Abstract
Background and Aims
Genome duplication is a central process in plant evolution and contributes to patterns of variation in genome size within and among lineages. Studies that combine cytogeography with genome size measurements contribute to our basic knowledge of cytotype distributions and their associations with variation in genome size.
Methods
Ploidy and genome size were assessed with direct chromosome counts and flow cytometry for 78 populations within the Claytonia perfoliata complex, comprised of three diploid taxa with numerous polyploids that range to the decaploid level. The relationship between genome size and temperature and precipitation was investigated within and across cytotypes to test for associations between environmental factors and nuclear DNA content.
Key Results
A euploid series (n = 6) of diploids to octoploids was documented through chromosome counts, and decaploids were suggested by flow cytometry. Increased variation in genome size among populations was found at higher ploidy levels, potentially associated with differential contributions of diploid parental genomes, variation in rates of genomic loss or gain, or undetected hybridization. Several accessions were detected with atypical genome sizes, including a diploid population of C. parviflora ssp. grandiflora with an 18 % smaller genome than typical, and hexaploids of C. perfoliata and C. parviflora with genomes 30 % larger than typical. There was a slight but significant association of larger genome sizes with colder winter temperature across the C. perfoliata complex as a whole, and a strong association between lower winter temperatures and large genome size for tetraploid C. parviflora.
Conclusions
The C. perfoliata complex is characterized by polyploids ranging from tetraploid to decaploid, with large magnitude variation in genome size at higher ploidy levels, associated in part with environmental variation in temperature.
Keywords: Claytonia perfoliata, Portulacaceae, flow cytometry, C-value, cytogeography, genome duplication, DNA content
INTRODUCTION
Whole-genome duplication, or polyploidy, is common and recurrent among flowering plants (Jiao et al., 2011). Nearly all extant angiosperm lineages exhibit genomic signatures consistent with one or more ancient whole-genome duplication event followed by genome reduction (Blanc and Wolfe, 2004; Cui et al., 2006; Soltis et al., 2009). Hypotheses for the ubiquity of polyploidy include advantages such as ecological benefits of increased heterozygosity, ability to invade novel habitats (Ramsey, 2011) and labile expression of divergent parental genomes (Stebbins, 1971; Grant, 1981; Levin, 1983; Comai, 2005; Dubcovsky and Dvorak, 2007), as well as potentially neutral explanations such as a high rate of polyploid formation relative to extinction (e.g. Meyers and Levin, 2006; but see Mayrose et al., 2011). Central to our understanding of the ecological aspects of polyploidy are cytogeographic studies of the distributions of diploids and polyploids across environments, allowing the association of cytotypes with environmental gradients such as altitude and latitude to be studied. Contemporary cytogeographic studies that combine direct chromosome counts with flow cytometric estimation of genome size are informative not only for geographic patterns of ploidy, but also for understanding patterns of genome size evolution within polyploid complexes. The extensive range of genome sizes observed within polyploid complexes makes them ideal systems for investigating patterns of genome size variation across ploidy levels and across environments.
The ease with which genome size can be estimated through flow cytometry (FCM) has contributed to our understanding of patterns of genome size evolution through the accumulation of genome size estimates across numerous taxa, allowing for comparative examinations. In recent years, advances in FCM methods that control for interference from plant secondary compounds (e.g. Bennett et al., 2008) have enabled rigorous documentation of intraspecific variation among individuals within populations (Šmarda et al., 2008; Marhold et al., 2010). The common trend among both auto- and allopolyploids is a reduction in genome size relative to the sum of the inferred parental diploids (Leitch and Bennett, 2004), although some examples of increased genome size have also been reported (Leitch et al., 2008). However, in autopolyploids, genome reduction appears to be associated with the restoration of diploid meiotic behaviour, as autopolyploids with polysomic inheritance may more commonly exhibit a pattern of genome size that is additive relative to the inferred parental genome sizes (Eilam et al., 2010; Parisod et al., 2010). Given what appears to be a general trend of selection for reduced genome size among neopolyploids, stochastic variation in the rate of DNA loss among populations may contribute to differences in genome size among polyploid populations.
Variation in the strength of selection across environments may also contribute to variation in genome size. Among the many genetic and ecological hypotheses suggested to explain patterns of variation in genome size, it has been proposed that larger genomes are adaptive in colder environments, where cell expansion rather than division is a more efficient means of growth (e.g. Grime and Mowforth, 1982; Knight and Ackerly, 2002). Thus, polyploids occurring in colder environments may experience reduced selection for genome reduction through the positive association between genome size and cell size. However, numerous other ecological factors have been proposed to influence genome size (Miller, 1978; Miller and Chambers, 2006), including selection for small genomes associated with rapid growth and short life spans, and smaller genomes associated with drought and other stressful environmental conditions (Kalendar et al., 2000; Jakob et al., 2004; Knight et al., 2005; Veselý et al., 2012). Some of these patterns may be complicated by extensive endopolyploidy of somatic cells, where cells achieve polyploid DNA content through duplication of the genome within the nucleus, without cellular division (Barow, 2006; Lee et al., 2009). With the intense interest in explaining patterns of genome size variation there is a need for further data on genome size variation, especially from groups with elevated variation such as polyploid complexes, to provide raw data for comparative studies across taxa.
