Abstract
Background and Aims
The holoparasitic flowering plant Balanophora displays extreme floral reduction and was previously found to have enormous rate acceleration in the nuclear 18S rDNA region. So far, it remains unclear whether non-ribosomal, protein-coding genes of Balanophora also evolve in an accelerated fashion and whether the genes with high substitution rates retain their functionality. To tackle these issues, six different genes were sequenced from two Balanophora species and their rate variation and expression patterns were examined.
Methods
Sequences including nuclear PI, euAP3, TM6, LFY and RPB2 and mitochondrial matR were determined from two Balanophora spp. and compared with selected hemiparasitic species of Santalales and autotrophic core eudicots. Gene expression was detected for the six protein-coding genes and the expression patterns of the three B-class genes (PI, AP3 and TM6) were further examined across different organs of B. laxiflora using RT-PCR.
Key Results
Balanophora mitochondrial matR is highly accelerated in both nonsynonymous (dN) and synonymous (dS) substitution rates, whereas the rate variation of nuclear genes LFY, PI, euAP3, TM6 and RPB2 are less dramatic. Significant dS increases were detected in Balanophora PI, TM6, RPB2 and dN accelerations in euAP3. All of the protein-coding genes are expressed in inflorescences, indicative of their functionality. PI is restrictively expressed in tepals, synandria and floral bracts, whereas AP3 and TM6 are widely expressed in both male and female inflorescences.
Conclusions
Despite the observation that rates of sequence evolution are generally higher in Balanophora than in hemiparasitic species of Santalales and autotrophic core eudicots, the five nuclear protein-coding genes are functional and are evolving at a much slower rate than 18S rDNA. The mechanism or mechanisms responsible for rapid sequence evolution and concomitant rate acceleration for 18S rDNA and matR are currently not well understood and require further study in Balanophora and other holoparasites.
Keywords: Balanophora, Balanophoraceae, B-class genes, LFY, RPB2, mitochondrial matR, substitution rate, phylogeny, Santalales, parasitic plants.
INTRODUCTION
Rates of nucleotide substitution are known to vary among species and genes, and the factors affecting such variation include population size, life history, metabolic rate and DNA repair efficiency (Britten, 1986; Nickrent and Starr, 1994; Gaut et al., 1996; Laroche et al., 1997; Ohta, 2000; Smith and Donoghue, 2008). Several studies have demonstrated that the substitution rates in nuclear small-subunit ribosomal DNA (18S rDNA) for many holoparasitic and mycoheterotrophic plants are usually high compared with other angiosperms (Nickrent and Starr, 1994; Lemaire et al., 2010). Such rate accelerations are found in the other two intracellular genomes of many holoparasites, including plastid 16S rDNA (Nickrent et al., 1997) and rps2 (dePamphilis et al., 1997) and mitochondrial small-subunit rDNA (Duff and Nickrent, 1997) and matR (Barkman et al., 2004). Among the cases with accelerated substitution rates, the holoparasitic plant Balanophora has one of the highest substitution rates in 18S rDNA among angiosperms (Nickrent and Starr, 1994; Nickrent et al., 2005).
Balanophora (Balanophoraceae) is a root parasite with unisexual flowers, the plants being either dioecious or monoecious. The floral organs of Balanophora are highly modified and reduced (Fig. 1A–D). The male flower (Fig. 1D) of Balanophora is composed of a single tepal whorl surrounding the synandrium formed by fused anthers. The female flower (Fig. 1C) completely lacks a perianth and consists only of a single pistil, and the female flowers (pistils) are clustered around numerous club-shaped structures called spadicles (Hansen, 1972) or claviform bodies (Eberwein et al., 2009).
Fig. 1.

Photographs of Balanophora laxiflora: (A, C) female flowers and inflorescences; (B, D) male inflorescence and flowers. Abbreviations: f, female flower; s, spadicle; ax, inflorescence axis; b-br, basal bract; f-br, floral bract; tep, tepal; syn; synandrium.
Based on phylogenetic studies using nuclear 18S rDNA and mitochondrial matR regions, the family Balanophoraceae is allied with Santalales (Nickrent et al., 2005; Barkman et al., 2007) and is included in that order in the Angiosperm Phylogeny Group classification (APG III, 2009), but its exact position remains unresolved. Considering that the extreme rate acceleration of 18S rDNA in Balanophora could impose problems for the inference of phylogenetic relationships (see Nickrent et al., 2005), additional gene markers with less rate heterogeneity would be useful in further clarifying phylogenetic relationships of Balanophoraceae. The MADS-box genes that have conserved functions for floral homoeotic patterning in angiosperms are good candidates for consideration.
The floral homoeotic genes for floral patterning under the current ABCDE model have been extensively studied in the two model species, Arabidopsis thaliana and Antirrhinum majus (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Theissen and Saedler, 2001). According to the model, five major classes of floral homoeotic genes work in different combinations to specify the identities of sepals, petals, stamens and carpels. Among these, B-class genes are known to be responsible for specifying the identities of petals and stamens. Two major duplication events occurred during the evolution of B-class genes in seed plants (Kramer et al., 1998). The first duplication event was before the origin of the angiosperms and this gave rise to the PISTILLATA/GLOBOSA (PI/GLO) and APELATA3/DEFICIENS (AP3/DEF) gene lineages (Kramer et al., 1998; Kramer and Irish, 2000; Theissen et al., 2000). The second major duplication event was in the AP3 lineage after the divergence of Trochodendraceae in extant core eudicots, giving rise to the euAP3 and TM6 gene sub-lineages (Kramer et al., 2006).
A gene duplication event also occurred in the evolution of RNA polymerase II (RPB2) gene which is responsible for transcription. A major duplication occurred shortly before the core eudicot divergence that leads to the RPB2-i and RPB2-d gene lineages (Oxelman et al., 2004; Luo et al., 2007). Because the i copies have been lost several times during the diversification of core eudicots, some lineages may only retain the d copies such as Arceuthobium (Santalaceae s.l. or Viscaceae) of Santalales and Liquidambar (Altingiaceae) of Saxifragales (Luo et al., 2007).
Compared with plastid and mitochondrial genes, fewer studies exist that have analysed evolutionary rates of nuclear protein-coding genes in parasitic plants. Extreme rate acceleration of 18S rDNA has been found in many heterotrophic taxa including Rafflesia keithii (Rafflesiaceae) and Balanophora spp. (Balanophoraceae), but not all holoparasitic plants, however, display such rate accelerations (Nickrent and Starr, 1994; Nickrent et al., 1998; Young and dePamphilis, 2005; Lemaire et al., 2010). One hypothesis to be tested for the holoparasitic Balanophora is to see if it is a general feature that all genes in the genome evolve more quickly than those from hemiparasitic relatives and autotrophic core eudicots.
The highly reduced and modified floral morphology of Balanophora provides an opportunity to examine the conservation and divergence of B-class genes. The B-class genes are expected to show a certain degree of conservation due to the functional constraint. Similarly for the LEAFY homologues, a single-copy gene is responsible for the initiation and patterning of flowers (Blázquez et al., 1997; Moyroud et al., 2009) and RPB2 homologues from Balanophora and other Santalales species. Comparison of the substitution rates of the B-class genes and other nuclear markers could help elucidate whether the rate acceleration is a common feature in the nuclear genome of Balanophora, compared with other santalalean relatives and non-parasitic core eudicots. Moreover, we examined the expression patterns of floral B-class genes in B. laxiflora to determine whether there are underlying genetic patterns that correlate with the unusual floral phenotypes.
