | 117–131 | |
| Mu23a | 8 | 0.79 | 0.81 | 0.06 | 0.36 | 142–156 | |
| Mu26b | — | — | — | — | — | <0.001 | 182–200 |
| Mu50 | 7 | 0.80 | 0.80 | 0.07 | 0.37 | 79–103 | |
| Mu51 | 5 | 0.56 | 0.59 | 0.26 | 0.52 | 0.036 | 115–127 |
| Mu59 | 9 | 0.86 | 0.85 | 0.04 | 0.34 | 97–121 | |
| Mu61 |
6 |
0.76 |
0.78 |
0.08 |
0.38 |
|
141–153 |
|
Mean
|
6.75
|
0.73
|
0.74
|
0.12
|
0.41
|
|
|
| s.d. | 1.77 | 0.08 | 0.07 | 0.06 | 0.05 | ||
| s.e. | 0.40 | 0.02 | 0.02 | 0.01 | 0.01 |
Abbreviations: A, number of observed alleles; He, expected heterozygosity; Ho, observed heterozygosity; P(HW), probability of deviation from Hardy–Weinberg equilibrium; PI, probability of identity; PIsib, probability of identity for siblings; s.d., standard deviation; s.e., standard error; Size, size range of alleles.
Values in bold under P(HW) remained statistically significant after a Holm–Bonferroni multiple test correction (corrected α=0.05).
Locus Mu23 has a single base pair deletion in this population and consequently an irregular repeat pattern. The locus was included in the calculation of averages in this table, but excluded from the reference population.
Locus Mu26 has null alleles, and has been excluded from the calculation of averages.
Comparison of genetic diversity of brown bear populations across the range of the species
The results of the range-wide comparison of genetic diversity in brown bears are summarized in Table 2, and show considerable differences between populations. On one extreme, the most diverse is the Carpathian population in Romania, followed by large populations in Canada and Alaska. At the other extreme, the lowest levels of diversity are observed for island populations and very small populations of high conservation concern (Gobi Desert, Cantabrian Mountains—Spain, Kodiak Island—Alaska).
Table 2. Comparison of genetic diversity between bear populations using bears in NW Dinaric Mountains (Slovenia, population Alps-Dinara-Pindos NW in bold face) as a reference to correct for different panels of loci and sample sizes.
|
Compared population
|
Reference pop. (resampled)
|
Ratio
|
||||||
|---|---|---|---|---|---|---|---|---|
| Population | N | Study | A (s.e.) | He (s.e.) | A (s.e.) | He (s.e.) | Art (s.e.) | Her (s.e.) |
| Carpathians–Romania (1) | 16 | 5 | 7.78 (0.81) | 0.81 (0.010) | 5.15 (0.56) | 0.70 (0.030) | 1.51 (0.23) | 1.16 (0.05) |
| Carpathians–Romania (2) | 109 | 10 | 8.46 (0.57) | 0.80 (0.014) | 6.33 (0.54) | 0.73 (0.023) | 1.34 (0.15) | 1.09 (0.04) |
| Alaska Range, Alaska | 28 | 1 | — | 0.78 (—) | 5.84 (0.68) | 0.72 (0.026) | — | 1.08(—) |
| Kluane, Yukon | 50 | 1,2 | 7.38 (0.56) | 0.76 (0.025) | 6.12 (0.70) | 0.73 (0.026) | 1.21 (0.17) | 1.04 (0.05) |
| Richardson Mountains, NWT | 119 | 2 | 7.50 (0.63) | 0.76 (0.030) | 6.48 (0.72) | 0.73 (0.025) | 1.16 (0.16) | 1.03 (0.05) |
| Brooks Range, Alaska | 148 | 2 | 7.63 (0.50) | 0.75 (0.019) | 6.56 (0.72) | 0.74 (0.025) | 1.16 (0.15) | 1.02 (0.04) |
| Croatia (Alps-Dinara-Pindos NW) | 156 | 9 | 7.58 (0.54) | 0.74 (0.028) | 6.48 (0.60) | 0.73 (0.025) | 1.17 (0.14) | 1.01 (0.05) |