In this paper, I report chromosome counts and genome size estimates from populations of the Claytonia perfoliata Don ex Wild (Portulacaceae) polyploid complex in western North America, and ask whether variation in genome size within a cytotype is associated with environmental variation in the form of temperature and precipitation. The C. perfoliata complex is an ideal system in which to investigate patterns of genome size variation with ploidy and environment, as classical cytogenetic studies document a range of ploidy from diploid to decaploid (Swanson, 1964; Miller, 1978; Miller and Chambers, 2006), and more recent work suggests ecological differentiation across cytotypes (McIntyre, 2012). The group is comprised of three allopatric diploid species, C. rubra (Howell) Tidestrom. ssp. rubra, C. perfoliata ssp. mexicana (Rydb.) John M. Miller & K.L. Chambers and C. parviflora ssp. grandiflora (Rydb.) John M. Miller & K.L. Chambers with a series of auto- and allopolyploid derivatives (Miller and Chambers, 2006; McIntyre, 2012). Apart from diploid C. parviflora ssp. grandiflora, cytotypes are highly selfing. The origins of many of the polyploid populations in the group are uncertain, but analysis based on chloroplast and internal transcribed spacer (ITS) sequences confirms the presence of C. rubra autotetraploids and allotetraploids between C. parviflora ssp. grandiflora and C. perfoliata ssp. mexicana, while higher ploidy levels contain diverse sequences of the diploids C. parviflora ssp. grandiflora and C. perfoliata ssp. mexicana, suggesting multiple formation of polyploid lineages (Rausch, 2008). Polyploids are variously ascribed to a number of sub-specific designations, reflecting complex patterns of morphological variation thought to be associated with hybridization and polyploidy (Miller and Chambers, 2006); however, for the purposes of this study, plants were assigned identity as one of the diploids described above or as a polyploid belonging to C. perfoliata ssp. perfoliata, C. parviflora ssp. parviflora or C. rubra ssp. depressa, based on morphology, as described in Miller and Chambers (2006). Morphological intermediates not easily assigned to a taxon are identified as Claytonia perfoliata sensu lato.
MATERIALS AND METHODS
Sampling
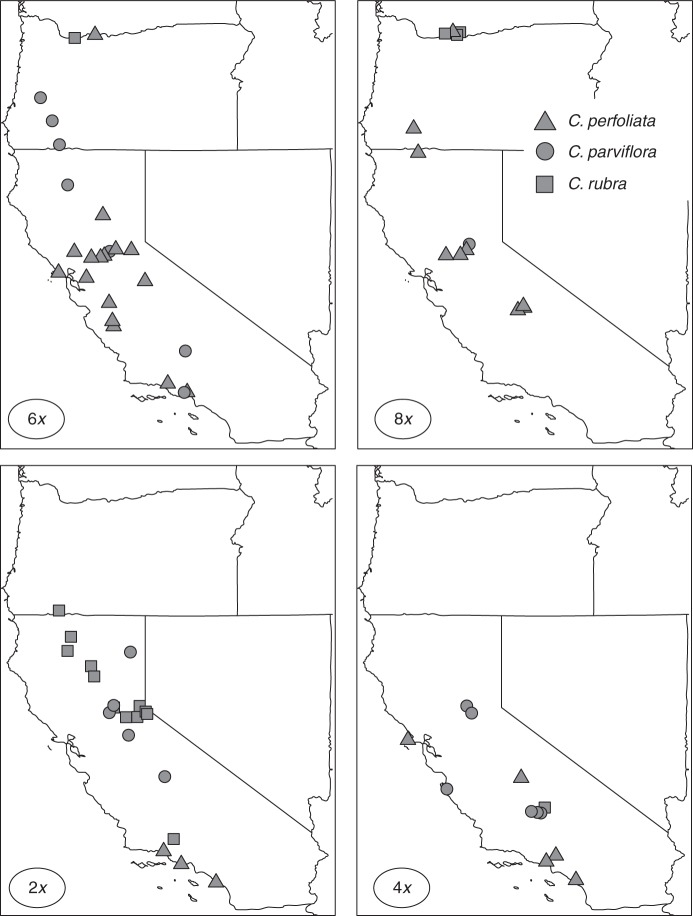
A total of 78 populations of Claytonia perfoliata were sampled along the west coast of North America, between Washington State in the north and southern California to the south (Fig. 1, Supplementary Data Table S1). Sampling was focused to capture the diversity of reported cytotypes documented for the group from previous cytogenetic studies, spanning the range from diploids to decaploids (Miller, 1978; Miller and Chambers, 2006), but does not represent the entire geographic distribution of the C. perfoliata complex. Due to logistical constraints, sampling did not include disjunct diploids and decaploids from Mexico or Guatemala.
Fig. 1.
Map of the western USA depicting locations of diploid, tetraploid, hexaploid and octoploid cytotypes of the Claytonia perfoliata complex identified in this study. Precise locations of all populations are provided in the Supplementary Data Table S1.
