Abstract
Background and Aims
In mountain plant populations, local adaptation has been described as one of the main responses to climate warming, allowing plants to persist under stressful conditions. This is especially the case for marginal populations at their lowest elevation, as they are highly vulnerable. Adequate levels of genetic diversity are required for selection to take place, while high levels of altitudinal gene flow are seen as a major limiting factor potentially precluding local adaptation processes. Thus, a compromise between genetic diversity and gene flow seems necessary to guarantee persistence under oncoming conditions. It is therefore critical to determine if gene flow occurs preferentially between mountains at similar altitudinal belts, promoting local adaptation at the lowest populations, or conversely along altitude within each mountain.
Methods
Microsatellite markers were used to unravel genetic diversity and population structure, inbreeding and gene flow of populations at two nearby altitudinal gradients of Silene ciliata, a Mediterranean high-mountain cushion plant.
Key Results
Genetic diversity and inbreeding coefficients were similar in all populations. Substantial gene flow was found both along altitudinal gradients and horizontally within each elevation belt, although greater values were obtained along altitudinal gradients. Gene flow may be responsible for the homogeneous levels of genetic diversity found among populations. Bayesian cluster analyses also suggested that shifts along altitudinal gradients are the most plausible scenario.
Conclusions
Past population shifts associated with glaciations and interglacial periods in temperate mountains may partially explain current distributions of genetic diversity and population structure. In spite of the predominance of gene flow along the altitudinal gradients, local genetic differentiation of one of the lower populations together with the detection of one outlier locus might support the existence of different selection forces at low altitudes.
Keywords: Gene flow, genetic variation, glacial refuge, local adaptation, mountain plant, altitudinal range, Silene ciliata, Caryophyllaceae
INTRODUCTION
Plant responses to environmental change include migration, local adaptation and phenotypic plasticity (Parmesan, 2006). However, the ability of species to adopt one or a combination of these strategies is largely unknown (Jump and Peñuelas, 2005). Rapid and extreme environmental changes, with diverse drivers acting simultaneously in the same or opposite directions (Nogués-Bravo et al., 2007), can determine plant responses in each oncoming scenario. Marginal populations located at the edge of their distribution range have become the focus of several ecological studies in a variety of habitats (Sagarin and Gaines, 2002, and references therein). These populations share features such as restricted habitat availability, more stressful conditions than in central populations and, usually, geographical isolation (Lawton, 1993).
Adaptation in marginal habitats plays a crucial role in the evolution and survival of populations growing in peripheral areas, making adaptive processes particularly important in species occurring in geographically disconnected areas (Kawecki, 2008). The ability of an organism to persist in these marginal habitats depends on several factors, including the mechanisms allowing sufficient genetic variation to be maintained (essential for retaining enough evolutionary potential in small populations, Hedrick, 1986), and the traits affected by natural selection (Hedrick, 2006) and their role of these factors in the diversification process (reviewed by Schluter, 2001). As adaptation is a complex response involving multiple factors (Kawecki and Ebert, 2004), its study requires the formulation of hypotheses which engage ecological and genetic factors that may promote or hinder such response (Mitchell-Olds et al., 2008).
Under any selective pressure scenario (like ongoing global warming), individuals may be the target of natural selection through the selection of phenotypes and the underlying genotypes that maximize fitness under new environmental conditions (Ennos, 2001). Genetic diversity plays a crucial role in this process by providing genotypes which would be tested by natural selection (Hughes et al., 2008). Inbreeding is another critical process that can potentially reduce the ability of plants to respond to selection pressures, especially in small populations (Armbruster and Reed, 2005; Kawecki, 2008). It can lead to a rapid excess of homozygous genotypes, dramatically reducing within-population genotypic diversity and increasing the frequency of deleterious genotypes (Frankham et al., 2002), if they have not been purged previously. This is especially true for populations that have a short inbreeding history or have recently experienced bottlenecks or fragmentation (Gautschi et al., 2002). Inbreeding depression can also make peripheral populations more prone to extinction (Nieminen et al., 2001). However, it is difficult to interpret inbreeding depression responses to stressful and marginal conditions, because the intensity of inbreeding depression could vary with environmental conditions (Armbruster and Reed, 2005).
Gene flow has long been recognized as one of the major factors potentially limiting responses in marginal habitats, because high levels of gene flow may swamp locally adapted genotypes (Kirkpatric and Barton, 1997). This makes the balance between gene flow and natural selection crucial (Kawecki and Ebert, 2004) in determining the potential level of local adaptation of a population. While some basal levels of gene flow could contribute to maintaining genetic diversity in populations where selection could act (Kawecki, 2008), high levels of gene flow would tend to genetically homogenize populations. This could erode local adaptation by diluting pre-adapted genes and reducing the frequency of locally adapted phenotypes, which would ultimately reduce overall population fitness (García-Ramos and Kirkpatrick, 1997). Nevertheless, populations may still manage to maintain locally adapted genotypes despite high levels of gene flow from other populations (Byars et al., 2009; Gonzalo-Turpin and Hazard, 2009), especially when selection pressures are also high, reaching an equilibrium between gene flow and selection (Jain and Bradshaw, 1966; Antonovics, 1968). It is worthy of note that the effective gene flow among populations is not solely dependent on dispersal capabilities, but also on the performance of immigrants, offspring fitness (Kawecki and Ebert, 2004) and the number of genes or traits that are involved in local adaptation (Kawecki, 2008).
High-mountain habitats are ideal natural laboratories for testing evolutionary hypotheses related to the balance between gene flow and local adaptation (Körner, 2003). Local altitudinal gradients allow for great environmental heterogeneity in a small geographical distance, making them suitable for conducting comparative observations or manipulative experiments in natural populations. High-mountain habitats have been identified among the most fragile environments in the world due to global change (Nogués-Bravo et al., 2007), and plant populations in mountain systems are especially vulnerable to the consequences of warming (Jump and Peñuelas, 2005, see also www.gloria.ac.at, Gottfried et al., 2012). Range shifts of plant populations along altitudinal gradients in mountains have been inferred for past and present times (Taberlet et al., 1998). Phylogeographical evidence has shown that population migrations along latitudinal ranges were frequent during the glacial–interglacial periods (Hewitt, 2001), that upward and downward migrations along the altitudinal range also occurred (Gutiérrez Larena et al., 2002), and that it is still an ongoing process (e.g. Gonzalo-Turpin and Hazard, 2009). These altitudinal range shifts of plant populations in a landscape of interspersed isolated high-mountain habitats may lead to an ecological scenario where gene flow is more likely to occur along altitudinal gradients than between mountains, thereby structuring genetic diversity in ‘mountain islands’ (Hewitt, 2001) and hindering local adaptation along the altitudinal gradient. In an alternative scenario, gene flow among populations at the same altitude but in neighbouring mountains may prevail over gene flow among populations along the altitudinal gradient of the same mountain (Taberlet et al., 1998; Byars et al., 2009) leading to the structuring of genetic diversity in ‘isolated belts’ (Fig. 1) and promoting local adaptation along the altitudinal gradient. In both scenarios, gene flow may not be sufficiently intense to restrict local adaptation responses (Byars et al., 2007; Gonzalo-Turpin and Hazard, 2009). Similar patterns have been shown by latitudinal gradients (e.g. Ward, 1969; Ingvarsson et al., 2006; Hall et al., 2007). Thus, this context fosters the study of the role of genetic diversity and gene flow in the process of local adaptation of plant populations in marginal habitats, such as those in high mountains (see Ohsawa and Ide, 2008, and references therein).
Fig. 1.