| Slovenia (Alps-Dinara-Pindos NW) | 513 | REF a | 6.68 (0.41) | 0.73 (0.020) | — | — | 1.00 (0.06) | 1.00 (0.03) |
| Greece(Alps-Dinara-Pindos s.e.) | 49 | 8 | 6.33 (0.42) | 0.76 (0.020) | 6.55 (0.52) | 0.77 (0.023) | 0.97 (0.10) | 0.99 (0.04) |
| Carphatians–Northern Slovakia | 71 | 10 | 6.08 (0.29) | 0.71 (0.025) | 6.20 (0.54) | 0.73 (0.023) | 0.98 (0.10) | 0.97 (0.05) |
| Scandinavia–NN | 29 | 3 | 5.59 (0.40) | 0.68 (0.024) | 5.59 (0.42) | 0.72 (0.020) | 1.00 (0.10) | 0.96 (0.04) |
| Flathead River, BC/MT | 40 | 2 | 6.50 (0.71) | 0.69 (0.027) | 6.01 (0.69) | 0.73 (0.026) | 1.08 (0.17) | 0.95 (0.05) |
| Carpathians–Central Slovakia | 96 | 10 | 6.00 (0.25) | 0.70 (0.031) | 6.30 (0.54) | 0.73 (0.023) | 0.95 (0.09) | 0.95 (0.05) |
| Scandinavia–NS | 108 | 3 | 6.18 (0.35) | 0.69 (0.027) | 6.10 (0.44) | 0.73 (0.019) | 1.01 (0.09) | 0.95 (0.04) |
| West Slope, Alberta | 41 | 2 | 6.38 (0.56) | 0.68 (0.036) | 6.03 (0.69) | 0.73 (0.026) | 1.06 (0.15) | 0.93 (0.06) |
| Kuskoskwim Range, Alaska | 55 | 1,2 | 6.13 (0.44) | 0.68 (0.026) | 6.15 (0.71) | 0.73 (0.025) | 1.00 (0.14) | 0.93 (0.05) |
| Scandinavia–M | 88 | 3 | 5.94 (0.40) | 0.68 (0.022) | 6.02 (0.44) | 0.73 (0.019) | 0.99 (0.10) | 0.93 (0.04) |
| Scandinavia–S | 155 | 3 | 5.47 (0.33) | 0.68 (0.020) | 6.20 (0.44) | 0.73 (0.019) | 0.88 (0.08) | 0.93 (0.04) |
| East Slope, Alberta | 45 | 2 | 7.00 (0.82) | 0.67 (0.062) | 6.07 (0.70) | 0.73 (0.026) | 1.15 (0.19) | 0.92 (0.09) |
| Carpathians–Eastern Slovakia | 16 | 10 | 5.23 (0.22) | 0.65 (0.028) | 5.47 (0.49) | 0.72 (0.025) | 0.96 (0.09) | 0.91 (0.05) |
| Paulatuk Alaska | 58 | 2 | 5.75 (0.88) | 0.65 (0.650) | 6.18 (0.71) | 0.73 (0.026) | 0.93 (0.18) | 0.89 (0.89) |
| Admiralty Island, Alaska | 30 | 1 | — | 0.63 (—) | 5.88 (0.68) | 0.73 (0.026) | — | 0.87 (—) |
| Coppermine, NWT | 36 | 2 | 5.75 (1.03) | 0.61 (0.073) | 5.96 (0.69) | 0.73 (0.026) | 0.96 (0.21) | 0.84 (0.10) |
| Pakistan | 28 | 4 | 3.92 (0.38) | 0.58 (0.043) | 5.45 (0.53) | 0.72 (0.025) | 0.72 (0.10) | 0.81 (0.07) |
| Yellowstone, MT/WY | 57 | 2 | 4.38 (0.60) | 0.55 (0.081) | 6.17 (0.7) | 0.73 (0.025) | 0.71 (0.13) | 0.75 (0.11) |
| Cantabrian (Spain)–W | 39 | 7 | 3.44 (0.30) | 0.48 (0.050) | 5.73 (0.49) | 0.71 (0.022) | 0.6 (0.07) | 0.67 (0.07) |
| Baranof and Chicgagof Is, Alaska | 35 | 1 | — | 0.49 (—) | 5.96 (0.69) | 0.73 (0.026) | — | 0.67 (—) |
| Apennines | 17 | 5 | 2.44 (0.24) | 0.44 (0.069) | 5.19 (0.56) | 0.70 (0.030) | 0.47 (0.07) | 0.63 (0.10) |
| Gobi (Mongolia) | 8 | 6 | 2.00 (—) | 0.29 (—) | 4.59 (0.62) | 0.68 (0.038) | 0.44 (—) | 0.43 (—) |
| Cantabrian (Spain)–E | 8 | 7 | 1.75 (0.17) | 0.28 (0.062) | 4.56 (0.38) | 0.68 (0.026) | 0.38 (0.05) | 0.41 (0.09) |
| Kodiak Island, Alaska | 34 | 1,2 | 2.13 (0.35) | 0.27 (0.098) | 5.94 (0.69) | 0.73 (0.026) | 0.36 (0.07) | 0.37 (0.14) |
Abbreviations: A, allelic richness; Art, allelic richness ratio between the compared population/resampling-corrected, marker-set specific values for bears in NW Dinaric Mountains; He, expected heterozygosity; Her, heterozygosity ratio; N, number of samples; s.e., standard error.