MATERIALS AND METHODS
Taxonomic sampling
Eleven members of Santalales representing eight (or six if Santalaceae s.l. is used) families were used in this study: Loranthus kaoi and L. delavayi (Loranthaceae), Champereia manillana (Opiliaceae), Thesium chinense (Santalaceae s.l. or Thesiaceae), Santalum album (Santalaceae s.s.), Viscum articulatum and V. alniformosanae (Santalaceae s.l. or Viscaceae), Olax imbricata Roxb. (Olacaceae), Schoepfia jasminodora (Schoepfiaceae) and Balanophora laxiflora and B. fungosa (Balanophoraceae). All plant materials were collected in Taiwan.
Gene sequence determination
The 18S rDNA and mt matR gene sequences were amplified by direct PCR of genomic DNA from the above taxa. Total genomic DNAs were extracted according to standard CTAB extraction methods (Doyle and Doyle, 1987) from fresh leaves or flowers. The PCR conditions and primers for amplification and sequencing of 18S rDNA followed Nickrent (1994) and the primers used for the matR region are listed in Supplementary Data Table S1.
For LFY, RPB2 and B-class MADS-box genes (PI, euAP3, TM6), coding sequences were determined by analysis of reverse transcriptase PCR (RT-PCR) products generated from mRNA. Total RNA was prepared from young inflorescences or flowers using either Concert Plant reagent (Invitrogen, Carlsbad, CA, USA) or a method developed for Pinus (Chang et al., 1993). The cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen). Degenerate primers for initial amplification of B-class, LFY and RPB2 gene homologues were based on previous studies (Frohlich and Meyerowitz, 1997; Oxelman et al., 2004; Stellari et al., 2004). PCR reactions were performed with 1 µL of Advantage 2 polymerase mix (Clontech, Palo Alto, CA, USA), 5 µL 10× Advantage 2 PCR buffer, 200 µm of each dNTP, 200 µm of each primer and 1 µL cDNA template in a final volume of 50 µL for the reaction solution. The PCR amplification programme was as follows: 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 45 s, 72 °C for 50 s and a final extension at 72 °C for 3 min. The PCR products were then cloned into the pGEM-T Easy Vector system (Promega, Madison, WI, USA) and 30–50 clones were sequenced for each taxon. Complete cDNA sequences of B-class gene homologues of B. laxiflora were obtained using 5'RACE kit (Invitrogen) and new gene specific primers were then designed for RT-PCR analysis; these primers are listed in Supplementary Data Table S1.
To determine the gene structure of Balanophora B-class genes (BalAP3, BalTM6, BafAP3 and BafTM6), PCR was performed with genomic DNA of B. laxiflora and B. fungosa. Gene specific primers were designed (see Supplementary Data Table S1) and the PCR amplifications were carried out using Blend Tag (Toyobo, Osaka, Japan) following the manufacturer's instructions. Intron position was determined by aligning the genomic fragments with the corresponding cDNA sequences.
Phylogenetic analyses
Phylogenetic analyses were conducted with Bayesian inference (BI) using MRBAYES 3·04b (Huelsenbeck and Ronquist, 2001) and with maximum likelihood using Genetic Algorithm for Rapid Likelihood Inference (GARLI) version 0·951 (Zwickl, 2006). The model of DNA substitution for later incorporation was estimated using Modeltest 3·06 (Posada and Buckley, 2004), and the GTR + I + G model was selected as the fittest for the 18S rDNA, PI, AP3/TM6 and RPB2 datasets according to the Akaike information criterion. For the mt matR dataset, the fittest TIM + G model suggested by Modeltest is not implemented in MRBAYES, and therefore we chose the closely related GTR + G model instead. Sequence(s) of Akebia (Lardizabalaceae) was chosen as outgroup in all analyses [following Shan et al. (2006) and Jaramillo and Kramer (2007)]. Trees were sampled every 500 generations from 2·5 million generations of Markov chain Monte Carlo searches in BI analysis. The first 500 trees were discarded and the remaining trees were used to calculate posterior probabilities. Maximum likelihood bootstrap percentages (MLBP) (Felsenstein, 1985) were evaluated from 200 iterations using GARLI version 0·951 (Zwickl, 2006).
Branch length estimates and rate heterogeneity test
Branch lengths representing the number of non-synonymous (dN) and synonymous (dS) substitutions per site were estimated for protein-coding genes in our study using HyPhy package (Pond et al., 2005). The Muse-Gaut (MG94) model of codon substitution was adopted to estimate dN and dS rates independently in each lineage. Likelihood ratio tests (Yang, 1998) were implemented to determine the significance of rate heterogeneity for dN and dS of protein-coding genes among holoparasitic Balanophora, hemiparasitic Santalales and autotrophic core eudicots. Both B. laxiflora and B. fungosa sequences were included to represent ‘Balanophora’ in the analysis. Each of the tests contained two hypotheses for the categories compared: (1) that they share the same rate parameters (null hypothesis) or (2) that they have different parameters (alternate hypothesis). If the likelihood log estimate of alterative hypothesis is significantly higher than that of the null hypothesis (P < 0·05), it is assumed that the two categories have significantly different evolutionary rates. Rate correlations between dN and dS were analysed using Spearman's rank correlation test under the statistical program R (R Development Core Team, 2008).
Gene expression analysis
Different fresh floral tissues of B. laxiflora were dissected and frozen in liquid nitrogen for gene expression assays. Tepals, synandria and pollen were collected from open male flowers, and floral bracts were collected from floral buds and mature flowers. The bracts that occur basally on the inflorescence and parts of the inflorescence axis were collected from young male and female inflorescences. Female flowers and spadicles were prepared in a mixture since they are difficult to separate. Total RNA was extracted from the dissected tissues separately and was reverse transcribed as described above. Gene-specific primers of B. laxiflora used in this study are listed in Supplementary Data Table S1. Fifty nanograms of RNA was used in each PCR reaction and the amplification was performed with 94 °C for 2 min, followed by 34 cycles of 94 °C for 30 s, 55 °C for 40 s and 72 °C for 50 s and a final extension at 72 °C for 3 min using Platinum Taq DNA Polymerase (Invitrogen). The amplified fragments were further confirmed by direct sequencing.
Scanning electron microscopy
Male inflorescences of B. laxiflora were cut into small pieces and preserved in 70 % ethanol and then dehydrated in an ethanol series to 100 % ethanol. The samples were then critical-point dried, mounted onto pin stubs, coated with gold and examined using a Hitachi S-520 scanning electron microscope.
RESULTS
Sequence data
In this study we identified 25 B-class, two LFY, nine 18S rDNA, nine RPB2 (d-copy; Supplementary Data Fig. S1) and nine mitochondrial matR gene homologues from two Balanophora and nine hemiparasitic species (Supplementary Data Table S2). All these taxa have one PI-like gene and one TM6-like gene except for V. articulatum, which has two PI homologues and two TM6 homologues. However, only two euAP3 genes were obtained in our screening for Balanophoraceae and other Santalales, i.e. BalAP3 from B. laxiflora and BafAP3 from B. fungosa. All the B-class gene homologues obtained display the conserved amino acid domains M, I, K and C and the typical motifs of the C-terminal region for PI/AP3 subgroups (Kramer et al., 1998; see Supplementary Data Fig. S2).