Flow cytometry
Genome size was estimated from a total of 121 individuals from the 78 localities using FCM. Many of these localities were previously surveyed for ploidy based on direct chromosome counts in earlier cytogenetic studies of the group (Swanson, 1964; Miller, 1978). Nuclear suspensions for FCM were prepared by grinding 3–6 seeds of Claytonia simultaneously with three seeds of an internal standard, Solanum lycopersicum, between two sheets of fine (27·9 grit cm−2) sandpaper in a 60 mm diameter Petri plate. Solanum lycopersicum cultivar Stupicke was used as an internal standard since it yielded clear 2C peaks from seed, and its 2C genome size of 1·96 pg (Dolezel et al., 1992) was relatively close in size to that of the diploid Claytonia taxa. Seed rather than leaf tissue was used for analysis due to extensive endopolyploidy found in leaf tissue making determination of the first 2C peak difficult, along with strong interference in estimates of genome size associated with variation in leaf pigmentation and secondary chemistry among accessions. Following co-grinding of the sample and standard, the sandpaper and plate were washed with two 600 µL aliquots of Galbraith's buffer (Dolezel et al., 2007), and the resulting suspension was filtered through two layers of Miracloth (CalBiochem, Pasadena, CA, USA). A 600 µL aliquot of filtered solution was transferred to a 5 mL tube and stained at a propidium iodide concentration of 50 µg mL−1 along with RNase at a concentration of 50 µg mL−1. Samples were kept on ice and protected from light for 30–60 min prior to analysis.
FCM measurements were performed on a Becton Dickinson FACScan flow cytometer equipped with a 488 nm argon laser maintained at the Optical Biology CORE Facility of the UC Davis Cancer Center. Peak means were identified using Cellquest software (Becton Dickson). Absolute DNA content was calculated for between one and three samples of seed from each of the 121 individual plants (accessions) from 78 localities. Replicate measurements of seeds from the same individual occurred on different days in order to obtain a more precise measure of average genome size of seeds from an individual while controlling for variation due to instrument drift and sample preparation. The average genome size across these replicates is reported for these plants. Readings with >5 % coefficient of variation (CV) or <1000 nuclei were discarded. Genome size was compared among taxa at the diploid, tetraploid and hexaploid levels using analysis of variance (ANOVA) in R 2.13.1 (R Development Core Team, 2010) followed by the Tukey HSD test for comparisons among groups.
Chromosome counts
Direct chromosome counts were made from meiotic or mitotic material from 65 accessions spanning the range of genome size estimates based on FCM. Mitotic counts were prepared from root tips while meiotic counts were prepared from whole immature flowers. Root tips were collected variously from rapidly growing greenhouse seedlings planted in small pots with the roots allowed to grow into sand, or from germinated seeds placed on filter paper in a refrigerator at 4 °C. Immature flowers were collected from leaf axils of mature plants. Both floral buds and root tips were fixed in 4:3:1 chloroform:ethanol:acetic acid for a minimum of 24 h. Prior to fixation, root tips were treated with 0·002 m 8-hydroxyquinoline for 3 h at 4 °C. Following fixation, plant material was stained in alcoholic hydrochloric acid–carmine for a minimum of 24 h (Snow, 1963), and heated to 60 °C for 1 h prior to preparation of squashes. Plant material was squashed onto a slide with 45 % acetic acid, with or without a drop of Hoyer's solution to make permanent slides.
Environmental variation and genome size
To test whether variation in genome size is associated with environmental variation, I compared genome size within a cytotype with minimum winter temperature and growing season precipitation (October–May, as members of the C. perfoliata complex within the study region typically germinate with late autumn or winter rains and flower between March and May). Climate data for accessions were obtained from monthly temperature and precipitation values extracted for each collection locality using the climate downscaling software ClimateWNA v.4.6.1 (Mbogga et al., 2009; Wang et al., 2011) based on the PRISM climate data set for North America (www.prism.oregonstate.edu; Daly et al., 2002). Individual regressions of genome size vs. temperature and precipitation were performed for all cytotypes with at least five accessions. In order to investigate patterns of genome size variation with environment across the entire complex, 2C genome size was standardized within cytotype (by subtracting the mean 2C genome size for a cytotype and dividing by the standard deviation for the cytotype). Environmental values were also standardized within cytotypes in order to account for relative differences in the range of environmental variation experienced by the different cytotypes. Linear and quadratic regression were used to examine the relationship between variation in standardized genome size and the standardized environmental variables for the entire complex. This approach is similar to that used in phenotypic selection analysis (Lande and Arnold, 1983; Kingsolver et al., 2001). A sequential Bonferroni correction was used to adjust for multiple tests. Regressions were done in R 2.13.1 (R Development Core Team, 2010).
RESULTS
Flow cytometry from seed samples resulted in clear fluorescence peaks (Fig. 2) and revealed 2C DNA content ranging from 1·42 to 7·46 pg for the complex as a whole (Fig. 3, Table 1). The three diploid taxa had distinct 2C genome sizes (F2,21 = 253·3, P < 0·001, Tukey HSD), with C. rubra ssp. rubra having the smallest genome and C. parviflora ssp. grandiflora the largest. One population of diploid C. parviflora ssp. grandiflora from the southern Sierra Nevada mountains (population ‘Trim-7’, Supplementary Data Table S1) had a genome size 18 % smaller than that of other populations. In contrast to a typical 2C genome size of 1·87 pg for diploid C. parviflora, this anomalous population had a 2C genome size of 1·55 pg based on FCM of seed from two individuals (FCM results on leaf tissue from three additional members of this population were consistent with this smaller genome size, but were not included in genome size estimates due to high CVs). Genome sizes were not significantly different between C. parviflora and C. perfoliata for tetraploids (F1,10 = 4·206, P < 0·067) or hexaploids (F1,23= 0·006, P < 0·937). Claytonia rubra was not included in comparisons of polyploid genome sizes due to small sample sizes for polyploids.