‘Reference pop. (resampled)' column shows the multiple subsampling corrected values from the reference population used for calculating Her and Art ratios. The studies referenced in the ‘Study' column are detailed in the Appendix 2.
Reference population. Values of some parameters in certain populations are missing as they were not available in the published data.
Discussion
The reference population approach provides a simple and easy to implement method of comparing genetic diversity between different populations of a species that were analysed in different studies using different loci, while collecting no or only minimal additional data. We demonstrate the application of this approach by evaluating the global distribution of genetic diversity of brown bears. Typically, there are two obstacles to comparing genetic diversity reported by different studies of the same species: different panels of genetic markers used and differences in sample sizes. The standard approach to addressing this problem is to shrink the genetic marker set to the largest common denominator of all studies, and use the smallest sample size in any population to correct for unequal sampling (El Mousadik and Petit, 1996; Leberg, 2002). This approach works only if similar sets of markers were used to study all populations or if marker sets are very large, which is often not the case. Also, by using a very small sample size to correct for unequal sampling, the power to detect differences in allelic richness is greatly reduced decreasing the power of all comparisons (Leberg, 2002).
The reference population approach overcomes many of these issues with a simple solution of scaling the genetic diversity of each considered population relative to the genetic diversity of a single well-studied population, effectively using this reference population as a calibration ‘yardstick'. Its main advantage is the ability to compare studies that would be otherwise impossible to compare—for example, studies that have no common genetic markers—if the markers they used are also used in the study of the reference population. The problem of low power of comparison will still remain when a study with a small sample size is compared, but this would not affect the power of pairwise comparisons of other populations.
Technical considerations, application and limitations of the reference population approach
Application of this method requires a reference population with a large sample size and a large number of genotyped loci. It is beneficial if a large population with high genetic diversity is used as a reference. If a study is designed specifically to provide reference population data, the panel of loci chosen should cover all or the majority of the loci used in other populations of interest. As more journals require genotype-level data to be deposited in online data repositories, reference population data should be increasingly easy to obtain. When suitable reference population data are available, it is straightforward to compare genetic diversity estimated in any new study of the same species with the existing data, provided that a large enough proportion of the marker set matches the marker set of the reference population.
We used multiple subsampling (Leberg, 2002) to correct for unequal sample sizes in different studies. Although it is argued that allelic diversity is a better predictor of a population's evolutionary potential than heterozygosity (Allendorf, 1986), it is also much more sensitive to sample size, and corrections for unequal sampling must be applied to calculate allelic richness if studies with different sample sizes are being compared (El Mousadik and Petit, 1996; Leberg, 2002). The most commonly used method is the rarefaction approach suggested by El Mousadik and Petit (1996). Simulations done by Leberg (2002) suggest that the multiple subsampling approach we used provides marginally better precision, but both methods perform adequately and without bias.