Genomic DNA sequences of BalAP3 and BafAP3 were determined by direct PCR of the genomic DNA using several pairs of primers (see Supplementary Data Table S1) amplifying partial genomic sequences. All exon–intron structure and sequences can be identified, except for intron 4 sequences in all four B-class genes of Balanophora, because we could not obtain a PCR product using flanking primers of intron 4. After comparing cDNA sequences with other known euAP3 homologues, an additional 39-bp new exon was found at the C-domain region (which we denote as 6a) between the ordinary exons 6 and 7 of B-class genes in both B. laxiflora and B. fungosa (Supplementary Data Fig. S3). An approx. 0·5-kb region between exons 2 and 3 of LFY and an approx. 1·7-kb region between exons 12 and 24 of RPB2 genes were amplified. The two LFY homologues from Balanophora and the nine RPB2 homologues from Balanophora and other Santalales are quite conserved and can be easily aligned to other eudicot homologues. All of the sequences obtained and gene accessions used in the phylogenetic analyses are provided in Supplementary Data Table S2.
Molecular phylogenetics
The BI phylogenetic trees resulting from analysis of 18S rDNA, mt matR, PI, AP3 and RPB2 sequences are shown in Fig. 2. These are generally congruent with the relationships of angiosperm phylogenies inferred from previously published data based on multiple plastid, mitochondrial and nuclear rDNA gene datasets (Soltis et al., 1999; Soltis et al., 2000). In general, the PI and AP3 trees received higher nodal support compared with the 18S rDNA and mt matR trees. All of the trees consistently showed that Santalales including Balanophoraceae formed a monophyletic group, which received high BI support (1·0 on the PI, AP3 and RPB2 tree) and low to moderate maximum likelihood support (MLBP = 66 % on RPB2, 55 % on PI, 87 % on the AP3 tree) in RPB2 and B-class gene trees (Fig. 2C–E), and still lower for 18S rDNA (BI = 0·87, MLBP < 50 %) and mt matR (BI = 0·89, MLBP < 50 %) gene trees (Fig. 2A, B). In contrast, the relationship of Balanophora to other members of Santalales is not resolved in any of the five gene trees. With 18S rDNA, matR and RPB2, Balanophora is related to Olax, Schoepfia and/or Loranthus (Fig. 2A–C). With PI (Fig. 2D), Balanophora is sister to Olax and this clade is sister to the remaining Santalales, whereas with AP3/TM6 it is sister to all other Santalales except Olax. The relationships of the other santalalean taxa also differed among the four gene trees. Some of the nodes received relatively high support (BI > 0·95, MLBP > 90 %, marked as black dots on Fig. 2), but none of those nodes in Santalales showed incongruent phylogenetic relationships among the four gene trees.
Fig. 2.
Phylogenetic relationships of Santalales and Balanophora spp. inferred from Bayesian analysis (BI) of 18S rDNA, matR, RPB2, PI and AP3 gene sequences. Numbers above the branches are Bayesian posterior probabilities, and values below are maximum likelihood branch support determined by GARLI. Mitochondrial matR trees are based on a 1968 nucleotide alignment, and nuclear gene trees are based on 1739 (18S rDNA), 765 (PI), 864 (AP3), 549 (LFY), 1719 (RPB2) nucleotide alignments, respectively. See Supplementary Data Fig. S1 for RPB2 gene tree showing the obtained RPB2 sequences belonging to the d clade.
Rates of molecular evolution
Extensive rate variation was detected among the six Balanophora gene sequences (18S rDNA, matR, LFY, PI, AP3 and RPB2) (Fig. 3). Balanophora 18S rDNA showed an extraordinarily high substitution rate compared with hemiparasitic Santalales and non-parasitic species. Long branches for Balanophora sequences were also detected on mitochondrial matR and some of the other gene trees constructed from sites with synonymous substitutions. Therefore we performed likelihood ratio tests to examine rate heterogeneity in non-synonymous (dN) and synonymous (dS) substitutions for the three B-class genes (PI, euAP3, TM6), as well as LFY, RPB2 and mt matR, among holoparasitic Balanophora, hemiparasitic Santalales and autotrophic core eudicots.
Fig. 3.
Rate divergence of nuclear and mitochondrial genes in Balanophora. BI trees based on dN and dS for protein-coding genes or rDNA gene. All protein-coding gene trees are constrained to the same scale, whereas the rDNA tree is on a different scale (see notes in figure).
The matR, PI, TM6 and RPB2 sequences of other Santalales showed significantly lower rates of dN and dS in all the comparisons to Balanophora or other core eudicots (Table 1). In contrast, Balanophora sequences generally showed an increase in dN or dS; however, only Balanophora matR showed significant rate accelerations (>10-fold). On the other hand, the B-class genes and LFY of Balanophora showed varying substitution rates compared with the core eudicots. Among these genes, significant rate accelerations were only found in dS of Balanophora PI, TM6 and RPB2 genes, and dN of Balanophora euAP3. In other comparisons, Balanophora B-class gene sequences have only slight increases in substitution rates. It is worth noting that the dN and dS values of Balanophora B-class and RPB2 genes are all less than two times higher than the core eudicot sequences, far less than seen with mt matR. Furthermore, Balanophora LFY sequences not only did not show a significant rate increase in dN, they actually showed significantly lower dS values compared with the core eudicots (0·290 vs. 0·732, Table 1). The analyses showed that dN and dS are significantly correlated within euAP3, TM6, LFY, RPB2 and matR among the groups, but the rates are poorly correlated within PI sequences.
TABLE S2.
