Abstract
Background and Aims
Prolonged storage generally reduces seed viability and vigour, although the rate of deterioration varies among species and environmental conditions. Here, we suggest a possible ageing molecular marker: At3g08030 mRNA. At3g08030 is a member of the DUF642 highly conserved family of cell-wall-associated proteins that is specific for spermatophytes.
Methods
At3g08030 expression was performed by RT-PCR and qRT-PCR analysis in seed samples differing in their rate of germination and final germination following a matrix priming and/or controlled deterioration (rapid ageing) treatment.
Key Results
The At3g08030 gene transcript was present during the entire Arabidopsis thaliana plant life cycle and in seeds, during maturation, the ripening period and after germination. Matrix priming treatment increased the rate of germination of control seeds and seeds aged by controlled deterioration. Priming treatments also increased At3g08030 expression. To determine whether the orthologues of this gene are also age markers in other plant species, At3g08030 was cloned in two wild species, Ceiba aesculifolia and Wigandia urens. As in A. thaliana, the At3g08030 transcript was not present in aged seeds of the tested species but was present in recently shed seeds. A reduction in germination performance of the aged seeds under salt stress was determined by germination assays.
Conclusions
At3g08030 mRNA detection in a dry seed lot has potential for use as a molecular marker for germination performance in a variety of plant species.
Keywords: Seed ageing, molecular marker, At3g08030, DUF642 gene family, Arabidopsis thaliana, Ceiba aesculifolia, Wigandia urens
INTRODUCTION
Seed ageing is a crucial issue for plants with economic importance, and also for wild plants used for restoration or germplasm conservation. Prolonged storage generally reduces seed viability and vigour, although the rate of deterioration varies among species and environmental conditions. Most agricultural plants have life spans between 1 and 5 years stored in an uncontrolled environment (e.g. lettuce, garlic, soy bean, sunflower) while others can stay viable for more than 20 years (e.g. Phaseolus vulgaris and Lycopersicon lycopersicum). For wild plants, there is little experimental information on seed longevity and ageing. In general, it has been suggested that seeds of plants from the tropics and from wet mature forest ecosystems have short life spans even under optimal storage conditions, while pioneer trees and species of seasonal forests have the longest life span under these conditions. However, under suboptimal storage conditions seed viability can be reduced drastically in a short time (Vázquez-Yanes and Orozco-Segovia, 1993; Khurana and Singh, 2001; Probert et al., 2009).
Seed ageing results in a delay in the beginning of germination and a reduction in germination rate, especially under stressful conditions, and eventually leads to a loss of seed viability (Priestley, 1986). Under identical storage conditions, there is also a wide variation in seed deterioration between seed lots. For wild species, this variation could play an important ecological role but poses a considerable challenge for seed management. Invigoration treatments of seeds, referred to as seed priming, enhance germination performance (Heydecker et al., 1973; Bradford, 1986; Rajjou et al., 2012). In these treatments, seeds are subjected to a controlled hydration that promotes the initiation of germination-related processes while preventing radicle emergence. Priming treatments [hydration–dehydration cycle(s)] may also improve survival during storage (Butler et al., 2009). Natural variations in soil water content may also result in natural priming of seeds present in the soil bank. For example, priming by burial promoted uniform and rapid germination of Wigandia urens seeds (González-Zertuche et al., 2001) and burial enhanced seed longevity of Artemisia tridentate (Wijayratne and Pyke, 2012). Seeds in soil are also exposed to biotic and abiotic stresses that induce damage to the different cell structures and macromolecules and consequently affect seed ageing (Butler et al., 2009). The balance between both natural priming and seed ageing in the soil will determine seed performance during germination and subsequent seedling establishment. Changes that damage metabolic and genetic processes during seed ageing have been widely described (Rajjou and Debeaujon, 2008) and biochemical markers for the deterioration in stored seeds have been proposed (Evensen and Loy, 1978; Bailly et al., 2000).