There was considerable variation in resampled allelic richness for the reference population (Table 2). This is a consequence of both subsampling to a smaller sample size, as rare alleles will get missed (see Leberg, 2002), as well as of the differences in locus panels that were subsampled to match the panels in the compared populations. The related standard error shows the standard error of allelic richness at the subsample size, providing the basis for comparison with the population of interest. Comparing calibrated expected heterozygosity to the values reported in original studies, it is clear that we would draw similar inferences using either the reported He or the calibrated indices (Table 2). Brown bears are studied with a relatively standard set of microsatellite markers, so all the studies included in this comparison had considerable overlap in markers. Although the reference population approach provides a formal framework for the bear case study, it should be even more useful in a species studied with a more diverse set of markers.
A logical precondition of the reference population approach is that it assumes the same type of genetic markers used in all studies that are to be compared. We implemented the approach using microsatellite data; however, the general idea of using a ‘yardstick' reference population could be transferred to other types of markers suitable for measuring genetic diversity (for example, single-nucleotide polymorphisms). Another potential problem for application is that sometimes only summary genetic diversity data are reported for a population, without any estimate of standard errors. Although such data are still useful, testing hypotheses about statistical significance of the observed differences between populations is impossible. This shows the importance of publishing standard error estimates in all genetic diversity studies, even if only a single population was studied. However, with recent changes to published data accessibility policies such cases should become increasingly rare.
The brown bear case study
The dramatic range of genetic diversity in brown bears that was observed by Paetkau et al. (1998b) in North America is also evident at the global scale (Table 2). Most of the observed patterns are expected—high genetic diversity in large populations (Alaska, Canada, Carpathians, Dinaric Mountains) and very low levels of genetic diversity in populations that have been isolated for a long time or have passed through severe demographic bottlenecks. The demographic history of many of these populations shows a large decline and a questionable future: the Gobi population in Mongolia (McCarthy et al., 2009), Cantabrian population in Spain (Perez et al., 2009) and the population in the Apennines in Italy (Ciucci and Boitani, 2008).
However, the genetic diversity in these populations is higher than the diversity of Kodiak Island bears in Alaska. This latter population is relatively large (>2500) and healthy, with low genetic diversity attributed to a long period of isolation from the bears on the continent (Paetkau et al., 1998a, 1998b). On the other hand, the demographic history of the other populations with low genetic diversity is presumed to be one of a recent contraction and isolation. For example, the Apennine population is estimated at around 50 remaining animals (Gervasi et al., 2008) and has been isolated for at least 400–600 years (Ciucci and Boitani, 2008). The story is similar with the Cantabrian bears in Spain, where the population suffered a dramatic decline in recent centuries and is now threatened with extinction (Perez et al., 2009).
Despite evidence from Kodiak bears that a brown bear population can exist and even prosper at very low levels of genetic diversity measured at neutral markers, this should not be generalized to the small populations that live in human-dominated landscapes. An island population may stabilize in a mutation-drift equilibrium at very low levels of genetic diversity, but it is possible that these bears survived against all odds through many generations of reduced fitness, all the time purging strongly deleterious alleles (Peatkau et al., 1998b). Although this may be a plausible scenario in Alaskan wilderness with favourable habitat and low human densities, the risk of inbreeding depression is likely to increase due to increased stress in degraded and human-dominated landscapes (Armbruster and Reed, 2005). For these populations, it is quite possible that they will need genetic rescue or restoration (Tallmon et al., 2004; Hedrick, 2005), or face extinction.
The highest genetic diversity levels were observed in the Carpathian brown bears. The population is relatively large, estimated to number around 8100 animals (Zedrosser et al., 2001), which may explain the high diversity. Another possible explanation for such high diversity might be historical mixing of animals from Eastern and Western glacial refugia as suggested by mitochondrial DNA data (Zachos et al., 2008). It would be interesting to compare genetic diversity levels of large bear populations in Russian Far East, but unfortunately there is no published research that would enable these comparisons.
Conclusions
Genetic diversity is a key component of long-term population viability (Allendorf and Ryman, 2002; Keller and Waller, 2002; O'Grady et al., 2006). By calibrating previously incompatible studies through comparisons with a reference population, we were able to directly compare neutral genetic diversity of brown bears from all previously studied populations. This method can easily be applied to other species and to test hypotheses about variables that influence genetic diversity across the range of a species. The method will also be helpful for identifying populations with low levels of diversity that have the greatest need for direct conservation actions, and can aid in providing the scientific justification needed to gain management and public support. The simplicity of the reference population approach should make it useful in future comparisons of genetic diversity estimates between previously incompatible studies and in improving our understanding of how genetic diversity is distributed along a species range.