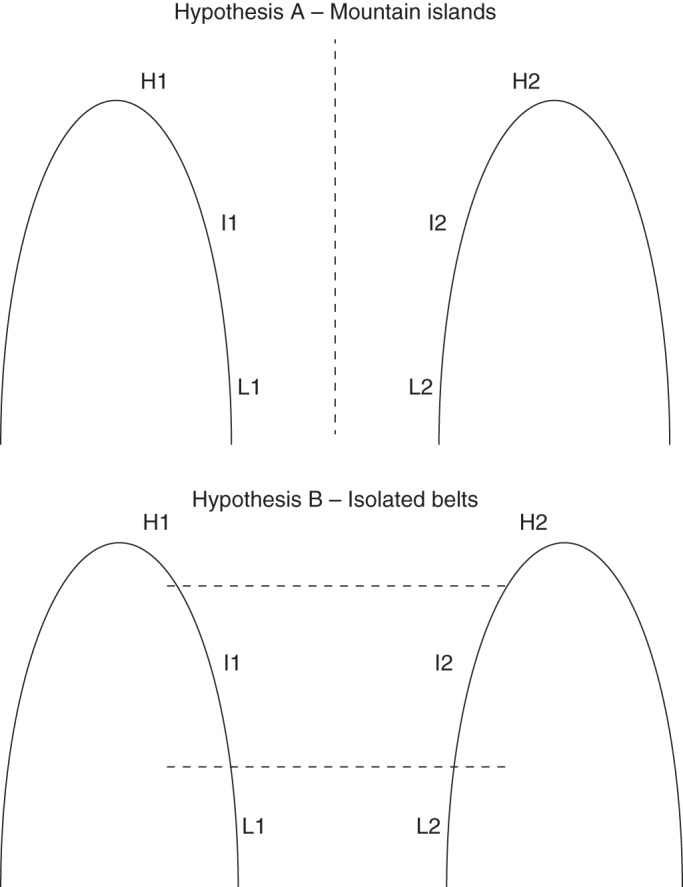
Hypothetical scenarios of predominant gene flow between mountains in the Sierra de Guadarrama. In each mountain (1, Peñalara; 2, Cabeza de Hierro), three populations were selected: High (H1 and H2), Intermediate (I1 and I2) and Low (L1 and L2). Discontinuous lines indicate hypothetical barriers for gene flow in each model.
Silene ciliata Pourr. (Caryophyllaceae) is a perennial high-mountain specialist that inhabits mountain ranges of the Mediterranean basin (Tutin et al., 1995). At the southern edge of the mountain systems of the central Iberian Peninsula, recent research has shown differences in phenology (Giménez-Benavides et al., 2007c), seed germination and seedling survival (Giménez-Benavides et al., 2007b) among populations located along an altitudinal gradient, suggesting the existence of local adaptation. In this species, populations inhabiting the lowest elevation are the most vulnerable, because they showed lower flower production (Giménez-Benavides et al., 2008) and lower recruitment rates (Giménez-Benavides et al., 2011). However, they also displayed greater evidence of local adaptation (Giménez-Benavides et al., 2007b). The aim of this study was to characterize the genetic patterns of S. ciliata populations located along two altitudinal gradients in two nearby mountains. For this purpose, we used expressed sequence tag (EST)–simple sequence repeat (SSR) markers transferred from the congener Silene latifolia and newly developed nuclear SSR loci from S. ciliata to genetically characterize S. ciliata populations. We addressed the following questions: (1) Do patterns of genetic diversity within and among populations depend on their location along the altitudinal gradient? (2) Are inbreeding coefficient values similar along the gradient? (3) Are levels of gene flow higher among populations within altitudinal gradients (‘mountain islands’) or among populations of similar altitudinal stages of nearby mountains (‘isolated belts’)? (4) Is the spatial genetic structure of S. ciliata correlated to either model (‘mountain islands’ or ‘isolated belts’)?
MATERIAL AND METHODS
Study species and population selection
Silene ciliata is a perennial cushion plant forming pulviniform rosettes of up to 2 cm in height and 15 cm in diameter. Self-pollination experiments (García-Fernández et al., 2012c) indicate that S. ciliata is potentially self-compatible. However, passive autogamy is partially restricted due to a pronounced protandry. S. ciliata inhabits the main mountain ranges in the northern half of the Iberian Peninsula, the Massif Central, the Apennines and the Balkan Peninsula (Tutin et al., 1995). In the Iberian Peninsula, S. ciliata reaches its southern latitudinal limit in the Sierra de Guadarrama (central Spain). These populations are considered relict because of their isolation from populations in other mountain systems further north. S. ciliata presents variable ploidy levels in natural populations (Tutin et al., 1995); however, all individuals from the studied Guadarrama populations were diploid (2n = 24, García-Fernández et al., 2012a). S. ciliata has nocturnal pollination interactions with Hadena consparcatoides Schawerda (Lepidoptera: Noctuidae) but can also be pollinated by diurnal insects (Giménez-Benavides et al., 2007a). Its seeds lack specialized structures to promote wind or animal dispersal and thus barochorous dispersal seems the most likely mechanism.
For this study, two altitudinal gradients were selected in the Sierra de Guadarrama (Figs 1 and 2). Altitudinal gradients (Gradient 1 and Gradient 2) were located on the two highest peaks of the mountain system (Peñalara at 2420 m a.s.l. and Cabeza de Hierro at 2380 m a.s.l., respectively), which are 6·1 km apart. The highest populations along each gradient (hereafter H1 and H2) were located near the summit of each mountain peak. The plant communities on these peaks and crests are dominated by patches of Festuca curvifolia Lag. ex Lange grassland, occasionally displaced by Nardus stricta L. grassland, and coexist with a high diversity of pulviniform plants. The intermediate populations were located at 2200 m a.s.l. in both gradients. We selected Dos Hermanas summit as the intermediate population in Gradient 1 (hereafter I1) and Bola del Mundo summit as the intermediate population in Gradient 2 (hereafter I2). These two intermediate populations are 6 km apart. Population I1 is 1·7 km from population H1, whereas the distance between the populations I2 and H2 is over 4·4 km. The dominant vegetation at both sites is F. curvifolia grassland with the occasional presence of Cytisus oromediterraneus Rivas-Mart. or Juniperus communis L. subsp. alpina (Suter) Čelak. The lowermost populations were located at 1980 m a.s.l. in each gradient. The two low populations are 5·7 km apart. The lowermost population in Gradient 1 (hereafter L1) is located in the lateral moraine deposit of the glacial cirque of Laguna de Peñalara and is about 1·9 km from population I1. The lowermost population in Gradient 2 (L2) is located on the margin of an old abandoned ski slope 1·5 km from population I2. In the lowermost populations, the plant community is dominated by a shrub matrix of C. oromediterraneus and J. communis with Scots pines (Pinus sylvestris L.) interspersed in an F. curvifolia pasture. These populations are not completely isolated as several populations are found between them and the selected intermediate populations. All the populations from Gradient 1 are located in the Peñalara Natural Park, whereas the high and intermediate populations of Gradient 2 (H2 and I2) are located within the boundaries of the Cuenca Alta del Manzanares Regional Park. Figure 1 shows the schematic representation of the populations and the hypothesized gene flow scenarios, and Fig. 2 shows their location in a topographical map. All studied sites have population sizes of over 100 individuals and have been the subject of other studies focused on different local adaptation responses between populations, floral asynchrony along the altitudinal gradient and the demographic bottleneck detected at certain locations. (e.g. Giménez-Benavides et al., 2007b, c, 2008).
Fig. 2.
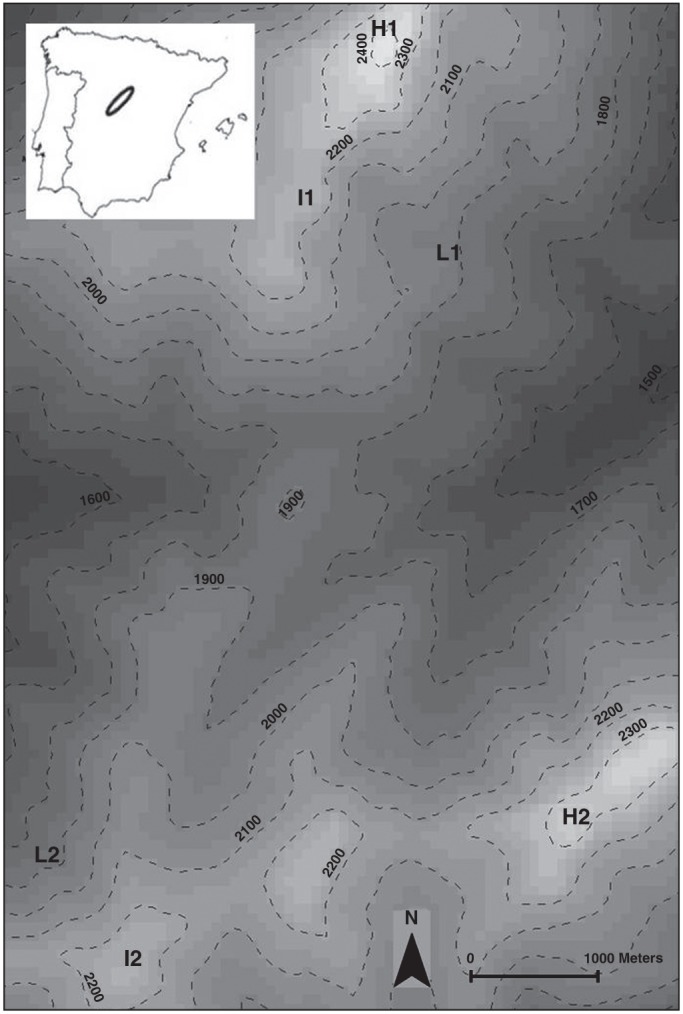
Location of the populations in the study zone. ‘High’ (H1 and H2), ‘Intermediate’ (I1 and I2) and ‘Low’ (L1 and L2) populations distributed over two mountains (1, Peñalara; 2, Cabeza de Hierro). Discontinuous lines represent 100-m contour intervals.