Seed-stored mRNAs play an important role in seed longevity and germination performance (Rajjou et al., 2012). Transcriptomic analyses from Arabidopsis thaliana seeds have revealed the presence of the At3g08030 transcript before, during and after seed development especially in ovules and post-mature green embryos. This gene is also expressed during reproductive and vegetative development (Le et al., 2010). In Brassica napus, orthologous expression of At3g08030 was found during priming treatments but the most important increase was detected during early germination (Soeda et al., 2005). At3g08030 encoded protein was detected in a proteome from seeds of A. thaliana ‘CVI’ ecotype (Chibani et al., 2006) and has also been described in cell-wall proteomes from different tissues (Jamet et al., 2008). The At3g08030 gene is a member of a highly conserved family, specific to spermatophytes, of cell-wall-associated proteins with a putative carbohydrate binding domain (DUF642). In particular, At3g08030 encoded protein has been described as a cellulose binding protein (Vázquez-Lobo et al., 2012). Another gene of the DUF642 family, At4g32460, is one of the after-ripening up-regulated genes (Carrera et al., 2008).
To determine whether At3g08030 transcript can be used as a marker for ageing of seeds from different plant species, we selected three species with high germination capacity, from three different ecosystems, that can remain in the seed bank for at least 2 years (D. Soriano and A. Orozco-Segovia, Universidad Nacional Autónoma de México, pers. comm.; Lundano et al., 2009). Arabidopsis thaliana is an annual weed from the Brassicaceae family that lives in mild to cold climates and is widely distributed in Europe, North America and Asia. Ceiba aesculifolia is a tree from the Bombacaceae family usually found in deciduous tropical forests. Wigandia urens is a perennial shrub or small tree from the Hydrophyllaceae family that grows in disturbed, open areas under mild to warm climates in America; in this species, burial treatment promoted a uniform and rapid germination and reserve mobilization (González-Zertuche et al., 2001; Gamboa-deBuen et al., 2006). A possible correlation between At3g08030 expression and ageing would provide evidence to support its use as a molecular marker for optimal germination performance in a variety of plant species.
MATERIALS AND METHODS
Biological material
Arabidopsis thaliana plants (Columbia ecotype) were grown in soil in controlled environmental chambers at 20 °C with a 16/8-h light–dark photoperiod. The harvested seeds were stored for 3 months at 20 °C.
Mature seeds of Wigandia urens (Ruiz & Pavón) Kunth. were collected from the trees growing in lava fields in the ‘El Pedregal de San Ángel’ Ecological Reserve. The Instituto de Ecología is also located in this zone. The seeds were separated from the fruits and placed in a closed container with 1 mL of 100 % ether for 24 h to avoid insect predation and were then aerated. Seeds were stored in jars non-hermetically sealed in the laboratory at room temperature and humidity [mean annual temperature 23 °C, mean annual relative humidity (RH) 40 %] for different times until use.
Mature seeds of Ceiba aesculifolia Kunth (Britten & Baker) were collected from plants in a tropical deciduous forest in Atocpan, Veracruz, México. Seeds were stored in jars non-hermetically sealed in the laboratory at room temperature and humidity (mean annual temperature 23 °C, mean annual RH 40%) for different times until use.
Hydropriming treatment (H)
Arabidopsis thaliana seeds were incubated in 1 mL water at 4 °C in the dark for 48 h. Seeds were not re-dried, so wet seeds were used for germination analysis or frizzed in liquid nitrogen and stored at –80 °C.
Matrix priming treatment (M)
Arabidopsis thaliana seeds were enclosed in sweet cellophane paper and buried to 2 cm in a field-capacity humid soil (hydric potential = –0·027 MPa) in a pot. The pot was covered with aluminium foil and was incubated at 22 °C for 24 h. Seeds were exhumed and air-dried at room temperature (25 °C). Subsequently, the seeds were used for germination analysis or frozen in liquid nitrogen and stored at –80 °C.