Data archiving
Data and all R code have been deposited at Dryad: doi:10.5061/dryad.qt3j5.
Acknowledgments
We would like to thank to Slovenia Forest Service personnel for providing sample collection, and especially to Marko Jonozovič for support to our research. We would also like to thank Franc Kljun for organizing samples, Craig Miller for a useful insight regarding statistical properties of the described method, Roman Luštrik for help with compiling the R package and two anonymous reviewers for their helpful comments and suggestions. Genotyping of samples was financed through Grants No. L1-6484, L1-2196 and 2523-07-100435 by the Environmental Agency of the Republic of Slovenia and Slovenian Research Agency, and co-financed by the Ministry of Agriculture of the Republic of Slovenia and the Institute of the Republic of Slovenia for Nature. Analysis was done as a part of the HUNT project of the 7th Framework Programme for Research and Technological Development, financed by the European Commission. Neither the European Commission nor any person acting on behalf of the Commission is responsible for the use made of the information.
Appendix 1
Sample collection, laboratory analysis and quality assurance protocols used for genotyping of brown bear tissue samples
About a cubic centimeter of muscle, liver and/or skin tissue was taken from each dead animal, stored in 96% ethanol and kept at −20 °C. Blood from live captures was stored in Vacutainer tubes with anticoagulant (BD Diagnostics, Franklin Lakes, NJ, USA), also at −20 °C. We extracted DNA using GeneElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer's recommendation. The extracted DNA was then kept at −20 °C until analysis.
PCR setup was performed in a pressurized PCR hood using dedicated pipettes. We implemented a strict one-way workflow, where PCR cycling and post-PCR sample handling was done in a different room and no PCR products were returned to the lab where DNA extraction and PCR setup took place. Aerosol barrier pipette tips were used for all liquid transfers. All critical pipetting steps were photographed and later rechecked to detect possible sample mixups.
Qiagen Multiplex PCR kit (Qiagen, Hilden, Germany) was used for all PCR. We prepared 10 μl reactions—5 μl of Qiagen Mastermix, 1 μl of Q solution, 1 μl of template DNA and 3 μl of water and primers mixture to obtain the appropriate concentration in the final solution.
PCR conditions of different multiplexes differ only in the annealing temperature. The initial 10 min denaturation step at 95 °C is followed by 29 cycles of denaturation at 94 °C for 30 s, annealing at the multiplex-specific temperature for 90 s and elongation at 72 °C for 60 s. Annealing temperatures were 58 °C for multiplexes M1 and M3, and 49.6 °C for multiplex M2. PCR was concluded with a 30-min final elongation step at 60 °C designed to add +A to all fragments and minimize the problem of split peaks.
Fragment analysis was done on an ABI 3130xl Genetic Analyzer (Applied Biosystems, USA). A mixture of 1 μl of the PCR product, 0.25 μl of GS500LIZ size standard (Applied Biosystems, Foster City, CA, USA) and 8.75 μl of formamide was loaded on the sequencer. We analyzed the output with GeneMapper software (version 4.0, Applied Biosystems).
Each electrophoregram was independently checked by two persons. If the genotype at any locus was unclear, the PCR and analysis of the entire multiplex were repeated.

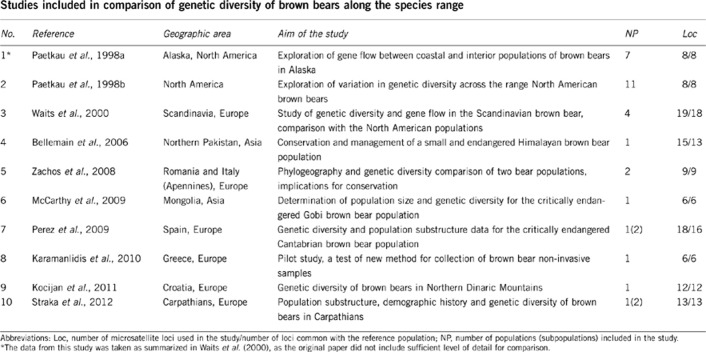
Appendix 2
The authors declare no conflict of interest.