In each population, 30 individuals separated by at least 5 m were sampled. Plant material consisting of green leaves was collected from all individuals, taking care that it showed no signs of parasites, fungal infection or drought injuries. The geographical coordinates of each individual were recorded using a Garmin GPS 12 receiver. Leaf material was cleaned and dried in silica gel (Chase and Hills, 1991) for storage until DNA extraction. DNA was extracted following the protocol of Elphinstone et al. (2003) in 96-well plates with 10–20 mg of dried S. ciliata tissue. DNA concentration was estimated with a Nanodrop 3300 fluorospectrometer (ThermoScientific, www.thermoscientific.com) and confirmed in agarose gels. Extracted DNA concentration was higher than 10 ng μL−1 for all samples.
EST-SSR transfer from S. latifolia to S. ciliata
Cross-species transferability of EST-SSR markers from S. latifolia Poir. was conducted from an initial screening of the EST-SSR markers developed by Moccia et al. (2009, 2010) to identify polymorphic markers with high reproducibility and reliable interpretation. These tests included some markers previously assayed in S. ciliata (Moccia et al., 2009). To conduct polymorphism tests and discard PCR amplification failures on these markers, we used three samples of S. ciliata from different populations and one sample of S. latifolia as a positive control.
Amplification of EST-SSRs followed the procedure of Schuelke (2000). PCR reactions consisted of a three-primer system: the forward primer with the 5′ end extended with a universal M13 tail, the reverse locus-specific primer and a fluorescently labelled M13 primer. PCR reactions were conducted in 10-μL reaction volume containing 10 ng template DNA, 2 mm MgCl2, 0·2 mm dNTPs, 0·2 µm fluorescently labelled (FAM or VIC) M13 primer, 0·2 µm reverse primer, 0·05 µm forward primer with M13 tail and 0·05 U GoTaq (Promega, Fitchburg, MA, USA). For this assay, the PCR cycling profile was an initial melting step for 5 min at 94 °C, followed by 30 cycles of 94 °C for 30 s, 60 °C for 45 s (annealing) and 72 °C for 45 s (extension), followed by eight additional cycles in which the annealing temperature was dropped to 52 °C, and a final extension at 72 °C for 10 min. One microlitre of PCR products was diluted with 9·1 µL of a loading mixture comprising 9 µL HiDi Formamide and 0·1 µL Genescan 500 LIZ internal size standard (Applied Biosystems, Madrid, Spain). Samples were run on an automated DNA sequencer (ABI PRISM 3730 Genetic Analyzer, Applied Biosystems). Fragment sizes were assigned to alleles using GeneMarker v. 1·75 (Softgenetics LLC, State College, PA, USA).
Genomic SSR marker development for S. ciliata
Microsatellite loci were isolated and characterized from three partial genomic libraries enriched for CT/GT/CTT repeats following the FIASCO protocol (Zane et al., 2002) with some modifications. Details of the protocol followed are included in the Supplementary Data.
A total of 122 clones were sequenced, 53 % of which contained microsatellite sequences. Primers were designated for 17 clones using PRIMER3 (Rozen and Skaletsky, 2000). Twelve primer pairs produced clear amplicons of the expected size in 2 % agarose gels and were subsequently selected for fluorescence labelling and further analysis. Forward primers were labelled with fluorescent dyes (6-FAM, VIC, PET and NED) for automated electrophoresis. PCRs were performed in a 20-μL mix containing 3 pmol each of the labelled (forward) and unlabelled (reverse) primers, 0·2 mm of each dNTP, 2 mm MgCl2, 1× Taq Buffer (Biotools, http://www.biotools.eu/), 1 U Taq DNA polymerase (Biotools) and 20 ng template DNA. The PCR program consisted of one step of 4 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 51 or 56 °C (depending on primer pair) and 1 min at 72 °C, and a final extension step of 7 min at 72 °C. The products were run on an ABI 373 automated sequencer (Applied Biosystems) and the amplified fragment lengths were sized with GENEMARKER v. 1·75 (Softgenetics LLC) using LIZ500 as the internal lane standard.
After an initial screening, eight of 12 loci selected showed monomorphic patterns or multiple amplification products, suggesting locus duplication or unspecific amplification, and were subsequently discarded (data not shown). The remaining four loci (Sci1224, Sci1208, Sci0106 and Sci1443) showed clear, interpretable amplification patterns and were polymorphic. They were thus selected for genotyping of all samples (Table 1).
Table 1.
Characterization of four nuclear microsatellites in six Silene ciliata populations
| Locus | Repeat motif | Primer sequences (5′–3′) | Ta (°C) | Size range (bp) | NA | GB |
|---|---|---|---|---|---|---|
| Sci-1224 | (CT)18 | F: NED-ACCTGATTAGAAGACACAGGAGGA | 56 | 140–196 | 26 | JF979125 |
| R: TTTATGTTGCCGCATCCTTATC | ||||||
| Sci-1208 | (CT)10 | F: PET-TGGAAACGATGTATGAGACGA | 56 | 158–182 | 11 | JF979126 |
| R: TGTGATTGAAGTAGCCAAACCT | ||||||
| Sci-0106 | (CT)7 | F: VIC-AAACAAACGAGCGATCATCTAA | 56 | 111–135 | 12 | JF979127 |
| R: TTCCGATGCTTCTGGTACTTCT | ||||||
| Sci-1443 | (CT)7 | F: FAM-GACGCCTTTCTCAAAATCACC | 56 | 126–160 | 11 | JF979128 |
| R: GGGAATCTAGGGTTTAGCAGTGA |
The repeat motif, forward (F) and reverse primer (R) sequence, annealing temperature (Ta), size range, total number of alleles (NA) and GenBank accession number (GB) are given for each locus.
Statistical analyses
Microsatellite polymorphism and genetic diversity
Genetic diversity indices, including total number of alleles (NA), observed (HO) and expected (HE) heterozygosities and inbreeding coefficients (FIS) were calculated with Genepop v. 4·1 (Rousset, 2008). This same software was used to estimate Wright's F-statistics according to Weir and Cockerham (1984), to check for departures from Hardy–Weinberg (HW) equilibrium at each locus and population using Fisher's exact tests, and to check for genotypic linkage disequilibrium between pairs of loci within each population using the log-likelihood ratio G statistic (based on 5000 permutations). The same parameters were estimated after grouping populations by mountain island origin (Gradient 1 vs. Gradient 2) or isolated belts (Low vs. Intermediate vs. High). FSTAT v. 2·9.3 (Goudet, 1995) was used to compare genetic diversity indices [allelic richness (A), HO, genetic diversity within populations (HS) and FIS] in the two hypothetical ecological scenarios (mountain island and isolated belts, Fig. 1) and significance was assessed based on 10 000 permutations. All parameters were calculated for each transferred EST-SSR and each genomic SSR locus. All tests which involved multiple comparisons were corrected with the Dunn–Šidák method (Sokal and Rohlf, 1995).
Population structure and differentiation
Pairwise FST values (Weir and Cockerham, 1984) were calculated for each population pair and between population groups considering both hypotheses (mountain islands or isolated belts) using FSTAT v. 2·9.3 (Goudet, 1995). The significance of between-site relationships was evaluated based on 10 000 permutations in each case. The number of migrants between populations was estimated with the maximum-likelihood method implemented in MIGRATE-N v. 3·0.3 (Beerli and Felsenstein, 1999, 2001). For these assumptions, ten short and three long chains were run, with sampling increments of 20 for each and a burn-in period set to 10 000.