Burial treatment
Burial treatment was as described by González-Zertuche et al. (2001). Briefly, 2 g of C. aesculifolia seeds were enclosed in nylon mesh bags and buried 3 cm deep in soil in Actopan forest for 1 month. After this period, the mesh bags were exhumed and air-dried at room temperature (25 °C). The seeds were used for germination analysis or stored in a glass jar for subsequent analysis. The same seed lot was used for control and treated seeds to avoid any variability in performance during germination that could be a result of environmental conditions that affect seeds during development and maturation. In addition, each set of treatments had a control of the same seed age.
Controlled deterioration treatment
The A. thaliana seeds deterioration/ageing treatment used was based on the controlled deterioration test (CDT) established by Tesnier et al. (2002) and used by Rajjou et al. (2008). Two seed samples were used for controls: control without treatment (C) and control with matrix priming treatment (M). Three seed samples were used for deterioration treatments (CD, MBD and MAD). CD corresponds to the sample of deteriorated seeds without matrix treatment, MBD is the sample of seeds placed in a matrix priming treatment before deterioration/ageing treatment and MAD is the sample of seeds given a matrix priming treatment after the deterioration treatment. All seeds were initially equilibrated at 85 % RH at 20 °C (hydric potential = –22 MPa, saturated KCl solution). Control seeds (C and M) were dried back at 32 % RH (hydric potential = –154 MPa, saturated CaCl2 solution) at 20 °C without any further treatment. The remaining seeds were deteriorated using the CDT (Tesnier et al., 2002) at 85 % RH and 40 °C for 3 days (CD, MBD and MAD). The MBD and CD seeds were then immediately dried back at 32 % RH at 20 °C. The MAD sample of seeds was given the matrix priming treatment for 24 h after deterioration/ageing and dried back at 32 % RH at 20 °C. All seed lots were sown at the same time under the same conditions.
Germination assays
Experiments were performed on 1 % pure agar plates placed in controlled environmental chambers at 20 °C with a 16/8-h light–dark photoperiod. From 50 to 100 seeds were used per replica (three) and germination was scored by counting seeds with radicle protrusion every 3 h for A. thaliana or daily for C. aesculifolia and W. urens. Three independent assays were performed. For salt stress experiments, agar plates containing 50 mm NaCl were used.
Statistical analysis
Cumulative germination data were presented graphically. These data were arcsine transformed, and the germination rate was calculated as the first maximal derivate of the exponential sigmoid curve fitted to the accumulated germination (TableCurve 2D, version 3·0). Mean germination time was also calculated from the fitted curves as the time to reach the first maximal derivate. ANOVAs were performed on the final germination percentages, the germination rates and the mean germination times. The Statgraphics program, version 5·0 (Statistical Graphics Co., Rockville, MD, USA), was used for statistical analysis. Post-hoc analysis was done by Tukey's test.
RT-PCR analysis
Total RNA from A. thaliana seedlings, leaves, flower and siliques, from C. aesculifolia seedlings and W. urens leaves were isolated using TRIZOL reagents (Invitrogen) according to the manufacturer's instruction. In all cases the sample was frozen in liquid nitrogen and ground up. For the homogenization step the volume of ground sample used did not exceed the 50 % of the volume of TRIZOL reagent utilized.
Total RNA in A. thaliana seeds was extracted according to the procedure described by Oñate-Sánchez and Vicente-Carbajosa (2008). This method is particularly suitable for A. thaliana, which has oily seeds. For the high-carbohydrate-content seeds (C. aesculifolia and W. urens), a method designed for high-starch-content tissues was used (Li and Trick, 2005). Around 3 g (6–8 seeds) of C. aesculifolia seeds and 100 mg (approx. 500 seeds) of W. urens and A. thaliana seeds was used.