Sequences used in phylogenetic construction and evolutionary rate analysis
| Taxa | Species | nuclear | mitochondrial | |||||
| PI | AP3 | FLO/LFY | 18S rDNA | matR | RPB2/RPB2-d copy | |||
| paleoAP3/TM6 | euAP3 | |||||||
| Basal eudicots | ||||||||
| Buxales | ||||||||
| Buxaceae | Pachysandra procumbens | AF094533 | AF197784 | |||||
| Pachysandra terminalis | GU357455 (PAtePI) | PAteAP3-1 (GU357454) | ||||||
| Proteales | PAteAP3-2 (GU357462) | |||||||
| Platanaceae | Platanus occidentalis | PloAP3-1 (AY162881) | AF106842 | U42794 | AF197793 | |||
| PloAP3-2 (AY162882) | ||||||||
| Platanus sp. | AY566618 | |||||||
| Ranunculales | ||||||||
| Lardizabalaceae | Akebia quinata | AkqPI (AY162837) | AkqAP3-1 (AY162835) | L31795 | AF197810 | |||
| AkqAP3-2 (AY162839) | ||||||||
| Akebia longeracemosa | AY566613 | |||||||
| Papaveraceae | Eschscholzia californica | AY188789 | ||||||
| Sabiales | ||||||||
| Sabiaceae | Meliosma dilleniifolia | MedMdPI (AY436712) | MedMdAP3-1 (AY436709) | |||||
| MedMdAP3-2 (AY436710) | ||||||||
| MedMdAP3-2 (AY436711) | ||||||||
| Meliosma squamulata | DQ008728 | DQ007426 | ||||||
| Trochodendrales | ||||||||
| Trochodendraceae | Trochodendron aralioides | TraPI-1 (EF436259) | TraAP3 (DQ453774) | AF230078 | U42816 | AF197792 | AY563269 | |
| TraPI-2 (EF436260) | ||||||||
| Core eudicots | ||||||||
| Apiales | ||||||||
| Apiaceae | Apium graveolens | AF206852 | ||||||
| Daucus carota | MADS2 (AJ271148) | MADS3 (AJ271149) | ||||||
| Petroselinum crispum | AY453081 | |||||||
| Aralidiaceae | Hedera helix | AJ563601 | ||||||
| Aquifoliales | ||||||||
| Aquifoliaceae | Ilex aquifolium | IxaGLO1 (GQ141105) | IxaTM6 (DQ479357) | IxaAP3 (DQ479356) | AY453090 | |||
| Ilex meseryeae | AJ557240 | |||||||
| Ilex opaca | AF161010 | |||||||
| Asterales | ||||||||
| Asteraceae | Chrysanthemum x morifolium | AB451217 | ||||||
| Gerbera hybrida | GGLO1 (GHY9726) | GDEF1 (AJ009724) | GDEF2 (AJ009725) | AF107576 | ||||
| PMADS2 (X69947) | ||||||||
| Helianthus annuus | Ham31 (AY173069) | Ham91 (AY173070) | Ham63 (EF612598) | AY453114 | ||||
| Brassicales | ||||||||
| Brassicacaeae | Arabidopsis thaliana | PI (D30807) | AP3 (D21125) | DQ447103 | X16077 | NC_001284 | z19121 | |
| Caricaceae | Carica papaya | CpPI (EF562500) | CpTM6-1 (EF562498) | U42514 | AF520141 | |||
| CpTM6-2 (EF562499) | ||||||||
| Caryophyllales | ||||||||
| Amaranthaceae | Spinacia oleracea | SoPI (AY604515) | SoAP3 (AY604514) | L24420 | AY453110 | AF020840 | ||
| Caryophyllaceae | Silene latifolia | SLM2 (X80489) | SlAP3A (AB090863) | SLM3 (X80490) | ||||
| SlAP3Y (AB090864) | ||||||||
| Silene coeli-rosa | AJ311804 | |||||||
| Phytolaccaceae | Phytolacca americana | PhaTM6-1 (DQ479365) | U42793 | |||||
| PhaTM6-2 (DQ479366) | ||||||||
| Polygonaceae | Homalocladium platycladum | DQ017114 | ||||||
| Cornales | ||||||||
| Cornaceae | Alangium platanifolium | AJ555916 | ||||||
| Davidia involucrata | AJ557002 | |||||||
| Hydrangeaceae | Hydrangea macrophylla | HmPI (AF230711) | HmTM6 (AF230703) | HmAP3 (AF230702) | U42781 | AY453091 | ||
| Nyssaceae | Nyssa sylvatica | NsGLO1 (GQ141111) | NsTM6 (GQ141155) | NsDEF (GQ141170) | X16603 | |||
| NsGLO2 (GQ141109) | ||||||||
| Styracaceae | Styrax japonicus | SjGLO (GQ141121) | SjTM6 (GQ141163) | SjDEF (GQ141185 ) | L49296 | |||
| Cucurbitales | ||||||||
| Coriariaceae | Coriaria sarmentosa | DQ017097 | ||||||
| Cucurbitaceae | Cucumis sativus | CsPI (AF043255) | CsMADS1 (AY944060) | AF059320 | AF206894 | |||
| Dilleniales | ||||||||
| Dilleniaceae | Dillenia indica | DiiPI-1 (EF549702) | DiiTM6 (EF549705) | DiiDEF (EF549703) | AF520095 | |||
| DiiPI-2 (EF549703) | ||||||||
| Dillenia alata | U38314 | |||||||
| Ericales | ||||||||
| Balsaminaceae | Impatiens auriculata | AF520117 | ||||||
| Impatiens alsamina | AJ888755 | |||||||
| Clethraceae | Clethra tomentosa | CtGLO (GQ141143) | CtTM6 (GQ141180) | CtDEF (GQ141166) | ||||
| Ericaceae | Rhododendron macrophyllum | AY566626 | ||||||
| Diapensiaceae | Galax urceolata | GuGLO1 (GQ141151) | GuTM6 (GQ141164) | GuDEF (GQ141187) | L49281 | |||
| Polemoniaceae | Phlox paniculata | PpGLO (GQ141129) | PpTM6 (GQ141157) | PpDEF (GQ141172) | ||||
| Theaceae | Camellia japonica | AY566627 | ||||||
| Fagales | ||||||||
| Betulaceae | Betula pendula | BpPI (AJ488589) | AY453121 | |||||
| Juglans nigra | AF206943 | |||||||
| Juglans regia | JrAP3 (AJ313089) | GU194836 | ||||||
| Fabales | ||||||||
| Fabaceae | Lotus corniculatus | LcPIa (AY770398) | LcTM6 (AY770401) | LcAP3 (AY770400) | AY770393 | |||
| LcPIb (AY770399) | ||||||||
| Pisum sativum | AF035163 | U43011 | AY453078 | |||||
| Tephrosia purpurea | AJ556782 | |||||||
| Vicia sativa | AJ556176 | |||||||
| Gunnerales | ||||||||
| Gunneraceae | Gunnera tinctoria | GtAP3-1 (AY337753) | ||||||
| GtAP3-2 (AY337754) | ||||||||
| GtAP3-3 (AY337755) | ||||||||
| GtAP3-4 (AY337756) | ||||||||
| GtAP3-5 (AY337757) | ||||||||
| Gunnera manicata | U43787 | AF520163 | ||||||
| Lamiales | ||||||||
| Oleaceae | Syringa vulgaris | SvPI (AF052861) | SvAP3 (AF052869) | |||||
| Plantaginaceae | Antirrhinum majus | GLOBOSA (X68831) | DEFA (X62810) | M55525 | AJ236047 | AY453102 | AJ566619 | |
| Malpighiales | ||||||||
| Euphorbiaceae | Ricinus communis | XM_002519014 | ||||||
| Hypericaceae | Hypericum canariense | AJ556993 | ||||||
| Salicaceae | Populus trichocarpa | PtPI (grail3·0002017601) | PTD (AF057708) | |||||
| Populus balsamifera | U93196 | AY652861 | ||||||
| Populus maximowiczii | AY674556 | |||||||
| Myrtales | ||||||||