Mantel and partial Mantel tests were used to check for the correlation between genetic, geographical and altitudinal distances, searching for isolation by distance (IBD). Slatkin's (1995) pairwise linearized FST values [i.e. FST/(1 – FST)] were calculated using ARLEQUIN v. 3·5 (Excoffier and Lischer, 2010) while geographical and altitudinal distances were generated from GPS coordinates in Arc-ViewTM (ESRI®, http://www.esri.com/) and subsequently log-transformed. Partial Mantel tests are similar to a partial correlation, as they can detect the correlation between two matrices of interest when the effect of a third matrix is kept constant (Legendre and Legendre, 1998). Mantel and partial Mantel tests were carried out in R v. 2·9.2 using the Vegan package with 10 000 permutations and Kendall and Pearson correlation methods.
Genetic structure was also analysed using Bayesian clustering methods implemented in STRUCTURE v. 2·3.1 (Falush et al., 2007). Our analyses were based on an admixture ancestral model with correlated allele frequencies (because of high FIS values; Falush et al., 2007) for a range of K values starting from 1 to the number of populations considered plus 2 (i.e. 8). The proportion of membership of each individual and population to the inferred K clusters was then calculated. We used a burn-in period and a run length for the Monte Carlo Markov chain (MCMC) of 5 × 105 and 1 × 106 iterations, respectively. Ten runs were carried out for each K to quantify the amount of likelihood variation. The number of K present in the data set was evaluated according to Evanno et al. (2005). This method uses an ad hoc parameter (ΔK) to estimate the rate of change of likelihood values between successive K values. Additional Bayesian analyses were conducted with BAPS v. 5·4 (Bayesian Analysis of Population Structure; Corander and Marttinen, 2006), which uses stochastic optimization instead of MCMC to find the optimal partition (with the highest estimated probability). In this case, five replicates for each possible K were run and ‘spatial clustering of groups’ analysis was performed. Both Bayesian analyses were performed with the combined matrix of EST-SSR and SSR markers.
To estimate genetic structure following an alternative non-Bayesian approach, analyses of molecular variance (AMOVA) were conducted to quantify the proportion of molecular variance within and among populations using ARLEQUIN v. 3·5. Nested AMOVAs were conducted to further decompose genetic variance among predefined population groups according to the two ecological scenarios considered (mountain islands or isolated belts) and the results provided by the Bayesian clustering analyses.
Outlier loci analysis: potential loci under selection
The outlier loci scan test procedure is based on FST comparisons, identifying the loci that present FST coefficients that are significantly different from expected under neutrality and a given demographic model. To identify possible outlier loci from all SSR loci detected, we used the approach proposed by Excoffier et al. (2009) based on Beaumont and Nichols (1996). We adopted a conservative approach, only considering the loci outside the estimated 99 % confidence interval to be outlier loci (i.e. Bonin et al., 2006; Mäkinen et al., 2008). Outlier loci were calculated using the ARLEQUIN v. 3·5 application (Excoffier and Lischer, 2010). To minimize the risk of bias due to false positives, BayeScan v. 2·0 analysis (Foll et al., 2010) was simultaneously carried out based on the microsatellite dataset to ensure that the outlier loci detected only reflect selection processes. In this genome scan, a Bayesian factor (BF) > 10 (strong, Jeffreys' scale of evidence) was considered as the threshold for proposing that a marker could be under selection.
RESULTS
Transferability analysis of EST-SSR loci from S. latifolia to S. ciliata
While most of the 42 assayed EST-SSR loci from S. latifolia produced amplicons of the expected size in S. ciliata, only eight of them were reproducible and consistently interpretable. Two of these eight loci were monomorphic, and as the absence of a microsatellite motif was confirmed after their sequencing, they were discarded. Six EST-SSR loci from S. latifolia were finally chosen to analyse the entire set of S. ciliata samples (Supplementary Data Table S1).
Genetic diversity of EST-SSR and genomic SSR loci in S. ciliata
A total of 88 and 60 different alleles were scored from the six EST-SSR and the four genomic SSR loci in the 180 individuals analysed, respectively. The mean (±s.d.) number of alleles was 14·8 ± 5·7 per locus. Summarized data for total number of alleles (NA), observed (HO) and expected (HE) heterozygosities, and inbreeding coefficients (FIS) are shown in Table 2 and detailed information for each marker is given in Supplementary Data Table S1. The largest mean number of alleles was found in the I1 population, 8·2 ± 4·2, whereas H1 and H2 had the lowest values (7·1 ± 3·5 and 7·2 ± 3·9, respectively). The L2 and L1 populations had the highest number of exclusive alleles (13 and 10, respectively), while the two highest altitude populations H2 and H1 had the lowest number of exclusive alleles (two and five, respectively). Observed heterozygosities ranged from 0·35 ± 0·29 (L1) to 0·21 ± 0·26 (H1), whereas expected heterozygosities varied from 0·56 ± 0·25 (L2) to 0·50 ± 0·28 (H1) (Table 2).
Table 2.
Genetic diversity indices in six Silene ciliata populations from two altitudinal gradients derived from six EST-SSR transferred loci from Silene latifolia and four genomic SSR loci developed in S. ciliata
| Population | Population code | N | NA | HO | HE | FIS |
|---|---|---|---|---|---|---|
| Morrena | L1 | 30 | 75 (10) | 0·352 | 0·506 | 0·351* (5) |
| Dos Hermanas | I1 | 30 | 82 (9) | 0·289 | 0·551 | 0·391* (5) |
| Peñalara | H1 | 30 | 71 (5) | 0·206 | 0·502 | 0·459* (4) |
| Ski slope | L2 | 30 | 81 (13) | 0·283 | 0·561 | 0·401* (5) |
| Bola Mundo | I2 | 30 | 77 (5) | 0·303 | 0·522 | 0·437* (4) |
| Cabeza | H2 | 30 | 72 (2) | 0·301 | 0·556 | 0·384* (4) |
N, sample size; NA, total number of alleles; HO and HE, average observed and expected heterozygosities, respectively; FIS, average inbreeding coefficient. Asterisks indicate significant values (P < 0·05). See Supplementary Data Table S1 for detailed results for each marker and population. Numbers in parentheses show the number of exclusive alleles (column NA), and the number of loci that deviate significantly from Hardy-Weinberg equilibrium (FIS column).
Inbreeding coefficients (FIS) ranged from 0·351 in the L1 population to 0·459 in H1 (Table 2). FIS estimated with the EST-SSR loci data set was similar to that obtained with genomic SSR markers, except in the L1 population where it was lower (see Supplementary Data Table S1). All populations showed significant departures from HW equilibrium towards heterozygote deficiency considering all loci. Of the 60 pairwise comparisons of loci and sites for HW equilibrium, 27 (45 %) were significant after correction for multiple comparisons (Supplementary Data Table S1). All populations had four or five loci showing significant heterozygote deficiency. No consistent linkage disequilibrium was found between any of the 60 pairwise comparisons across loci and sites, and none of the results was significant after correction for multiple comparisons.
No significant differences were found for any of the genetic diversity estimators (including allelic richness, observed heterozygosity, genetic diversity within populations and FIS) when populations were grouped according to the mountain islands or ‘isolated belts’ models (Table 3). Thus, genetic diversity values within populations were similar among altitudinal ranges (Table 3). However, allelic richness and observed heterozygosity tended to decrease with increasing altitude, while the inbreeding coefficient tended to increase (Table 3).
Table 3.
Comparison of mean genetic diversity values in the two ecological models tested in Silene ciliata based on six EST-SSR and four genomic SSR loci
| Mountain islands |
Isolated belts |
||||||
|---|---|---|---|---|---|---|---|
| Gradient 1 | Gradient 2 | P | Low | Intermediate | High | P | |
| A | 7·275 | 7·292 | 0·991 | 7·353 | 7·591 | 6·892 | 0·133 |
| HO | 0·358 | 0·356 | 0·899 | 0·370 | 0·355 | 0·349 | 0·773 |
| HE | 0·597 | 0·600 | 0·698 | 0·593 | 0·602 | 0·602 | 1 |
| FIS | 0·400 | 0·406 | 0·897 | 0·377 | 0·410 | 0·420 | 0·672 |
| FST | 0·041 | 0·031 | 0·719 | 0·071 | 0·132 | 0·063 | 0·337 |
A, allelic richness calculated after the rarefaction method of El Mousadik and Petit (1996) and based on a minimum sample size of 30 individuals; HO, HE, average observed and expected heterozygosity within populations, respectively. Mountain islands: Gradient 1 and 2, n = 3 populations each; Isolated belts: Low, Intermediate and High, n = 2 populations each. Grouping comparisons were tested for significance using 10 000 permutations.