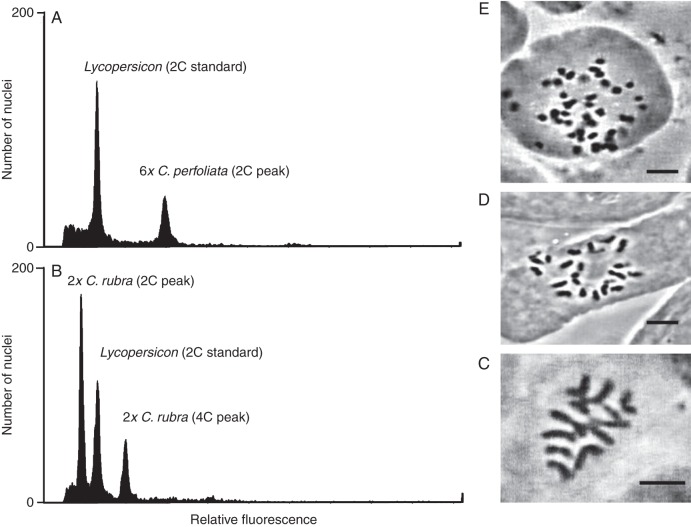
Fig. 2.
Representative chromosome counts (mitotic) and flow cytometry results for the C. perfoliata complex. (A) Hexaploid C. perfoliata ssp. perfoliata. (B) Diploid C. rubra ssp. rubra. (C) Diploid C. parviflora ssp. grandiflora, 2n = 12. (D) Tetraploid C. parviflora ssp. parviflora, 2n = 24. (E) Octoploid C. perfoliata ssp. perfoliata, 2n = 48. Scale bars = 10 µm.
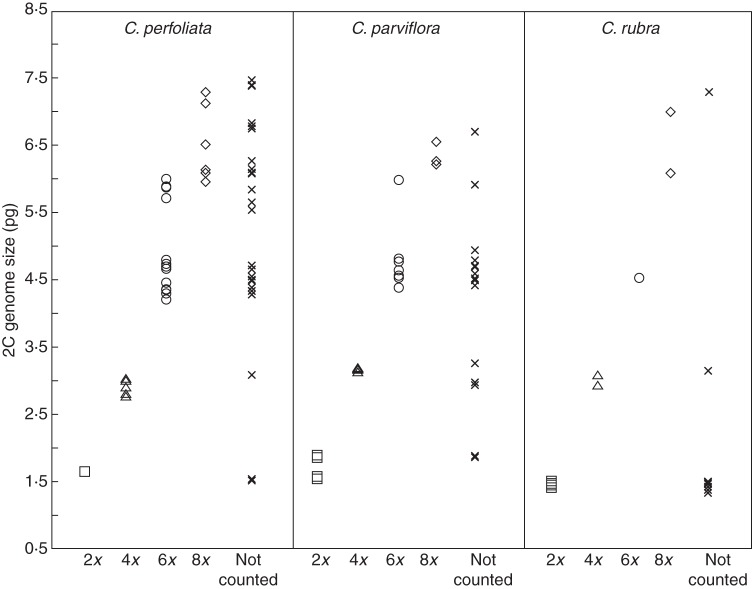
Fig. 3.
Genome size (2C) estimates from flow cytometry for all individuals in this study, for C. perfoliata, C. parviflora and C. rubra. Symbols depict ploidy status as determined by direct chromosome counts, or whether values are from individuals without direct chromosome counts. Note the overlap in genome size between hexaploids and octoploids among individuals, preventing clear determination of ploidy from flow cytometry at these ploidy levels.
Table 1.