References
- Allendorf FW. Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol. 1986;5:181–190. [Google Scholar]
- Allendorf FW, Luikart G. Conservation and the Genetics of Populations. Blackwell Publishing: Malden, USA; 2007. [Google Scholar]
- Allendorf FW, Ryman N.2002The role of genetics in population viability analysisIn: Beissinger SR, Mccullough DR (eds). Population Viability Analysis The University of Chicago Press: Chicago; 50–85. [Google Scholar]
- Armbruster P, Reed DH. Inbreeding depression in benign and stressful environments. Heredity. 2005;95:235–242. doi: 10.1038/sj.hdy.6800721. [DOI] [PubMed] [Google Scholar]
- Bellemain E, Nawaz MA, Valentini A, Swenson JE, Taberlet P. Genetic tracking of the brown bear in northern Pakistan and implications for conservation. Biol Conserv. 2006;134:537–547. [Google Scholar]
- Broquet T, Petit E. Quantifying genotyping errors in noninvasive population genetics. Mol Ecol. 2004;13:3601–3608. doi: 10.1111/j.1365-294X.2004.02352.x. [DOI] [PubMed] [Google Scholar]
- Ciucci P, Boitani L. The Apennine brown bear: a critical review of its status and conservation problems. Ursus. 2008;19:130–145. [Google Scholar]
- Clark JD, Huber D, Servheen C. Bear reintroductions: lessons and challenges: invited paper. Ursus. 2002;13:335–345. [Google Scholar]
- El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor Appl Genet. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Frankham R.2009Genetic considerations in reintroduction programmes for top-order, terrestrial predatorsIn: Hayward MW, Somers M (eds). Reintroduction of Top-Order Predators Blackwell Publishing Ltd; 372–287. [Google Scholar]
- Gervasi V, Ciucci P, Boulanger J, Posillico M, Sulli C, Focardi S, et al. A preliminary estimate of the Apennine brown bear population size based on hair-snag sampling and multiple data source mark-recapture Huggins models. Ursus. 2008;19:105–121. [Google Scholar]
- Guo S, Thompson E. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Hedrick P. ‘Genetic restoration:' a more comprehensive perspective than ‘genetic rescue'. Trends Ecol Evol. 2005;20:109. doi: 10.1016/j.tree.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Karamanlidis A, Drosopoulou E, de Gabriel Hernando M, Georgiadis L, Krambokoukis L, Pllaha S, et al. Noninvasive genetic studies of brown bears using power poles. Eur J Wildlife Res. 2010;56:693–702. [Google Scholar]
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–241. [Google Scholar]
- Kocijan I, Galov A, Ćetković H, Kusak J, Gomerčić T, Huber Ð. Genetic diversity of Dinaric brown bears (Ursus arctos) in Croatia with implications for bear conservation in Europe. Mamm Biol. 2011;76:615–621. [Google Scholar]
- Leberg PL. Estimating allelic richness: Effects of sample size and bottlenecks. Mol Ecol. 2002;11:2445–2449. doi: 10.1046/j.1365-294x.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- Linnell JDC, Promberger C, Boitani L, Swenson JE, Breitenmoser U, Andersen R.2005The linkage between conservation strategies for large carnivores and biodiversity: the view from the “half-full” forests of EuropeIn: Ray JC, Redford KH, Steneck RS, Berger J (eds). Large Carnivores and the Conservation of Biodiversity Island Press: Washington, DC; p526 [Google Scholar]
- McCarthy TM, Waits LP, Mijiddorj B. Status of the Gobi bear in Mongolia as determined by noninvasive genetic methods. Ursus. 2009;20:30–38. [Google Scholar]
- O'Grady JJ, Brook BW, Reed DH, Ballou JD, Tonkyn DW, Frankham R. Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol Conserv. 2006;133:42–51. [Google Scholar]
- Ostrander EA, Sprague GF, Rine J. Identification and characterization of dinucleotide repeat (ca)n markers for genetic mapping in dog. Genomics. 1993;16:207–213. doi: 10.1006/geno.1993.1160. [DOI] [PubMed] [Google Scholar]