Population genetic structure and spatial analyses
All pairwise values of differentiation between populations (FST) were significantly different from zero except for population pair L2–I2 (Table 4). Significant FST values ranged between 0·021 (I1–H1) and 0·169 (H1–I2) with an overall mean FST of 0·087 ± 0·044. In most cases FST values were lower when populations of the same mountain gradient were compared (Table 4). When populations were grouped according to the two proposed ecological scenarios (‘mountain islands’ and ‘isolated belts’), no significant differences were found between overall FST comparisons (Table 3). The mean number of migrants among population pairs was 1·2 ± 0·2. The highest values were obtained between populations along the altitudinal gradient (L2 – H2 = 1·6; L1 – I1 = 1·4), whereas the lowest values were obtained between populations located in different mountains (e.g. L1 – L2 = 0·7) (Table 4).
Table 4.
Matrix of pairwise differentiation: FST (lower diagonal) values among populations
| Low 1 | Intermediate 1 | High 1 | Low 2 | Intermediate 2 | High 2 | |
|---|---|---|---|---|---|---|
| Low 1 | 1·419 | 0·901 | 0·725 | 0·962 | 1·008 | |
| Intermediate 1 | 0·057 | 1·027 | 0·913 | 0·884 | 1·063 | |
| High 1 | 0·080 | 0·021 | 0·876 | 0·794 | 1·39 | |
| Low 2 | 0·083 | 0·114 | 0·125 | 0·994 | 1·568 | |
| Intermediate 2 | 0·120 | 0·151 | 0·169 | 0·018 | 0·77 | |
| High 2 | 0·092 | 0·085 | 0·078 | 0·038 | 0·068 |
FST values significantly different from zero (after Bonferroni correction) are in bold. Migration rate (Nem; upper diagonal) was estimated following the maximum-likelihood algorithm in MIGRATE.
We found a significant correlation between genetic and log-transformed geographical distances (r = 0·72, P < 0·01). However, no significant correlation was found between genetic distances and log-transformed altitudinal distances (r = 0·18, P = 0·72). The partial Mantel test between genetic and log-transformed geographical distances was significant (r = 0·71, P < 0·01) when the log-transformed altitudinal distance matrix was included as the covariable. By contrast, non-significant results were obtained when the spatial distance matrix was used as the covariable.
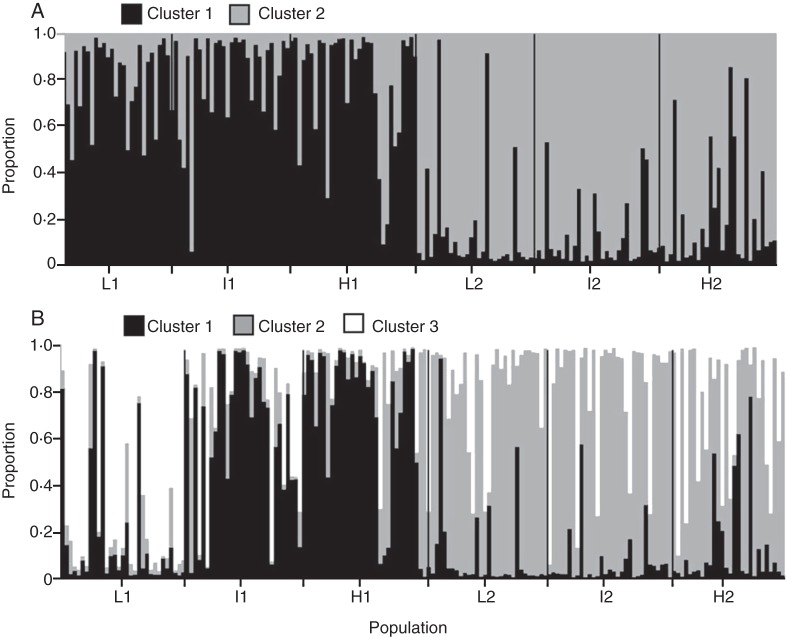
Results from the Bayesian clustering analyses conducted with BAPS considering the whole set of loci converged at K = 3 as the most likely number of genetic clusters. For K = 3, L2 – I2 – H2 populations had a higher proportion of membership to cluster 1, populations I1 and H1 had a higher proportion of membership to cluster 2, and L1 had a higher proportion of membership to a third independent genetic cluster (BAPS K = 3, cluster probability = 0·99).
When Bayesian clustering analyses were carried out with STRUCTURE, another highly plausible clustering scenario was obtained. The populations from each altitudinal gradient were assigned to a different genetic cluster (K = 2, L1–I1–H1 in cluster 1 and L2–I2–H2 in cluster 2) with a high proportion of membership (Fig. 1, hypothesis A, Fig. 3A). According to the method of Evanno et al. (2005), a modal value of ΔK = 398·53 was obtained for K = 2, whereas for K = 3 the value was lower (ΔK = 121·71) (Fig. 3).
Fig. 3.
Membership proportion of 180 individuals in the Bayesian analysis of population structure, carried out with STRUCTURE in S. ciliata. Analyses were based on the whole data set (six EST-SSRs and four genomic SSRs) for two (K = 2, A) and three (K = 3, B) predefined genetic clusters.
AMOVAs conducted for S. ciliata attributed 8·83 % of the total variation between populations. Hierarchical AMOVAs revealed highly congruent patterns of explained variability with the Bayesian clustering analyses (Table 5). The hierarchical AMOVA for the mountain island scenario obtained a larger proportion of variation among groups (6·94 %) as well as a lower proportion of variation among populations within groups (4·42 %), i.e. more homogeneous groups, than the AMOVA for the isolated belt scenario (Table 5). These results were consistent with the primary number of genetic clusters (K = 2) detected by the STRUCTURE analyses. Further hierarchical AMOVA with population arranged into three population groups (independent L1 vs. I1 + H1 vs. complete altitudinal gradient 2) revealed an even greater proportion of variation among groups (7·36 %) and higher homogeneity among populations within groups (3·26 %), as obtained for the secondary number of genetic clusters (K = 3) detected by STRUCTURE and BAPS analyses. AMOVAs conducted for isolated belts showed that differences among groups were not significantly different from zero (Table 5).
Table 5.
Analyses of molecular variance (AMOVA) of Silene ciliata based on 10 microsatellite markers
| Source of variation | d.f. | Sum of squares | Variance components | Fixation indices | Proportion of variation | P |
|---|---|---|---|---|---|---|
| 1. Silene ciliata | ||||||
| Among populations | 5 | 86·4 | 0·25 | 0·09 | 8·83 | <0·01 |
| Within populations | 354 | 898·3 | 2·53 | 91·17 | <0·01 | |
| 2. Mountain islands: Low1 + Int.1 + High1 vs. Low2 + Int.2 + High2 (K = 2) | ||||||
| Among groups | 1 | 45·87 | 0·19 | 0·06 | 6·94 | 0·1 |
| Among populations within groups | 4 | 40·53 | 0·13 | 0·04 | 4·42 | <0·01 |
| Within populations | 354 | 898·3 | 2·53 | 0·11 | 88·64 | <0·01 |
| 3. Mountain islands with Low1 isolated: Low1 vs. Int.1 + High1 vs. Low2 + Int.2 + High2 (K = 3) | ||||||
| Among groups | 2 | 62·13 | 0·21 | 0·07 | 7·36 | 0·01 |
| Among populations within groups | 3 | 24·27 | 0·09 | 0·04 | 3·26 | <0·01 |
| Within populations | 354 | 898·3 | 2·53 | 0·11 | 89·38 | <0·01 |
| 4. Isolated belts: Low1 + Low2 vs. Int.1 + Int.2 vs. High1 + High2 (K = 3) | ||||||
| Among groups | 2 | 25 | –0·06 | 0·08 | –2·40 | 0·73 |
| Among populations within groups | 3 | 61·4 | 0·3 | 0·1 | 10·79 | <0·01 |
| Within populations | 354 | 898·3 | 2·54 | −0·2 | 91·61 | <0·01 |
The four possible scenarios considered were: no population structure (1), mountain islands (2), mountain islands with Low1 population isolated (3) and isolated belts (4).