Mean 2C genome sizes for cytotypes of the Claytonia perfoliata complex, with standard errors for genome size estimates, and inferred monoploid genome size (for a haploid set of six chromosomes). Taxa may be listed multiple times due to different ploidy levels or genome sizes within the same taxon
| Taxon | Populations (individuals) | Inferred ploidy | 2C genome size (pg) | s.e. | Inferred monoploid (1Cx) genome size (pg) |
|---|---|---|---|---|---|
| C. perfoliata subsp. mexicana | 3 (7) | 2x | 1·53 | 0·01 | 0·77 |
| C. parviflora subsp. grandiflora | 5 (11) | 2x | 1·87 | 0·01 | 0·94 |
| C. parviflora subsp. grandiflora – atypical | 1 (3) | 2x | 1·55 | 0·01 | 0·78 |
| C. rubra subsp. rubra | 9 (12) | 2x | 1·44 | 0·01 | 0·72 |
| C. perfoliata subsp. perfoliata | 4 (5) | 4x | 2·94 | 0·05 | 0·73 |
| C. rubra subsp. depressa | 4 (5) | 4x | 3·06 | 0·06 | 0·76 |
| C. parviflora subsp. parviflora | 6 (7) | 4x | 3·12 | 0·05 | 0·78 |
| C. perfoliata subsp. perfoliata | 17 (23) | 6x | 4·52 | 0·07 | 0·75 |
| C. perfoliata subsp. perfoliata – atypical | 3 (3) | 6x | 5·81 | 0·04 | 0·97 |
| C. parviflora subsp. parviflora | 15 (18) | 6x | 4·62 | 0·04 | 0·77 |
| C. parviflora subsp. parviflora – atypical | 2 (2) | 6x | 5·99 | 0·06 | 1·00 |
| C. rubra subsp. depressa | 1 (2) | 6x | 4·58 | 0·06 | 0·76 |
| C. perfoliata subsp. perfoliata | 5 (6) | 8x | 6·44 | 0·20 | 0·81 |
| C. parviflora subsp. parviflora | 2 (2) | 8x | 6·42 | 0·11 | 0·80 |
| C. rubra subsp. depressa | 2 (2) | 8x | 6·53 | 0·45 | 0·82 |
| C. perfoliata sensu latu -higher polyploids | 11 (13) | 8x or 10x | 6·97 | 0·14 | 0·70–0·87 |
Sample sizes are given as the number of populations sampled, with the total number of individuals measured in parentheses.
The majority of hexaploid plants had a 2C genome size between 4·2 and 4·7 pg; however, a subset of hexaploids in C. perfoliata and C. parviflora had a 2C genome size of approx. 5·9 pg, about 30 % larger than typical of most hexaploids, and within the range of genome sizes for octoploids (Fig. 3, Table 1). These hexaploids were confirmed through multiple chromosome counts, and the elevated 2C content confirmed through multiple bouts of flow cytometry. These hexaploids contributed to a pattern of increased variation in genome content with ploidy level. Variation in 2C genome size was significantly greater for hexaploid cytotypes than diploids (Fig. 3), Levene's test (F1,78 = 13·1, P = 0·005). There was no general pattern of reduced monoploid (1C×) genome content with higher ploidy, as mean monoploid genome content ranged from 0·72 to 0·94 pg among diploids and from 0·73 to 1·00 pg among polyploids. Several accessions had a genome content consistent with decaploidy, and decaploids are well documented from previous studies of this complex (Miller, 1978). However, chromosome squashes in the present study from accessions with putative decaploid levels of DNA suffered from such extensive overlap among chromosomes that a precise count was not possible (estimates ranged between 52 and 64). Chromosome counts documented a range of ploidy levels from diploid (2n = 12) to octoploid (2n = 48) based on a haploid set of six chromosomes as previously documented for the C. perfoliata complex (Fig. 2, Supplementary Data Table S1) (Swanson, 1964; Miller, 1978). No decaploid counts were confirmed in this study, although several accessions were consistent with a decaploid genome size in FCM, but with such poor spread and extensive overlap among chromosome preparations that an accurate count was not possible. The distribution of ploidy across the landscape was similar to that documented by previous researchers. Hexaploids of C. parviflora and C. perfoliata were widespread, while diploids of C. rubra were restricted to high latitudes or elevations, diploids of C. perfoliata were restricted to southern and coastal regions, and diploids of C. parviflora were restricted to the Sierra Nevada foothills (Fig. 1). Other cytotypes were sporadic in occurrence. In contrast to previous work, a mixed population of diploid and tetraploid C. perfoliata ssp. mexicana was documented in Cleveland National Forest in southern California. Although meiotic behaviour of populations was not generally recorded in this study (as the focus was ploidy determination), the meiotic behaviour of tetraploids from this population was distinctly aberrant, with the majority of observed meioses (seven of 12) exhibiting behaviours such as asynchronous meiosis, lagging chromosomes and multiple spindle orientations (e.g. Vasek, 1962) leading to unbalanced gametes. Nearly all meitoic events from other populations were normal, apart from occasional lagging chromosomes, as reported from previous observations of C. perfoliata polyploids (Swanson, 1964; Miller, 1978).
Comparisons of genome size with environmental variables revealed a significant but weakly negative linear relationship between variation in genome size and minimum winter temperature for the complex as a whole (R2 = 0·049, B = –0·26, P = 0·003; Table 2). However, when individual cytotypes were examined, this relationship was significant only for tetraploid C. parviflora, for which minimum winter temperature was strongly negatively correlated with genome size (R2 = 0·939, B = 0·974, P = 0·006; Table 3). The relationship for the complex as a whole was not driven solely by tetraploid C. parviflora, as the results changed only slightly when these were excluded from the analysis (R2 = 0·023, B = –0·21, P = 0·033). No significant correlations were found between genome size and precipitation for the complex as a whole (Table 2) or for individual cytotypes (Table 3).
Table 2.
Regression and partial regression coefficients (values from simple and multiple regression, respectively) for the linear and quadratic relationships between variation in genome size and variation in environmental factors across the Claytonia perfoliata complex
| Regression coefficient |
Partial regression coefficent |
|||
|---|---|---|---|---|
| Linear | Quadratic | Linear | Quadratic | |
| Winter temperature | –0·26** | –0·12 | –0·23* | –0·088 |
| Growing season precipitation | 0·19 | –0·127 | 0·129 | –0·11 |
Genome size and environmental variables were standardized within cytotypes prior to regression analysis.