| Myrtaceae | Callistemon subulatus | DQ017092 | ||||||
| Eucalyptus grandis | EGM2 (AF029976) | AY640314 | ||||||
| Eucalyptus lehmannii | AM235528 | |||||||
| Melaleuca quinquenervia | MeqPI (EU004206) | |||||||
| Myrtus communis | EU281109 | AJ556164 | ||||||
| Rosales | ||||||||
| Rosaceae | Malus x domestica | MadMdPI (AJ291490) | MadMdTM6 (AB081093) | AB162028 | ||||
| MadMADS13 (AJ25116) | ||||||||
| Prunus persica | - | - | EU375891 | L28749 | ||||
| Taihangia rupestris | TrPI (DQ248947) | TrTM6 (DQ248946) | AF520201 | |||||
| Saxifragales | ||||||||
| Altingiaceae | Liquidambar sp. | AY566623 | ||||||
| Grossulariaceae | Ribes aureum | - | L28143 | |||||
| Ribes sanguineum | RsPI (AY337742) | RsAP3-1 (AY337758) | ||||||
| RsAP3-2 (AY337758) | ||||||||
| Ribes sp. | EF370723 | |||||||
| Hamamelidaceae | Corylopsis pauciflora | CopTM6-1 (DQ479354) | CopAP3 (DQ479353) | AF094548 | ||||
| CopTM6-2 (DQ479355) | ||||||||
| Saxifragaceae | Astillbe sp. | AY566622 | ||||||
| Saxifraga careyana | SacTM6 (DQ479368) | SacAP3 (DQ479367) | ||||||
| Santalales | ||||||||
| Balanophoraceae | Balanophora fungosa | BafPI (JQ613262) | BafTM6 (JQ613231) | BafAP3 (JQ613229) | JQ613242 | JN392868 | JQ613244 | JQ613269 |
| Balanophora laxiflora | BalPI (JQ613261) | BalTM6 (JQ613232) | BalAP3 (JQ613230) | JQ613243 | JN392870 | JQ613245 | JQ613270 | |
| Loranthaceae | Loranthus delavayi | LodPI (JQ613264) | LodTM6 (DQ453773) | JQ613220 | JQ613248 | |||
| Loranthus kaoi | LokPI (JQ613263) | LokTM6 (JQ613241) | JQ613221 | JQ613249 | JQ613272 | |||
| Olacaceae | Olax imbricata | OliPI (JQ613259) | OliTM6 (JQ613239) | JQ613222 | JQ613246 | JQ613268 | ||
| Opilliaceae | Champereia manillana | ChmPI (JQ613258) | ChmTM6 (JQ613237) | JQ613223 | JQ613247 | JQ613271 | ||
| Santalaceae | Santalum album | SaaPI (JQ613256) | SaaTM6 (JQ613238) | JQ613224 | JQ613250 | JQ613266 | ||
| Thesiaceae | Thesium chinense | ThcPI (JQ613257) | ThcTM6 (JQ613235) | JQ613225 | JQ613251 | JQ613267 | ||
| Schoepfiaceae | Schoepfia jasminodora | ScjPI (JQ613260) | ScjTM6 (JQ613240) | JQ613226 | JQ613252 | JQ613273 | ||
| Viscaceae | Viscum articulatum | ViArPI-1 (JQ613255) | ViArTM6-1 (JQ613233) | JQ613228 | ||||
| ViArPI-2 (JQ613254) | ViArTM6-2 (JQ613234) | |||||||
| Viscum anilformosanae | ViAlPI (JQ613253) | ViAlTM6 (JQ613236) | JQ613227 | JQ613265 | ||||
| Solanales | ||||||||
| Solanaceae | Petunia hybrida | FBP1 (m91190) | PhTM6 (AF230704) | PhGP (X69946) | AF030171 | DQ020641 | ||
| Nicotiana sylvestris | AY453113 | |||||||
| Lycopersicon esculentum | TPI (DQ674531) | TDR6 (X60759) | LeAP3 (AF052868) | X51576 | U28403 | |||
| Vitales | ||||||||
| Vitaceae | Vitis vinifera | MADS9 (DQ988043) | VvTM6 (DQ979341) | VvAP3 (EF418603) | AF450278 | NC_012119 | ||
| Vitis sp. | AF207053 | |||||||
Note: sequences newly identified in this study are shown in bold.
Table 1.
The estimated dN, dS values and P-values of the likelihood ratio tests between taxa
| mt matR | PI | TM6 | AP3 | LFY | RPB2 | |
|---|---|---|---|---|---|---|
| dN values | ||||||
| Core eudicots | 0·015 | 0·078 | 0·072 | 0·083 | 0·037 | 0·009 |
| Balanophora | 0·134 | 0·091 | 0·088 | 0·118 | 0·039 | 0·010 |
| Other Santalales* | 0·011 | 0·059 | 0·056 | – | – | 0·005 |
| P-values of dN comparison | ||||||
| Core eudicots vs. other Santalales* | <0·01 | <0·001 | <0·001 | – | – | <0·001 |
| Balanophora vs. other Santalales* | <0·001 | <0·001 | <0·001 | – | – | <0·001 |
| Balanophora vs. core eudicots | <0·001 | 0·14 (n.s.) | 0·05 (n.s.) | <0·001 | 0·85 (n.s.) | 0·61 (n.s.) |
| dS values | ||||||
| Core eudicots | 0·021 | 0·378 | 0·315 | 0·369 | 0·732 | 0·293 |
| Balanophora | 0·220 | 0·563 | 0·560 | 0·450 | 0·290 | 0·572 |
| Other Santalales* | 0·013 | 0·237 | 0·308 | – | – | 0·266 |
| P-values of dS comparison | ||||||
| Core eudicots vs. other Santalales* | <0·01 | <0·001 | <0·001 | – | – | <0·05 |
| Balanophora vs. other Santalales* | <0·001 | <0·001 | <0·001 | – | – | <0·001 |
| Balanophora vs. core eudicots | <0·001 | <0·001 | <0·001 | 0·10 (n.s.) | <0·001 | <0·001 |
| dN/dS | ||||||
| Core eudicots | 0·714 | 0·206 | 0·228 | 0·225 | 0·050 | 0·030 |
| Balanophora | 0·609 | 0·161 | 0·157 | 0·262 | 0·135 | 0·017 |
| Other Santalales* | 0·846 | 0·248 | 0·181 | – | – | 0·018 |
| Correlation coefficient of dN and dS | 0·881 | −0·272 | 0·739 | 0·653 | 0·614 | 0·672 |
| P-values of dN and dS correlation | <0·01 | 0·06 (n.s.) | <0·01 | <0·01 | <0·01 | <0·01 |
n.s., not significant; –, data not available.
*Santalales here includes hemiparasitic taxa and excludes Balanophoraceae.
Expression in the six protein-coding genes of Balanophora laxiflora
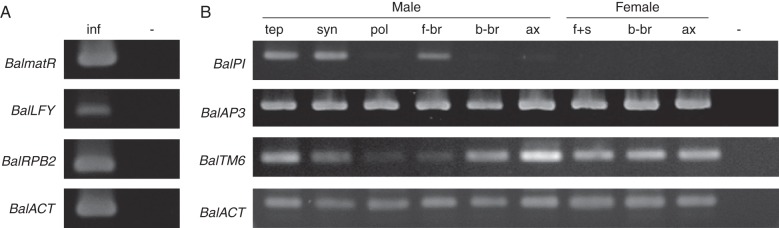
To find out whether the six protein-coding genes identified in B. laxiflora are actually expressed, RT-PCR was carried out using gene-specific primers on RNA samples. The results showed that all six protein-coding gene products were present in inflorescences (Fig. 4). The three B-class homoeotic genes displayed differential expression patterns in different parts including both reproductive and vegetative organs (Fig. 4B). BalAP3 is expressed at high levels in male and female inflorescences, and the expression can be detected in all parts examined. Similarly, BalTM6 is also expressed in male and female inflorescences, but the expression is low in the pollen and floral bracts compared with BalAP3. In contrast, BalPI is not expressed in the female inflorescence, and it is mainly restricted to the tepals, synandria and the floral bracts of male inflorescences.
Fig. 4.