Outlier loci
The outlier test conducted with ARLEQUIN showed that one of the microsatellites (Sci. 0106, SSR marker) was detected as a potential outlier locus, using a 99 % confidence interval. The same locus was found under Bayesian analysis, showing a Bayes factor > 2 (considered ‘decisive’). This locus showed high allelic diversity in I2 and L1 populations (ten and nine different alleles, respectively, Table S1) and one of these alleles was exclusive to the L1 population.
DISCUSSION
Genetic diversity in S. ciliata along altitude gradients
Genetic diversity within populations is determined by opposing evolutionary forces that may increase or decrease net levels of diversity. While mutation and migration on the one hand and drift on the other hand represent the primary evolutionary forces with opposite effects on levels of genetic diversity, a variety of species-specific factors may shift the mutation-migration/drift equilibrium in one direction or another. Thus, several life-history traits related to longevity (i.e. annuals vs. perennials), reproductive traits (i.e. the presence of self-incompatibility systems and types of pollination) and seed dispersal mechanisms have been identified among the most relevant in determining the levels and distribution of genetic diversity in plant populations (Hamrick and Godt, 1996). Furthermore, historical factors, especially in alpine populations which were most likely affected by glacial retreats and expansions, cannot be disregarded in having shaped the genetic diversity of current populations (Segarra-Moragues et al., 2007). Understanding the relative contribution of each of these factors may help to identify the key forces driving population dynamics, evolution and current responses to global climate change.
As our study is focused on a single species in a narrow geographical area, we did not expect to find substantial differences in genetic diversity (or in other genetic variables) among populations. Similar genetic diversity, inbreeding coefficients or numbers of loci under HW disequilibrium were observed in all populations (Table 2). These values were slightly lower than those found in other alpine species studied along altitudinal gradients which also exhibited local adaptation (e.g. Ohsawa and Ide, 2008; Byars et al., 2009; Gonzalo-Turpin and Hazard, 2009), although in all these cases, populations were separated by greater geographical distances or showed completely different life-history and reproductive traits. An additional factor explaining this genetic similarity may be historical population movements related to ice contraction and expansion during glaciations in the Quaternary period (Hewitt, 2001), which might have significantly contributed to genetically homogenizing populations. These periods of ice expansion, which forced the migration of northern species to warmer southern European refugia (the tabula-rasa hypothesis of Hewitt, 2001), could also have caused local migrations to lower altitudes in this mountain system, similar to those suggested in the southern Sierra Nevada Massif (Gutiérrez Larena et al., 2002). The existence of one (or several) refuges at low altitudes which preserve genetic diversity in periods of ice expansion has also been proposed for these mountain systems in the Iberian peninsula (Nieto Feliner, 2011). We observed that low-altitude populations had higher numbers of exclusive alleles compared with higher altitude sites (Table 2). Our results suggest that a stepping-stone migration pattern towards higher altitudes during the interglacial periods may have caused this slight decrease in allelic richness in the higher altitude populations (Table 2). Similarly, the rejoining of higher and lower altitude populations at lower altitudinal ranges could have contributed to the increase in allelic diversity in this area during the glacial periods. Repeated contraction and expansion of population ranges during the climatic oscillations of the Quaternary period could have contributed to the high genetic similarity among populations within altitudinal gradients (Tables 2 and 3).
Gene flow and altitude: mountain islands vs. isolated belts
Gene flow along altitudinal gradients has been frequently detected in other alpine systems (e.g. Stöcklin et al., 2009), including plants from other Mediterranean mountains (e.g. Medrano and Herrera, 2008). Furthermore, horizontal gene flow between populations located at the same altitude has also been described in other mountain species (Byars et al., 2009). Thus, alpine and arctic plants are able to cover long distances both through seed (Alsos et al., 2007) and pollen dispersal (Schulke and Waser, 2001; García-Camacho and Totland, 2009, and references therein).
Bayesian clustering and AMOVA analyses (Fig. 3A, Table 5) showed that the genetic structure of S. ciliata best fits a mountain island scenario. Populations from the two altitudinal gradients clustered separately into the two main genetic groups detected in the Bayesian analysis with the highest ΔK values (Fig. 3A, Table 5). Moreover, higher FST values were obtained in the comparison of populations from the same altitude than when populations from the same altitudinal gradient were compared (Table 4). These results suggest that the valley between the mountains, which reaches its lowest altitude at 1700 m, probably acts as a topographical barrier severely hindering current gene flow between mountains (Widmer et al., 2009). Estimates for the number of migrants support this idea, showing a high number of effective migrants between populations located in the same mountain island (Table 4).
Gene flow may also be partially restricted by factors affecting seed dispersal and pollination in this particular species. Silene species generally show a short average seed dispersal distance (Young, 2002). Thus, S. ciliata seeds, which lack a specialized mechanism for long-distance dispersal, have been estimated to disperse at distances of approx. 1 m from the mother plant (C. Lara-Romero & J. J. Robledo-Arnuncio, Rey Juan Carlos University and Forest Research Centre, respectively, Madrid, Spain, pers. comm.) and are therefore expected to contribute little to gene flow among populations. Geographical location along the altitudinal range can also condition accessibility to larger pollinator guilds and promote a more extended flowering period in the lower altitude populations compared with the higher altitude populations, thus contributing to differences in gene flow, migration rates and genetic diversity levels along the altitudinal gradient. Yet flowering asynchrony, which is typically associated with altitudinal gradients (Körner, 2003) is also frequent in rear-edge populations of S. ciliata (Giménez-Benavides et al., 2007c). The temporally restricted flowering period of S. ciliata, which occurs late in the summer (Giménez-Benavides et al., 2011), may contribute to shortening pollen dispersal distances. However, in especially warm years flowering asynchrony is drastically reduced, because there seems to be a photoperiodic limit to how much flowering start can be anticipated (Giménez-Benavides et al., 2007c), potentially allowing for higher levels of gene flow via pollen dispersal among populations along the altitudinal gradient. On the other hand, pollination of S. ciliata is carried out not only by diurnal pollinators such as syrphid flies, wasps and bees, but also by the nocturnal moth H. consparcatoides with which S. ciliata seems to have developed a specific insect–plant association (Giménez-Benavides et al., 2007a). These moths can easily cover the geographical distances separating populations within an altitudinal gradient (Pasquet et al., 2008), as suggested by the obtained estimates of gene flow (Table 4) and as shown in other Silene species (e.g. Young, 2002; Barluenga et al., 2011). Furthermore, the presence of the valley, whose altitude decreases to 1700 m a.s.l., and the existence of a pine forest between both mountains, could play important roles as geographical barriers for seed and/or pollen dispersal for horizontal gene flow (Kreyer et al., 2004). Considering these current barriers, the relevant horizontal gene flow detected in this study suggests that populations from the two mountains have been in contact in the past, possibly as a result of downward shifts originated at extended ice expansion periods that allowed the occurrence of continuous populations between the two mountains.
The role of selection forces: evidence of local adaptation in genetic structure and outlier loci
Local adaptation occurs as a response to highly contrasting environmental conditions, with selective pressures acting differently in each population. Moreover, low levels of gene flow have been traditionally considered essential for such adaptive differentiation to occur (Kawecki, 2008). Nonetheless, evidence of local adaptation has been found in other studies examining populations along altitudinal gradients in the presence of high levels of gene flow (e.g. Kawecki, 2008; Byars et al., 2009). On the other hand, self-pollination has been considered to be evolutionarily advantageous to preserve preselected loci under local adaptation (Allard, 1975; Lenormand, 2002).