* P < 0·05; ** P < 0·01.
Table 3.
Results of regressions of genome size on winter temperature and growing season precipitation
| Minimum winter temperature |
Growing season precipitation |
||||
|---|---|---|---|---|---|
| Cytotype | n | B | r2 | B | r2 |
| C. parviflora 2x | 13 | –0·470 | 0·225 | 0·164 | 0·027 |
| C. parviflora 4x | 7 | –0·974** | 0·939 | 0·152 | 0·023 |
| C. parviflora 6x | 16 | –0·060 | 0·004 | 0·403 | 0·163 |
| C. perfoliata 2x | 6 | –0·686 | 0·470 | 0·633 | 0·401 |
| C. perfoliata 4x | 8 | –0·562 | 0·317 | 0·240 | 0·058 |
| C. perfoliata 6x | 30 | –0·019 | 0·001 | –0·168 | 0·028 |
| C. rubra 2x | 14 | –0·056 | 0·003 | 0·196 | 0·039 |
| 8x (combined) | 25 | –0·105 | 0·011 | –0·247 | 0·060 |
Slope values (B) are standardized coefficients, in order to compare values of slope across the two environmental factors. Significance tests were adjusted using the sequential Bonferroni procedure.
** P < 0·01.
DISCUSSION
Chromosome counts revealed a similar geographic distribution of cytotypes to that previously reported (Swanson, 1964; Miller, 1978), with geographically widespread and co-occurring hexaploids of C. parviflora and C. perfoliata, scattered octoploids, and allopatric diploids of C. rubra, C. perfoliata and C. parviflora, restricted, respectively, to northern and montane, southern and coastal, and Sierran foothill environments (Fig. 2D). FCM, however, suggested previously undocumented, large-magnitude variation in genome size within several cytotypes. In addition, the presence of tetraploids within a diploid population of C. perfoliata ssp. mexicana suggests the possibility of ongoing formation of polyploids, and highlights the potential for contemporary gene flow from diploid to polyploid populations via newly formed polyploids. Previous investigations of C. perfoliata ssp. mexicana have not documented mixed diploid–tetraploid populations for this taxa, nor meiotic irregularities among tetraploids (Swanson, 1964; Miller, 1978; Miller and Chambers, 2006). The irregular meiosis observed from these tetraploids suggests the possibility that they are recently formed autotetraploids. Although irregular gamete formation is often a poor indicator of autopolyploidy vs. allopolyploidy and time since ploidy formation (Ramsey and Schemske, 2002), normal meiotic behaviour is typical of C. perfoliata polyploids, suggesting that recent autopolyploidy should be investigated as an explanation.
Extensive variation in genome size within cytotypes was found for hexaploids of C. parviflora and C. perfoliata, and for diploids of C. parviflora ssp. grandiflora. For diploid C. parviflora ssp. grandiflora, members of one population were found to have a genome size 18 % smaller than typical (1·55 pg. vs. 1·87 pg), while exhibiting typical diploid chromosome counts (n = 6). This population was located in the southern Sierra Nevada foothills and was morphologically distinct from other populations of C. parviflora ssp. grandiflora in that the plants possessed strongly pigmented red foliage and paler than typical pink floral colour. There are several possibilities that may explain the markedly reduced genome size of this population. Individuals at this site may be a cryptic species, may represent individuals from an otherwise typical C. parviflora ssp. grandiflora genomic background which has experienced genome reduction, or may be hybrids between C. parviflora ssp. grandiflora and a congener with a smaller genome. Alternatively, hybridization among other populations of this taxa might have led to an increase in genome size (Baack et al., 2005; Ungerer et al., 2009). Phylogenetically informative DNA sequence information from this population and other members of the C. perfoliata complex and closely related lineages would allow discrimination among these hypotheses. Although secondary compounds associated with red pigmentation such as anthocyanins or betalins may reduce fluorescence readings and lead to spuriously small genome size estimates (Dolezel et al., 2007; Bennett et al., 2008), readings were consistently lower for this population from both leaf and seed tissue and for an alternative buffer solution containing β-mercaptoethanol to reduce interference from cytosolic compounds (Ramsey, 2007).