For semi-quantitative RT-PCR, cDNA was synthesised using SuperScript II Reverse Transcriptase (Invitrogen) and oligo dT, starting from up to 2 µg of RNA for each sample. For A. thaliana, the gene-specific primers to amplify At3g08030 are AT-8030F and AT-8030R (Table 1) and ATC7 expression was used as an internal standard (Graeber et al., 2011). The thermal conditions were 2 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 62 °C and 60 s at 72 ºC, and a final step of 2 min. The PCR products were visualized on a 1·5 % (w/v) agarose gel stained with ethidium bromide.
Table 1.
List of primers
| Name | Sequence |
|---|---|
| AT-8030 F | 5′-GGAGTCATGAAGAAAACAGTTCTACTAG-3′ |
| AT-8030 R | 5′-GAGCTCCTAAGCGACATGAGAAAAC-3′ |
| qRT-8030F | 5′-ATGGCGGTTCCCAAAGCCA-3′ |
| qRT-8030F | 5′-CGGTTTTCTTTGGTGACTC-3′ |
| DEG8030F | 5′-GGDRTTSAAGARGAYCSHRCYTGTGGWCC-3′ |
| DEG8030R | 5′-AGARAGYGCMRWHGCHCA-3′ |
| 8030CeibaF | 5′-GAGAAGCGCGCTTGTGGTCCA-3′ |
| 8030CeibaR | 5′-AGGGAGAGAGAGCGCCGTCG-3′ |
| 8030WigF | 5′-GGAGCGGTGCTTGTGGACCAC-3′ |
| 8030WigR | 5′-TGGGCGACGGCGCTCTCTC-3′ |
To obtain the At3g08030 orthologous gene sequence from W. urens and C. aesculifolia, degenerated primers were designed (DEG8030F, DEG8030R; Table 1). An alignment of the At3g08030 A. thaliana gene and the orthologous sequences of Populus trichocarpa (accession no. EF146051), Medicago sativa (XM_0036056745), Brassica rapa (AC189368), Vitis vinifera (XM_002283904) and Oryza sativa (NM_001058179) was performed, and two 30-bp regions with high similarities were found. The RT-PCR product obtained for W. urens leaves (JX115004) and C. aesculifolia seedlings (JX115005) were cloned. To confirm the At3g08030 orthologous identity, the nucleotide sequence was compared with that of A. thaliana (Fig. 4). With these sequences, specific primers were designed to analyse the gene expression in seeds from C. aesculifolia (8030CeibaF, 8030CeibaR; Table 1) and W. urens (8030WigF, 8030WigR; Table 1) by RT-PCR, using ATC7 as internal standard.
Fig. 4.
Clustal W alignment (BioEdit) of the At3g08030 and C. aesculifolia and W. urens At3g08030 orthologues. *Conserved nucleotides between the three sequences.
For quantitative RT-PCR, RNA extracted from 100 mg of treated A. thaliana seeds (see above) was digested with DNase I (Qiagen) and cleaned using an RNeasy Plant Mini Kit (Qiagen). RNA was then subjected to a Power SYBR Green RNA-to-CTTM 1-Step Kit (Applied Biosystems) using an Applied Biosystems StepOne System (Applied Biosystems). All PCR mixtures contained 100 ng RNA and final concentration of 2 µm of each primer, 1× of Power SYBR® Green RT-PCR Mix and 0·8× of RT Enzyme Mix. The thermal conditions were 30 min at 48 °C and 10 min at 95 °C for retrotranscription reaction, followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C, and a final reaction of 15 s at 95 °C, 15 s at 60 °C and 15 s at 95 °C for melting-curve analysis. RT-PCR was performed in duplicate for each of two independent biological replicates using a specific primer set for the At3g08030 gene (Table 1) and ACT7 as the endogenous control. CT and slope values were used to compare data from different PCR runs and RNA samples were analysed using the mathematical model established by Pfaffl (2001) to obtain the relative expression ratio. For ANOVA, data were natural log-transformed. Post-hoc analysis was via Tukey's test (significance at P < 0·05). The significance of the difference between C and T were confirmed using Student's t-test (t = 15·07, P = 0·00000538583).