Our study has revealed substantial but limited gene flow between the two altitudinal gradients of S. ciliata, at least in the recent past (Fig. 3, Table 4), and a relevant number of migrants between populations located in the same mountain along an altitudinal gradient (Table 4). The prevalence of the ‘mountain island’ gene flow pattern over the ‘isolated belt’ pattern would seem to hinder local adaptation processes at the lowest populations which show the strongest vulnerability to high temperature and drought. However, previous studies have provided significant evidence of local adaptation in S. ciliata populations inhabiting the same ranges as in this study, especially in the case of the L1 population. Individuals from this population showed higher percentages of in situ seed germination in reciprocal transplant experiments and higher resistance to drought stress (Giménez-Benavides et al., 2007b, García-Fernández et al., 2012b). Although this population is closely related to other populations along the same altitudinal gradient, it showed a higher proportion of membership to an exclusive genetic cluster in the Bayesian analysis (Fig. 2B) and also increased the differences among groups in the AMOVA when this population was considered independently from both gradients (Table 5). The detection of one outlier locus containing an exclusive allele in the L1 population also supports the idea of selective pressures acting differently on the S. ciliata genome along the altitudinal gradient, although this should be interpreted with caution. These results are congruent with previously detected local adaptation and with other differences (number of exclusive alleles, genome size; García-Fernández et al., 2012a) in this population. The relatively high levels of FIS detected in all populations (Table 2) confirm previous evidence of the mixed mating system of this species (García-Fernández et al., 2012c) and the occurrence of some self-pollination. Thus, the occurrence of self-pollination in these populations may be a significant factor in facilitating the persistence of local adaptation in spite of substantial gene flow from other populations along the altitudinal gradient.
Concluding remarks
Substantial levels of gene flow among S. ciliata populations within altitudinal gradients suggest that seed and especially pollen dispersal is more likely to occur along altitudinal gradients than between populations located at the same altitude on different mountains, although occasional horizontal movements cannot be discarded. Even though current gene flow between populations may be partially limited, the weak genetic differentiation found may be due to the similar colonization history, with upward–downward migrations of populations along mountains following the last glaciations of the Quaternary period and the existence of common refuges at lower altitudes. The genetic differentiation observed in one of the lowest altitude populations and the detection of one outlier locus, along with previous evidence of local adaptation, suggest that current levels of gene flow are not high enough to prevent local adaptation in this high mountain species.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Massimiliano Gnesotto, Aria Minder, Claudia Michel and Maria D. Moccia and the Genetic Diversity Centre (GDC) at ETH Zurich for their help with the laboratory work. We also thank the staff of Parque Natural de las Cumbres, Circo y Lagunas de Peñalara and Parque Regional de la Cuenca Alta del Manzanares for permission to work in the field area. This work was supported by the projects ISLAS (CGL2009-13190-C03-02), LÍMITES (CGL2009-07229) and REMEDINAL2. A.G. was supported by a F.P.I. fellowship (CGL2006-09431/BOS) and a short visit grant (Ref. 2825) from the ConGen programme of the European Science Foundation. J.G.S-M. was supported by a postdoctoral research contract ‘Ramón y Cajal’ from the Ministerio de Ciencia e Innovación (MICINN).
LITERATURED CITED
- Alsos IG, Eidesen PB, Ehrich D, et al. Frequent long-distance plant colonization in the changing Arctic. Science. 2007;316:1606–1609. doi: 10.1126/science.1139178. [DOI] [PubMed] [Google Scholar]
- Allard RW. The mating system and microevolution. Genetics. 1975;79(Suppl):115–126. [PubMed] [Google Scholar]
- Antonovics J. Evolution in closely adjacent plant populations VI. Manifold effects of gene flow. Heredity. 1968;23:508–524. [Google Scholar]
- Armbruster P, Reed DH. Inbreeding depression in benign and stressful environments. Heredity. 2005;95:235–242. doi: 10.1038/sj.hdy.6800721. [DOI] [PubMed] [Google Scholar]
- Barluenga M, Austerlitz F, Elzinga JA, Teixeira S, Goudet J, Bernasconi G. Fine-scale spatial genetic structure and gene dispersal in Silene latifolia. Heredity. 2011;106:13–24. doi: 10.1038/hdy.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London. Series B: Biological Sciences. 1996;263:1619–1626. [Google Scholar]
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proceedings of the National Academy of Sciences. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A, Taberlet P, Miaud C, Pompanon F. Explorative genome scan to detect candidate loci for adaptation along a gradient of altitude in the common frog (Rana temporaria) Molecular Biology and Evolution. 2006;23:773–783. doi: 10.1093/molbev/msj087. [DOI] [PubMed] [Google Scholar]
- Byars SG, Papst W, Hoffmann AA. Local adaptation and cogradient in the alpine plant, Poa hiemata along a narrow altitudinal gradient. Evolution. 2007;61:2925–2941. doi: 10.1111/j.1558-5646.2007.00248.x. [DOI] [PubMed] [Google Scholar]
- Byars SG, Parsons Y, Hoffmann AA. Effect of altitude on the genetic structure of an Alpine grass, Poa hiemata. Annals of Botany. 2009;103:885–899. doi: 10.1093/aob/mcp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multilocus molecular markers. Molecular Ecology. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Chase MW, Hills HH. Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon. 1991;40:215–220. [Google Scholar]
- El Mousadik A, Petit RJ. Chloroplast DNA phylogeography of the argan tree of Morocco. Molecular Ecology. 1996;5:547–555. doi: 10.1111/j.1365-294x.1996.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Elphinstone MS, Hinten GN, Anderson MJ, Nock CJ. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Molecular Ecology Notes. 2003;3:317–320. [Google Scholar]
- Ennos RA. Inferences about spatial patterns in plant populations from the analysis of molecular markers. In: Silvertown J, Antonovics J, editors. Ecology and evolution in a spatial context. London: Blackwell Science; 2001. pp. 45–72. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3·5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Hofer T, Foll M. Detecting loci under selection in a hierarchically structured population. Heredity. 2009;103:285–298. doi: 10.1038/hdy.2009.74. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Fischer MC, Heckel G, Excoffier L. Estimating population structure from AFLP amplification intensity. Molecular Ecology. 2010;19:4638–4647. doi: 10.1111/j.1365-294X.2010.04820.x. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. New York: Cambridge University Press; 2002. [Google Scholar]
- García-Camacho R, Totland Ãr. Pollen limitation in the Alpine: a meta-analysis. Arctic, Antarctic, and Alpine Research. 2009;41:103–111. [Google Scholar]
- García-Fernández A, Iriondo J, Vallés J, Orellana J, Escudero A. Ploidy level and genome size of locally adapted populations of Silene ciliata across an altitudinal gradient. Plant Systematics and Evolution. 2012a;298:139–146. [Google Scholar]
- García-Fernández A, Iriondo JM, Bartels D, Escudero A. Response to artificial drying until drought-induced death in different elevation populations of a high-mountain plant. Plant Biology. 2012b doi: 10.1111/j.1438-8677.2012.00638.x. in press. http://dx.doi.org/10.1111/j.1438-8677.2012.00638.x . [DOI] [PubMed] [Google Scholar]
- García-Fernández A, Iriondo JM, Escudero A. Inbreeding at the edge: does inbreeding depression increase under more stressful conditions? Oikos. 2012c;9:1435–1445. [Google Scholar]
- García-Ramos G, Kirkpatrick M. Genetic models of adaptation and gene flow in peripheral populations. Evolution. 1997;51:8. doi: 10.1111/j.1558-5646.1997.tb02384.x. [DOI] [PubMed] [Google Scholar]
- Gautschi B, Widmer A, Joshi J, Koella JC. Increased frequency of scale anomalies and loss of genetic variation in serially bottlenecked populations of the dice snake, Natrix tessellata. Conservation Genetics. 2002;3:235–245. [Google Scholar]
- Giménez-Benavides L, Dotterl S, Jurgens A, Escudero A, Iriondo JM. Generalist diurnal pollination provides greater fitness in a plant with nocturnal pollination syndrome: assessing the effects of a Silene – Hadena interaction. Oikos. 2007a;116:1461–1472. [Google Scholar]