The typical range of genome size for hexaploid C. perfoliata (approx. 4·4–4·7 pg) agrees with the previously reported genome size of 4·35 pg for a plant with a hexaploid chromosome count (Bennett and Smith, 1976). However, a portion of hexaploid plants confirmed by chromosome counts were documented with a genome size 30 % larger than typical (approx. 5·9 pg), placing them within the range of octoploid populations. These plants were widespread, occurring between the Southern Coast Ranges of California and the northern Sierra Nevada foothills. These plants were extreme examples of a general pattern of increased genome size variation at higher ploidy levels (Fig. 3). A potential explanation for varying genome size among higher polyploids includes differential genomic contributions of diploid C. parviflora ssp. grandiflora, C. perfoliata ssp. mexicana and C. rubra ssp. Rubra, which have monoploid genomes of 0·94, 0·77 and 0·72 pg, respectively, to distinct hexaploid lineages. However, the monoploid genome content of atypical hexaploids averaged 0·97 pg, slightly larger than the monoploid content of C. parviflora ssp. grandiflora (0·94), while an intermediate monoploid genome content would be expected if differential contributions of the diploid genomes of other species were accounting for hexaploids with high 2C values. One possibility is that other species of Claytonia with larger genomes may hybridize with members of the C. perfoliata complex. Claytonia sibirica represents a potential source of introgression, as it is hypothesized to have hybridized with C. perfoliata to form the allotetraploid C. washingtoniana (Fellows, 1976), and is sympatric in portions of California, Washington and Oregon with members of the C. perfoliata complex (Miller et al., 1984). In addition, genomic contributions from putatively extinct diploid lineages have been documented in polyploid complexes (e.g. Brokaw and Hufford, 2010; Jakob and Blattner, 2010), representing an additional source of historic introgression. An additional possibility would be within-lineage factors associated with genome size, such as the spread of retrotransposons within some populations leading to increased genome size (e.g. Bennetzen et al., 2005; Hawkins et al., 2006; Ungerer et al., 2009). The use of seeds rather than adults in this study might result in an increase in measured variation within lines, and might result in differences between genome size estimates from adult tissue, as individual seeds may vary in genome size and also differ in their likelihood of survival. However, variation within lines would be unlikely to affect the comparisons in this study which are based on differences among maternal lines and across ploidy levels, unless there were very strong biases in viability of seeds or fitness of offspring among maternal lines with different genome size, which was not observed in field experiments involving these same maternal lines (McIntyre, 2011).
Environment and genome size
Environmental variation in the form of mean winter temperature was weakly negatively correlated with genome size across the complex, and strongly so for C. parviflora tetraploids, but not for any other individual cytotype. It is unclear why such a strong relationship would be found for only one cytotype; however, the sampling distribution of cytotypes across the landscape may influence the potential to detect such a correlation. Relative to other cytotypes, sampling for C. parviflora covered a wide latitudinal gradient, with little variation in whether populations were inland or coastal. It is possible that variation in whether a population is inland or coastal may reflect other environmental conditions altering selection on genome size. Alternatively, C. parviflora might be subject to stronger selection for genome size in relation to winter temperature than other members of the complex. Previous analysis of the distributions of cytopyes on the landscape suggests niche differentiation across ploidy levels (McIntyre, 2012), and it would be interesting to investigate whether variation in genome size is also associated with ecological differentiation, or if similar ecological factors are associated with variation in both ploidy and genome size. More extensive measurements of genome size from across the distribution of the C. perfoliata complex would be informative in determining whether genome size is associated with other environmental factors for individual cytotypes, as the number of individuals sampled in the study (121) did not allow for fitting multivariate relationships within cytotypes.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
Joyce Jiao assisted with cytometry measurements. John Miller provided collecting notebooks and advice on chromosome counts in Claytonia. N. Ivalú Cacho, Dena Grossenbacher, Jon Haloin, Michelle Afkhami and Jean Burns provided feedback on the topic of genome size variation among polyploids. Carol Oxford of the UC Davis Cancer Center Optical Biology Laboratory provided flow cytometry support. This research was funded by student grants from the Botanical Society of America, California Native Plant Society, Davis Botanical Society and UC Davis Center for Population Biology. Sharon Strauss oversaw this research as my dissertation advisor. This work was performed in partial fulfilment of the requirements for the PhD in Population Biology at UC Davis.
LITERATURE CITED
- Baack EJ, Whitney KD, Rieseberg LH. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytologist. 2005;167:623–630. doi: 10.1111/j.1469-8137.2005.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barow M. Endopolyploidy in seed plants. Bioessays. 2006;28:271–281. doi: 10.1002/bies.20371. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Price HJ, Johnston JS. Anthocyanin inhibits propidium iodide DNA fluorescence in Euphorbia pulcherrima: implications for genome size variation and flow cytometry. Annals of Botany. 2008;101:777–790. doi: 10.1093/aob/mcm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society B: Biological Sciences. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread polyploidy in model organisms inferred from age distributions of duplicate genomes. The Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw JM, Hufford L. Origins and introgression of polyploid species in Mentzelia section Trachyphytum (Loasaceae) American Journal of Botany. 2010;97:1457–1473. doi: 10.3732/ajb.0900388. [DOI] [PubMed] [Google Scholar]