- Paetkau DW, Shields GF, Strobeck C. Gene flow between insular, coastal and interior populations of brown bears in Alaska. Mol Ecol. 1998a;7:1283–1292. doi: 10.1046/j.1365-294x.1998.00440.x. [DOI] [PubMed] [Google Scholar]
- Paetkau DW, Waits LP, Clarkson PL, Craighead L, Vyse E, Ward R, et al. Variation in Genetic Diversity across the Range of North American Brown Bears. Conserv Biol. 1998b;12:418–429. [Google Scholar]
- Pérez T, Vázquez F, Naves J, Fernández A, Corao A, Albornoz J, et al. Non-invasive genetic study of the endangered Cantabrian brown bear (Ursus arctos) Conserv Genet. 2009;10:291–301. [Google Scholar]
- Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping Errors: Causes, Consequences and Solutions. Nat Rev Genet. 2005;6:847–846. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2011R: A Language And Environment For Statistical Computing R Foundation for Statistical Computing: Vienna, Austria; ISBN 3-900051-07-0, URL. . http://www.R-project.org/ . [Google Scholar]
- Reed DH, Frankham R. Correlation between Fitness and Genetic Diversity. Conserv Biol. 2003;17:230–237. [Google Scholar]
- Servheen C, Herrero S, Peyton B. Bears: Status Survey and Conservation Action Plan. IUCN: Gland, Switzerland; 1999. [Google Scholar]
- Skrbinšek T, Jelenčič M, Waits LP, Kos I, Trontelj P. Highly efficient multiplex PCR of noninvasive DNA does not require preamplification. Mol Ecol Resour. 2010;10:495–501. doi: 10.1111/j.1755-0998.2009.02780.x. [DOI] [PubMed] [Google Scholar]
- Skrbinšek T, Jelenčič M, Waits L, Kos I, Jerina K, Trontelj P. Monitoring the effective population size of a brown bear (Ursus arctos) population using new single-sample approaches. Mol Ecol. 2012;21:862–875. doi: 10.1111/j.1365-294X.2011.05423.x. [DOI] [PubMed] [Google Scholar]
- Straka M, Paule L, Ionescu O, Štofík J, Adamec M. Microsatellite diversity and structure of Carpathian brown bears (Ursus arctos): consequences of human caused fragmentation. Conserv Genet. 2012;13:153–164. [Google Scholar]
- Swenson JE, Taberlet P, Bellemain E. Genetics and conservation of European brown bears Ursus arctos. Mammal Rev. 2011;41:87–98. [Google Scholar]
- Taberlet P, Camarra JJ, Griffin S, Uhres E, Hanotte O, Waits LP, et al. Noninvasive genetic tracking of the endangered Pyrenean brown bear population. Mol Ecol. 1997;6:869–876. [PubMed] [Google Scholar]
- Tallmon DA, Luikart G, Waples RS. The alluring simplicity and complex reality of genetic rescue. Trends Ecol Evol. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. [DOI] [PubMed] [Google Scholar]
- UNEP [United Nations Environment Programme] Convention on Biological Diversity. United Nations Environment Programme, Environmental Law and Institutions Programme Activity Centre: Nairobi; 1992. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills D, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Waits LP, Luikart G, Taberlet P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol Ecol. 2001;10:249–256. doi: 10.1046/j.1365-294x.2001.01185.x. [DOI] [PubMed] [Google Scholar]
- Waits LP, Taberlet P, Swenson JE, Sandegren F, Franz R. Nuclear DNA microsatellite analysis of genetic diversity and gene flow in the Scandinavian brown bear (Ursus arctos) Mol Ecol. 2000;9:421–431. doi: 10.1046/j.1365-294x.2000.00892.x. [DOI] [PubMed] [Google Scholar]
- Zachos FE, Otto M, Unici R, Lorenzini R, Hartl GB. Evidence of a phylogeographic break in the Romanian brown bear (Ursus arctos) population from the Carpathians. Mamm Biol. 2008;73:93–101. [Google Scholar]
- Zedrosser A, Dahle B, Swenson JE, Gerstl N. Status and Management of the Brown Bear in Europe. Ursus. 2001;12:9–20. [Google Scholar]