Expression of the six protein-coding genes in Balanophora laxiflora: (A) expression of mitochondrial matR, RPB2 and LFY from male inflorescences; (B) expression of floral B-class genes from different parts of male and female inflorescences. Actin primers were used as an internal control. Abbreviations: inf, inflorescence; tep, tepal; syn, synandrium; pol, pollen; f-br, floral bract; b-br, basal bract; ax, inflorescence axis; f + s, female flowers and spadicles; –, negative control.
Surface morphology of bracts and tepals in Balanophora laxiflora
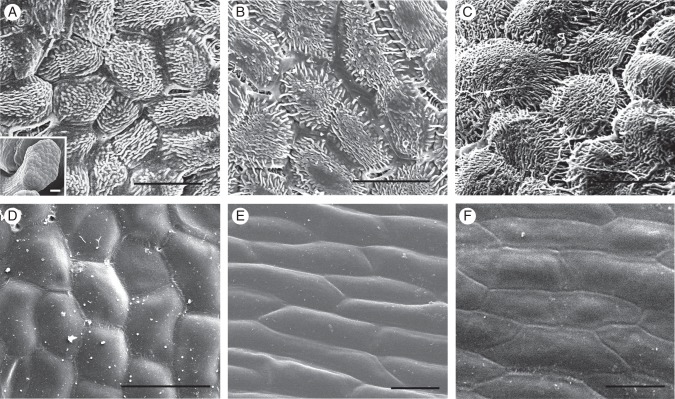
Surface morphology was examined in tepals, floral bracts and basal bracts in B. laxiflora. There are up to 14 bracts (sometimes referred to as ‘leaves’) at the bases of the male and female inflorescences of B. laxiflora (Fig. 1A, B). Each of the male flowers has two small subtending floral bracts (Fig. 1B, D). The epidermal cells of these floral bracts are oval in shape and the surface is papillate covered with short ridges (Fig. 5A). Although the basal bracts are showy, their epidermal cells are flattened and elongated with smooth surfaces on both adaxial and abaxial sides (Fig. 5E, F). In contrast, the epidermal cells of mature tepals are oval in shape and their surface is smooth on the adaxial side (Fig. 5D). They also show irregular cuticular sculpturing on their abaxial sides (Fig. 5B, C), similar to what was observed on the floral bracts (Fig. 5A). No stomata were observed on either side of the tepals, floral bracts or basal bracts.
Fig. 5.
Scanning electron micrographs of tepals and bracts of Balanophora laxiflora: (A) epidermal cells of the floral bract; (B) abaxial epidermal cells of the young tepals; (C) abaxial epidermal cells of the tepals at maturity; (D) adaxial epidermal cells of the tepals at maturity; (E) adaxial epidermal cells of the basal bract at maturity; (F) abaxial epidermal cells of the basal bract at maturity. Scale bars = 50 µm.
DISCUSSION
Rate variation in Balanophora species and other Santalales
It has previously been shown that Balanophora and other taxa of Balanophoraceae have significantly increased rates of nucleotide substitution in the 18S rDNA (Nickrent et al., 2005) and matR (Barkman et al., 2007) genes. In this study, we demonstrated that extreme rate acceleration is not ubiquitous in the Balanophora genome. The six B-class genes of Balanophora identified, three each from B. laxiflora and B. fungosa, show more modest nucleotide substitution rates as compared with rates for 18S rDNA and matR. Non-synonymous (dN) and synonymous (dS) substitutions per site were both estimated for protein-coding genes. In general, a lower dN : dS ratio indicates a conserved sequence or an increased rate of substitutions at silent sites, whereas a higher dN : dS ratio indicates a relaxation of purifying selection. Therefore the dS value is an indication of non-selective substitution rate, i.e. similar to the background substitution of a genome (Yang, 1998). In our results, both PI and TM6 display significant dS rate accelerations compared with dN rate, whereas the euAP3 paralogues are significantly elevated in dN rate (but not dS rate), suggesting that Balanophora euAP3 has evolved under a relaxed purifying selection, but they do not show congruent patterns among B-class genes.
In general, positive correlations between dN and dS are found in nuclear genes of mammals, prokaryotes and Drosophila (Graur, 1985; Li et al., 1985; Wolfe and Sharp, 1993; Akashi, 1994). In plants, dN and dS of the nuclear paralogues of Arabidopsis thaliana (Zhang et al., 2002) and the plastid genes of Geraniaceae (Guisinger et al., 2008) are also reported to be positively correlated. Our result also found significant correlations between dN and dS in the four protein-coding genes (except for PI, see Table 1), suggesting that the intensity of selective constraints on both dN and dS are similar in these gene regions. The reason for the incongruence of PI data requires further investigation.
Although the rates of sequence evolution detected from dN and dS in Balanophora are generally higher than those of other plants, the dN : dS ratios of the six protein-coding genes examined for the three groups (Balanophora, hemiparasitic Santalales and core eudicots) are all <0·3, with the exception of mitochondrial matR (Table 1), suggesting the B-class, LFY and RPB2 genes are being subjected to strong purifying selection. However, functional constraints alone cannot explain the rate heterogeneity in Balanophora since rDNA should be also under strong selection against nucleotide changes. The 18S rDNA of Balanophora has at least a 5-fold higher substitution rate compared with other flowering plants and it is even more extreme (10-fold higher) for its matR gene.
Unusual acceleration of substitution rate in genes or genomes of certain species or lineages have been documented in several plants, mostly for mitochondria and plastids. Mitochondrial-specific increases in substitution rate were found in some Plantago (Cho et al., 2004), Pelargonium hortorum (Parkinson et al., 2005) and several Silene spp. (Sloan et al., 2009). The nuclear and plastid genomes of these taxa are largely unaffected and possess typical substitution rates. On the other hand, plastid genes in parasitic plants sometimes show rate variations due to different degrees of selective constraints on photosynthetic genes. For example, the synonymous rate of Cuscuta obtusiflora plastid genes is five to eight times higher than the rates in C. exaltata because of a higher functional constraint for photosynthesis in the former (McNeal et al., 2007). Such rate increases are usually found to be lineage-specific and probably affecting the whole plastid genome, as in the case of Orobanchaceae (Young and dePamphilis, 2005).
The pattern of rate variation in 18S rDNA is more complicated since there is no clear relationship between elevated substitution rates and functional constraints. Although the extreme rate acceleration cases of 18S rDNA are all found in heterotrophic taxa such as Rafflesia keithii (Rafflesiaceae) and Balanophora spp., not all holoparasitic plants display such rate accelerations (Nickrent and Starr, 1994; Nickrent et al., 1998; Young and dePamphilis, 2005; Lemaire et al., 2010). Cynomorium coccineum (Cynomoriaceae) and Orobanche fasciculata (Orobanchaceae) do not show significantly accelerated substitution rates in 18S rDNA (dePamphilis et al., 1997; Wolfe and dePamphilis, 1997; Lemaire et al., 2010). Similarly, in mycoheterotropic plants, rate accelerations in 18S rDNA can be found in Rhizanthella gardneri (Orchidaceae) and Thismia aseroe (Thismiaceae); others like Monotropa uniflora (Ericaceae) and Corallorhiza maculata (Orchidaceae) lack such rate increases (Lemaire et al., 2010).