- Giménez-Benavides L, Escudero A, Iriondo JM. Local adaptation enhances seedling recruitment along an altitudinal gradient in a high mountain mediterranean plant. Annals of Botany. 2007b;99:723–734. doi: 10.1093/aob/mcm007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Benavides L, Escudero A, Iriondo JM. Reproductive limits of a late-flowering high-mountain Mediterranean plant along an elevational climate gradient. New Phytologist. 2007c;173:367–382. doi: 10.1111/j.1469-8137.2006.01932.x. [DOI] [PubMed] [Google Scholar]
- Giménez-Benavides L, Escudero A, Iriondo JM. What shapes the altitudinal range of a high mountain Mediterranean plant? Recruitment probabilities from ovule to seedling stage. Ecography. 2008;31:731–740. [Google Scholar]
- Giménez-Benavides L, García-Camacho R, Iriondo JM, Escudero A. Selection on flowering time in Mediterranean high-mountain plants under global warming. Evolutionary Ecology. 2011;25:777–794. [Google Scholar]
- Gonzalo-Turpin H, Hazard L. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. Journal of Ecology. 2009;97:742–751. [Google Scholar]
- Gottfried M, Pauli H, Futschik A, et al. Continent-wide response of mountain vegetation to climate change. Nature Climate Change. 2012;2:111–115. [Google Scholar]
- Goudet J. FSTAT (Version 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Gutiérrez Larena B, Fuertes Aguilar J, Nieto Feliner G. Glacial-induced altitudinal migrations in Armeria (Plumbaginaceae) inferred from patterns of chloroplast DNA haplotype sharing. Molecular Ecology. 2002;11:1965–1974. doi: 10.1046/j.1365-294x.2002.01594.x. [DOI] [PubMed] [Google Scholar]
- Hall D, Luquez V, Garcia VM, St Onge KR, Jansson S, Ingvarsson PK. Adaptative population differentiation in phenology across a latitudinal gradient in European aspen (Populus tremula): a comparison of neutral markers, candidate genes and phenotypic traits. Evolution. 2007;61:2849–2860. doi: 10.1111/j.1558-5646.2007.00230.x. [DOI] [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1996;351:1291–1298. [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: a decade later. Annual Review of Ecology and Systematics. 1986;17:535–566. [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: the age of genomics. Annual Review of Ecology, Evolution, and Systematics. 2006;37:67–93. [Google Scholar]
- Hewitt GM. Speciation, hybrid zones and phylogeography — or seeing genes in space and time. Molecular Ecology. 2001;10:537–549. doi: 10.1046/j.1365-294x.2001.01202.x. [DOI] [PubMed] [Google Scholar]
- Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M. Ecological consequences of genetic diversity. Ecology Letters. 2008;11:609–623. doi: 10.1111/j.1461-0248.2008.01179.x. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK, García MV, Hall D, Luquez V, Jansson S. Clinal variation in phyB2, a candidate gene for day-length-induced growth cessation and bud set, across a latitudinal gradient in European aspen (Populus tremula) Genetics. 2006;172:1845–1853. doi: 10.1534/genetics.105.047522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain SK, Bradshaw AD. Evolutionary divergence among adjacent plant populations I. The evidence and its theoretical analysis. Heredity. 1966;21:407–441. [Google Scholar]
- Jump A, Peñuelas J. Running to stand still: adaptation and the response of plants to rapid climate change. Ecology Letters. 2005;8:1010–1020. doi: 10.1111/j.1461-0248.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ. Adaptation to marginal habitats. Annual Review of Ecology, Evolution, and Systematics. 2008;39:321–342. [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Kirkpatric M, Barton NH. Evolution of species range. American Naturalist. 1997;150:1–23. doi: 10.1086/286054. [DOI] [PubMed] [Google Scholar]
- Körner C. Alpine plant life: functional plant ecology of high mountain ecosystems. Berlin: Springer-Verlag; 2003. [Google Scholar]
- Kreyer D, Oed A, Walther-Hellwig K, Frankl R. Are forests potential landscape barriers for foraging bumblebees? Landscape scale experiments with Bombus terrestris agg. and Bombus pascuorum (Hymenoptera, Apidae) Biological Conservation. 2004;116:111–118. [Google Scholar]
- Lawton JH. Range, population abundance and conservation. Trends in Ecology and Evolution. 1993;8:409–413. doi: 10.1016/0169-5347(93)90043-O. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre L. Numerical ecology. Amsterdam: Elsevier Science BV; 1998. [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends in Ecology & Evolution. 2002;17:183–189. [Google Scholar]
- Mäkinen HS, Cano JM, Merilä J. Identifying footprints of directional and balancing selection in marine and freshwater three-spined stickleback (Gasterosteus aculeatus) populations. Molecular Ecology. 2008;17:3565–3582. doi: 10.1111/j.1365-294X.2008.03714.x. [DOI] [PubMed] [Google Scholar]
- Medrano MN, Herrera CM. Geographical structuring of genetic diversity across the whole distribution range of Narcissus longispathus, a habitat-specialist, mediterranean narrow endemic. Annals of Botany. 2008;102:183–194. doi: 10.1093/aob/mcn086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Feder M, Wray G. Evolutionary and ecological functional genomics. Heredity. 2008;100:101–102. doi: 10.1038/sj.hdy.6801015. [DOI] [PubMed] [Google Scholar]
- Moccia M, Oger-Desfeux C, Marais G, Widmer A. A white campion (Silene latifolia) floral expressed sequence tag (EST) library: annotation, EST-SSR characterization, transferability, and utility for comparative mapping. BMC Genomics. 2009;10:243. doi: 10.1186/1471-2164-10-243. http:dx.doi.org/10.1186/1471-2164-10-243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia MD, Widmer A Molecular Ecology Resources Primer Development C. Permanent genetic resources added to Molecular Ecology Resources Database 1 October 2009–30 November 2009. Molecular Ecology Resources. 2010;10:404–408. doi: 10.1111/j.1755-0998.2009.02827.x. [DOI] [PubMed] [Google Scholar]
- Nieminen M, Singer MC, Fortelius W, Schops K, Hanski I. Experimental confirmation that inbreeding depression increases extinction risk in butterfly populations. American Naturalist. 2001;157:237–244. doi: 10.1086/318630. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G. Southern European glacial refugia: a tale of tales. Taxon. 2011;60:365–372. [Google Scholar]
- Nogués-Bravo D, Araujo MB, Errea MP, Martinez-Rica JP. Exposure of global mountain systems to climate warming during the 21st Century. Global Environmental Change. 2007;17:420–428. [Google Scholar]
- Ohsawa T, Ide Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Global Ecology and Biogeography. 2008;17:152–163. [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology, Evolution, and Systematics. 2006;37:637–669. [Google Scholar]
- Pasquet RMS, Peltier A, Hufford MB, et al. Long-distance pollen flow assessment through evaluation of pollinator foraging range suggests transgene escape distances. Proceedings of the National Academy of Sciences. 2008;105:13456–13461. doi: 10.1073/pnas.0806040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sagarin R, Gaines D. The ‘abundant centre’ distribution: to what extent is it a biogeographical rule? Ecology Letters. 2002;5:137–147. [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends in Ecology & Evolution. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Schulke B, Waser NM. Long-distance pollinator flights and pollen dispersal between populations of Delphinium nuttallianum. Oecologia. 2001;127:239–245. doi: 10.1007/s004420000586. [DOI] [PubMed] [Google Scholar]
- Segarra-Moragues JG, Palop-Esteban M, González-Candelas F, Catalán P. Nunatak survival vs. tabula rasa in the Central Pyrenees: a study on the endemic plant species Borderea pyrenaica (Dioscoreaceae) Journal of Biogeography. 2007;34:1893–1906. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. New York: W. H. Freeman; 1995. [Google Scholar]
- Stöcklin J, Kuss P, Pluess A. Genetic diversity, phenotypic variation and local adaptation in the alpine landscape: case studies with alpine plant species. Botanica Helvetica. 2009;119:125–133. [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, Valentine DH, Walters SM, Webb DA. Flora Europaea. Cambridge: Cambridge University Press; 1995. [Google Scholar]
- Ward RT. Ecotypic variation in Deschampsia caespitosa (L.) Beauv. from Colorado. Ecology. 1969;50:519–522. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Widmer A, Lexer C, Cozzolino S. Evolution of reproductive isolation in plants. Heredity. 2009;102:31–38. doi: 10.1038/hdy.2008.69. [DOI] [PubMed] [Google Scholar]
- Young HJ. Diurnal and nocturnal pollination of Silene alba (Caryophyllaceae) American Journal of Botany. 2002;89:433–440. doi: 10.3732/ajb.89.3.433. [DOI] [PubMed] [Google Scholar]
- Zane L, Bargelloni L, Patarnello T. Strategies for microsatellite isolation: a review. Molecular Ecology. 2002;11:1–16. doi: 10.1046/j.0962-1083.2001.01418.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.