RESULTS
At3g08030 expression in A. thaliana
At3g08030 was detected throughout the life cycle of A. thaliana (Fig. 1). The transcript was present in seedlings, rosettes, cauline leaves and flowers (Fig. 1A) and in siliques, recently shed seeds and germinating seeds (Fig. 1B).
Fig. 1.
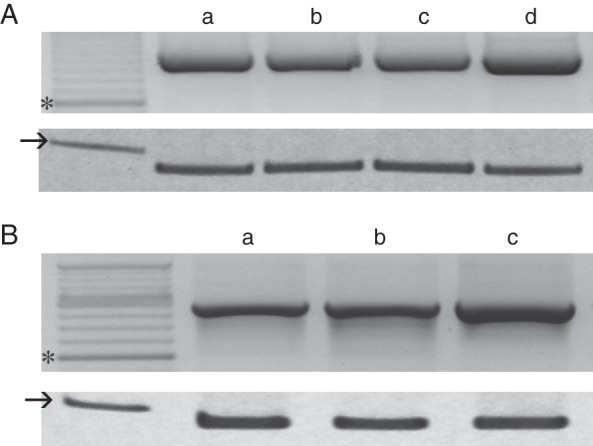
At3g08030 expression in different Arabidopsis thaliana tissues. (A) RT-PCR analysis was performed in (a) seedlings, (b) rosette leaves, (c) cauline leaves and (d) flowers. Expression of ACT7 was analysed simultaneously as an internal standard (lower panel). (B) RT-PCR analysis was performed in (a) siliques, (b) recently shed seeds (4–6 weeks) and (c) 2-day germinating seeds. Expression of ACT7 was analysed simultaneously as an internal standard (lower panel). At least three replicates were analysed for each tissue. *600-bp DNA standard; → 100-bp DNA standard.
Matrix priming treatment enhanced A. thaliana germination and At3g08030 expression
The effect of hydropriming and matrix priming on A. thaliana seed germination performance (Fig. 2A) was examined to determine the best priming method for A. thaliana. The lag time for germination was significantly shorter for matrix primed seeds (M) than for hydroprimed (H) and control (C) seeds (F2,6 = 14·33, P = 0·0052). However, maximal germination and germination rate showed no significant differences between control and primed seeds (F2,6 = 32·69, P = 0·022). Mean germination times to reach maximal first derivate are shown in Fig. 2B. A reduced mean germination time was seen for matrix primed seeds. At3g08030 expression was detected in both control and primed seeds (Fig. 2C), but the priming treatment greatly enhanced At3g08030 expression.
Fig. 2.