It was suggested that the functionality of 18S rDNA with accelerated substitution rates is retained in those heterotrophic plants, given the pattern of compensatory mutations, and that few mutations occur in the functionally and structurally important regions of the ribosomal molecule (Nickrent and Starr, 1994; dePamphilis, 1995; Lemaire et al., 2010). Therefore the increased substitution rate in 18S rDNA might reflect an overall elevated mutation rate in the nuclear genome in these plants. Several possible causes of the elevated substitution rates have been proposed, such as defective DNA repair efficiency, rapid generational time, higher speciation rates and small effective population size (Nickrent and Starr, 1994; Nickrent et al., 1998; Lemaire et al., 2010). However, such a hypothesis would suggest an overall elevated substitution rate in the entire genome for that species.
Here we provided evidence of rate heterogeneity in nuclear genomes of the two Balanophora spp. The substitution rates in 18S rDNA in B. laxiflora and B. fungosa are five times higher than other flowering plants, whereas the synonymous substitutions in PI, AP3/TM6, LFY and RPB2 show a comparable rate (0·5–2·0 times higher) to the autotrophic core eudicots. These results suggest that the molecular evolutionary factors affecting substitution rates in 18S rDNA might be different from nuclear protein-coding genes, at least for the B-class, LFY and RPB2 genes demonstrated in this study. Three models of rate variation have been proposed for explaining nucleotide substitution patterns: (1) gene-specific, (2) organism-specific and (3) genome-specific (Nickrent et al., 1998). Our preliminary data on the six protein-coding genes indicate that the rate variation in Balanophora probably evolved in a gene-specific manner. The mechanism for such rate heterogeneity in Balanophora is unknown and a thorough screening for other nuclear genes will reveal if it is ubiquitous in the Balanophora genome. It also remains to be determined if a rate discrepancy exists between 18S rDNA and other nuclear genes for the heterotrophic plants that are known to have highly elevated 18S rDNA substitutions.
B-class genes in Santalales and the novel exon in Balanophora AP3
Despite the fact that many PI and TM6 homologues were successfully identified from the selected members of Santalales including Balanophora, only two euAP3 homologues were found in the two Balanophora spp. examined, and no euAP3 homologues were detected in other members of Santalales. Different pairs of degenerate primers, including those most commonly used in previous studies (Kramer et al., 1998; Stellari et al., 2004) and primers designed specifically for the Balanophora euAP3 genes (BalAP3 and BafAP3), were employed under several PCR conditions. The lack of euAP3 sequences from these taxa of Santalales indicates that their homologues are either too divergent to be detected by PCR-based screening or their genes might express at a low levels, thus escaping detection using RT-PCR.
Most of the B-class genes in angiosperms consist of seven exons and six introns. Our alignment of euAP3 homologues showed that AP3 from both Balanophora species have relatively conserved M, I and K domains. Exons 1–5 in the Balanophora B-class genes have the same length as those in Vitis, but the length of the C-domain is quite variable (Supplementary Data Fig. S3). An additional AP3 exon in both Balanophora species is found between the C1 and C2 domains, which are known to involve transcriptional activation (Riechmann and Meyerowitz, 1997). However, we were unable to find other cases of similarly modified C-domains in other plants, and thus the issue of its origin is unclear at this time. Moreover, an intensive screening of Santalales and/or other members of Balanophoraceae is required to understand better the evolution of these B-class genes and to make inferences on their function.
Differential expression of B-class genes in Balanophora laxiflora
The expression patterns of Balanophora AP3 and TM6 suggest a largely redundant role for these two genes since they are co-expressed at comparable levels in all organs, except that TM6 expression is lower in pollen and floral bracts (Fig. 4B). Given the fact that AP3 has distinct C-terminal sequences, it is reasonable to propose a certain degree of functional differentiation from TM6 in Balanophora. In contrast, PI might have a conserved B-class gene function since it has strong expressions in tepals and synandria, but not in any reproductive organs of the female Balanophora plant. This result is congruent with the floral ABCDE model prediction, since no perianth and androecium structures can be found in the female plant of B. laxiflora. The tepals in male B. laxiflora have cobblestone-like epidermal cells that are similar to those seen in the petals of other plants which have short cuticular ridges on the surface (Kay and Daoud, 1981). Similar cuticular modification can also be found in the epidermal cells of floral bracts in the male B. laxiflora. It remains to be determined if the expression of PI in the floral bracts of Balanophora is correlated with epidermal cell morphology in these organs (tepals, synandria and floral bracts).
Phylogenetic utility of nuclear RPB2 and floral B-class genes in Balanophoraceae
Although 18S rDNA and matR are widely used markers in reconstructing angiosperm phylogeny, the extreme rate acceleration of nucleotide substitution in these genes in Balanophora presents problems for accurate phylogenetic reconstruction. Our results clearly showed that Balanophora is a member of Santalales based on both RPB2 and B-class gene phylogenies, even though the exact relationships are still problematic owing to low support for some nodes. We demonstrated that the evolutionary rates of the nuclear protein-coding genes are relatively conserved compared with 18S rDNA and matR; thus they represent promising markers for further phylogenetic analyses. Future work will be to increase the sampling of the highly diverse Olacaceae s.l. (Malecot and Nickrent, 2008).
Conclusions
For the holoparasite Balanophora, nucleotide substitution rates in the B-class genes, LFY and RPB2 homologues show that these nuclear protein-coding genes are relatively conserved compared with 18S rDNA and mitochondrial matR. These sequences are thus potential candidates for future phylogenetic analyses of Balanophoraceae. There is rate heterogeneity between 18S rDNA, mitochondrial matR and the five nuclear protein-coding genes, hinting at an unusual and currently unexplored mechanism of sequence evolution for the mitochondrial and nuclear ribosomal genes of Balanophora.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We appreciate Daniel Nickrent and another anonymous reviewer for making valuable comments on the manuscript and Shao-Lun Liu for helpful discussions. We appreciate the help provided by Cheng-Chiang Wu, Jiunn-Yih Huang, Shu-hui Wu and Shiu-Chung Wu in obtaining specimens. We are also grateful for the technical assistance with SEM by the staff of Technology Commons, College of Life Science, NTU. This study was supported by grants from the National Science Council, Taiwan (NSC 96-2621-B-002-008-MY3 and 99-2918-I-002-020).