- Comai L. The adavantages and disadvantages of being polyploid. Nature Reviews Genetics. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. Widespread genome duplications throughout the history of flowering plants. Genome Research. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly C, Gipson WP, Taylor GH, Johnson GL, Pasteris P. A knowledge-based approach to the statistical mapping of climate. Climate Research. 2002;22:99–113. [Google Scholar]
- Dolezel J, Sgorbati S, Lucretti S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiologia Plantarum. 1992;85:625–631. [Google Scholar]
- Dolezel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1865. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilam T, Anikster Y, Millet E, Manisterski J, Feldman M. Genome size in diploids, allopolyploids, and autopolyploids of Mediterranean triticeae. Journal of Botany. 2010;2010:341380. http://dx.doi.org/10.1155/2010/341380 . [Google Scholar]
- Fellows CE. Chromosome counts and a new combination in Claytonia sect. Limnia (Portulacaceae) Madrono. 1976;23:296–297. [Google Scholar]
- Grant V. Plant speciation. New York: Columbia University Press; 1981. [Google Scholar]
- Grime JP, Mowforth MA. Variation in genome size – an ecological interpretation. Nature. 1982;299:151–153. [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Research. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob SS, Blattner FR. Two extinct diploid progenitors were involved in allopolyploid formation in the Hordeum murinum (Poaceae: Triticeae) taxon complex. Molecular Phylogenetics and Evolution. 2010;55:650–659. doi: 10.1016/j.ympev.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Jakob SS, Meister A, Blattner FR. The considerable genome size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology, and speciation rates. Molecular Biology and Evolution. 2004;21:860–869. doi: 10.1093/molbev/msh092. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences, USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver JG, Hoekstra HE, Hoekstra JM, et al. The strength of phenotypic selection in natural populations. American Naturalist. 2001;157:245–261. doi: 10.1086/319193. [DOI] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters. 2002;5:66–76. [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany. 2005;95:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes and Development. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, et al. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) Annals of Botany. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. Polyploidy and novelty in flowering plants. American Naturalist. 1983;122:1–25. [Google Scholar]
- Marhold K, Kudoh H, Pak J-H, Watanabe K, Španiel S, Lihová J. Cytotype diversity and genome size variation in eastern Asian polyploid Cardamine (Brassicaceae) species. Annals of Botany. 2010;105:249–264. doi: 10.1093/aob/mcp282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ, et al. Recently formed polyploid plants diversify at lower rates. Science. 2011;333:1257. doi: 10.1126/science.1207205. [DOI] [PubMed] [Google Scholar]
- Mbogga M, Hamann A, Wang T. Historical and projected climate data for natural resource management in western Canada. Agricultural and Forest Meteorology. 2009;149:881–890. [Google Scholar]
- McIntyre PJ. Effects of whole genome duplication on adaptation, plasticity and niche variation in the Claytonia perfoliata (Portulacaceae) polyploid complex. 2011 doi: 10.1093/aob/mcs187. PhD thesis. University of California, Davis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre PJ. Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. American Journal of Botany. 2012;99:655–662. doi: 10.3732/ajb.1100466. [DOI] [PubMed] [Google Scholar]
- Meyers LA, Levin DA. On the abundance of polyploids in flowering plants. Evolution. 2006;60:1198–1206. [PubMed] [Google Scholar]
- Miller JM. Phenotypic variation, distribution and relationships of diploid and tetraploid populations of the Claytonia perfoliata complex (Portulacaceae) Systematic Botany. 1978;3:322–341. [Google Scholar]
- Miller JM, Chambers KL. Systematics of Claytonia (Portulaceae) Systematic Botany Monographs. 2006;78:1–236. [Google Scholar]
- Miller JM, Kenton LC, Fellows CE. Cytogeographic patterns and relationships in the Claytonia sibirica complex (Portulacaceae) Systematic Botany. 1984;9:266–271. [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. Evolutionary consequences of autopolyploidy. New Phytologist. 2010;186:5–17. doi: 10.1111/j.1469-8137.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. www.r-project.org . [Google Scholar]
- Ramsey J. Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae) Heredity. 2007;98:143–150. doi: 10.1038/sj.hdy.6800912. [DOI] [PubMed] [Google Scholar]
- Ramsey J. Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences, USA. 2011;108:7096–7101. doi: 10.1073/pnas.1016631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics. 2002;33:589–639. [Google Scholar]
- Rausch JH. The evolution of selfing, inbreeding depression, and polyploidy in the Claytiona perfoliata complex (Portulacaceae). 2008 PhD thesis, Washington State University, Pullman. [Google Scholar]
- Šmarda P, Bureš P, Horová L, Rotreklová O. Intrapopulation genome size dynamics in Festuca pallens. Annals of Botany. 2008;102:599–607. doi: 10.1093/aob/mcn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow R. Alcoholic hydrochloric acid–carmine as a stain for chromosomes in squash preparations. Stain Technology. 1963;38:9–13. doi: 10.3109/10520296309061161. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold Publishers; 1971. [Google Scholar]
- Swanson JR. Claytonia (Montia) perfoliata: a genecological and evolutionary study. 1964 PhD thesis, University of California at Berkeley. [Google Scholar]
- Ungerer MC, Strakosh SC, Stimpson KM. Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biology. 2009;7:40. doi: 10.1186/1741-7007-7-40. http://dx.doi.org/10.1186/1741-7007-7-40 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasek FC. ‘Multiple spindle’ – ameiotic irregularity in Clarkia exilis. American Journal of Botany. 1962;49:536–539. [Google Scholar]
- Veselý P, Bureš P, Šmarda P, Pavlíček T. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annals of Botany. 2012;109:65–75. doi: 10.1093/aob/mcr267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Hamann A, Spittlehouse DL, Murdock TQ. ClimateWNA—high-resolution spatial climate data for Western North America. Journal of Applied Meteorology and Climatology. 2011;51:16–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.