LITERATURE CITED
- APG III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkman TJ, Lim SH, Salleh KM, Nais J. Mitochondrial DNA sequences reveal the photosynthetic relatives of Rafflesia, the world's largest flower. Proceedings of the National Academy of Sciences of the USA. 2004;101:787–792. doi: 10.1073/pnas.0305562101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkman TJ, McNeal JR, Lim SH, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evolutionary Biology. 2007;7:248. doi: 10.1186/1471-2148-7-248. http://dx.doi.org/10.1186/1471-2148-7-248 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Britten RJ. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986;231:1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Chirney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Cho Y, Mower JP, Qiu Y-L, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proceedings of the National Academy of Sciences of the USA. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- dePamphilis CW. Genes and genomes. In: Press MC, Graves JD, editors. Parasitic plants. London: Chapman & Hall; 1995. pp. 177–205. [Google Scholar]
- dePamphilis CW, Young ND, Wolfe AD. Evolution of plastid gene rps2 in a lineage of hemiparasitic and holoparasitic plants: many losses of photosynthesis and complex patterns of rate variation. Proceedings of the National Academy of Sciences of the USA. 1997;94:7367–7372. doi: 10.1073/pnas.94.14.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Duff RJ, Nickrent DL. Characterization of mitochondrial small-subunit ribosomal RNAs from holoparasitic plants. Journal of Molecular Evolution. 1997;45:631–639. doi: 10.1007/pl00006267. [DOI] [PubMed] [Google Scholar]
- Eberwein R, Nickrent DL, Weber A. Development and morphology of flowers and inflorescences in Balanophora papuana and B. elongata (Balanophoraceae) American Journal of Botany. 2009;96:1055–1067. doi: 10.3732/ajb.0800289. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies with a molecular clock. Systematic Zoology. 1985;34:152–161. [Google Scholar]
- Frohlich MW, Meyerowitz EM. The search for flower homeotic gene homologs in basal angiosperms and Gnetales: a potential new source of data on the evolutionary origin of flowers. International Journal of Plant Sciences. 1997;158:S131–S142. [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear Adh parallel rate differences at the plastid gene rbcL. Proceedings of National Academy of Sciences of the USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D. Amino acid composition and the evolutionary rates of protein-coding genes. Journal of Molecular Evolution. 1985;22:53–62. doi: 10.1007/BF02105805. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JNV, Boore JL, Jansen RK. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proceedings of the National Academy of Sciences of the USA. 2008;105:18424–18429. doi: 10.1073/pnas.0806759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B. The genus Balanophora J. R. & G. Forster, a taxonomic monograph. Dansk Botanisk Arkiv. 1972;28:1–188. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jaramillo MA, Kramer EM. Molecular evolution of the petal and stamen identity genes, APETALA3 and PISTILLATA, after petal loss in the Piperales. Molecular Phylogenetics and Evolution. 2007;44:598–609. doi: 10.1016/j.ympev.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Kay QON, Daoud HS. Pigment distribution, light reflection and cell structure in petals. Botanical Journal of the Linnean Society. 1981;83:57–84. [Google Scholar]
- Kramer EM, Irish VF. Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. International Journal of Plant Sciences. 2000;161:S29–S40. [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Su HJ, Wu CC, Hu JM. A simplified explanation for the frameshift mutation that created a novel C-terminal motif in the APETALA3 gene lineage. BMC Evolutionary Biology. 2006;6:30. doi: 10.1186/1471-2148-6-30. http://dx.doi.org/10.1186/1471-2148-6-30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche J, Li P, Maggia L, Bousquet J. Molecular evolution of angiosperm mitochondrial introns and exons. Proceedings of the National Academy of Sciences of the USA. 1997;94:5722–5727. doi: 10.1073/pnas.94.11.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire B, Huysmans S, Smets E, Merckx V. Rate accelerations in nuclear 18S rDNA of mycoheterotrophic and parasitic angiosperms. Journal of Plant Research. 2010;124:561–576. doi: 10.1007/s10265-010-0395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Molecular Biology and Evolution. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Luo J, Yoshikawa N, Hodson MC, Hall BD. Duplication and paralog sorting of RPB2 and RPB1 genes in core eudicots. Molecular Phylogenetics and Evolution. 2007;44:850–862. doi: 10.1016/j.ympev.2006.11.020. [DOI] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biology. 2007;7:57. doi: 10.1186/1471-2229-7-57. http://dx.doi.org/10.1186/1471-2229-7-57 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecot V, Nickrent DL. Molecular phylogenetic relationships of Olacaceae and related Santalales. Systematic Botany. 2008;33:97–106. [Google Scholar]
- Moyroud E, Tichtinsky G, Parcy F. The LEAFY floral regulators in angiosperms: conserved proteins with diverse roles. Journal of Plant Biology. 2009;52:177–185. [Google Scholar]
- Nickrent DL, Starr EM. High rates of nucleotide substitution in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. Journal of Molecular Evolution. 1994;39:62–70. doi: 10.1007/BF00178250. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Duff RJ, Colwell AE, et al. Molecular phylogenetic and evolutionary studies of parasitic plants. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants. II. DNA sequencing. Boston, MA: Kluwer Academic Publishers; 1998. pp. 211–241. [Google Scholar]
- Nickrent DL, Der JP, Anderson FE. Discovery of the photosynthetic relatives of the ‘Maltese mushroom. Cynomorium. BMC Evolutionary Biology. 2005;5:38. doi: 10.1186/1471-2148-5-38. http://dx.doi.org/10.1186/1471-2148-5-38 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, Duff RJ, Konings DAM. Structural analyses of plastid-derived 16S rRNAs in holoparasitic angiosperms. Plant Molecular Biology. 1997;34:731–743. doi: 10.1023/a:1005808615763. [DOI] [PubMed] [Google Scholar]
- Ohta T. Mechanisms of molecular evolution. Philosophical Transactions of the Royal Society of London – Series B: Biological Sciences. 2000;355:1623–1626. doi: 10.1098/rstb.2000.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxelman B, Yoshikawa N, McConaughy BL, Luo J, Denton AL, Hall BD. RPB2 gene phylogeny in flowering plants, with particular emphasis on asterids. Molecular Phylogenetics and Evolution. 2004;32:462–479. doi: 10.1016/j.ympev.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Parkinson CL, Mower JP, Qiu Y-L, et al. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evolutionary Biology. 2005;5 doi: 10.1186/1471-2148-5-73. 73. http://dx.doi.org/10.1186/1471-2148-5-73 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2008. www.r-project.org . [Google Scholar]
- Riechmann JL, Meyerowitz EM. MADS domain proteins in plant development. Biological Chemistry. 1997;378:1079–1118. [PubMed] [Google Scholar]
- Shan HY, Su KM, Lu WL, Kong HZ, Chen ZD, Meng Z. Conservation and divergence of candidate class B genes in Akebia trifoliata (Lardizabalaceae) Development Genes and Evolution. 2006;216:785–795. doi: 10.1007/s00427-006-0107-2. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Oxelman B, Rautenberg A, Taylor DR. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evolutionary Biology. 2009;10 doi: 10.1186/1471-2148-9-260. 12. http://dx.doi.org/10.1186/1471-2148-10-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Chase MW, et al. Angiosperm phylogeny inferred from 18S rRNA, rbcL, and atpB sequences. Botanical Journal of the Linnean Society. 2000;133:381–461. [Google Scholar]
- Soltis PS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- Stellari GM, Jaramillo MA, Kramer EM. Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Molecular Biology and Evolution. 2004;21:506–519. doi: 10.1093/molbev/msh044. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. Floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T. A short history of MADS-box genes in plants. Plant Molecular Biology. 2000;42:115–149. [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Wolfe AD, de Pamphilis CW. Alternate paths of evolution for the photosynthetic gene rbcL in four nonphotosynthetic species of Orobanche. Plant Molecular Biology. 1997;33:965–977. doi: 10.1023/a:1005739223993. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM. Mammalian gene evolution: nucleotide sequence divergence between mouse and rat. Journal of Molecular Evolution. 1993;37:441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]
- Yang ZH. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Young ND, dePamphilis CW. Rate variation in parasitic plants: correlated and uncorrelated patterns among plastid genes of different function. BMC Evolutionary Biology. 2005;5:16. doi: 10.1186/1471-2148-5-16. http://dx.doi.org/10.1186/1471-2148-5-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LQ, Vision TJ, Gaut BS. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Molecular Biology and Evolution. 2002;19:1464–1473. doi: 10.1093/oxfordjournals.molbev.a004209. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin, TX, USA: University of Texas; 2006. PhD Dissertation. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.