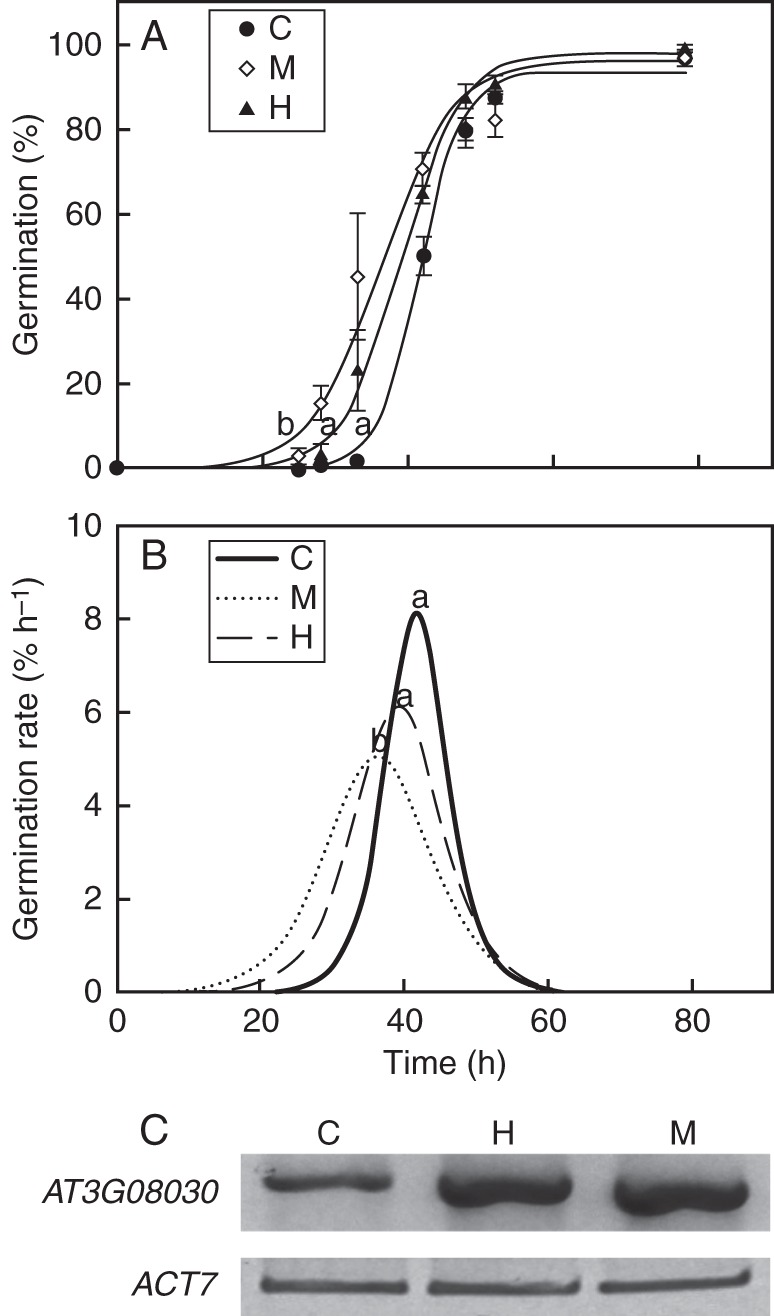
Germination curves and At3g08030 expression of control (C), matrix primed (M) and hydroprimed (H) seeds: (A) cumulative germination curve, (B) maximal germination rate. Priming treatments of 3-month-old seeds are as described in the Materials and Methods. Germination assays were carried out in triplicate at 20 °C. Different letters indicate significant differences. (C) RT-PCR analysis was performed using dry or primed seeds. ACT7 expression was used as an internal control. All experiments were performed at least three times with different seed lots.
Design of the experimental system for A. thaliana
Arabidopsis thaliana seeds were subjected to different treatments that affect seed germination performance (germination percentage, lag time, mean germination time and germination rate) (Fig. 3). Seeds were aged using a rapid ageing treatment, Controlled Deterioration (Tesnier et al., 2002). Germination performance was determined in control seeds (C), controlled deteriorated seeds (CD) and matrix primed control seeds (M). Matrix priming enhanced control seed germination as described in Fig. 2. To invigorate seeds subjected to CD treatment, the matrix priming treatment was performed before (MBD) and after (MAD) the CD treatment. (Fig. 3A, B). Controlled deterioration significantly reduced the rate of germination and the final proportion from almost 100 % for the control seeds to 42 % (Fig. 3A, B). MBD and MAD seeds showed a better germination performance (short lag and mean germination times and high germination rate and percentage germinating) than CD seeds. The final germination percentage for MBD seeds was 72 % whereas for MAD seeds it was 60 %. Germination rate of the two different matrix treatments was similar to control seeds although there were differences between the two treatments. MAD seeds germinated faster than MBD seeds. Significant differences in mean germination times were found between M, C and CD seeds (Fig. 3B).
Fig. 3.
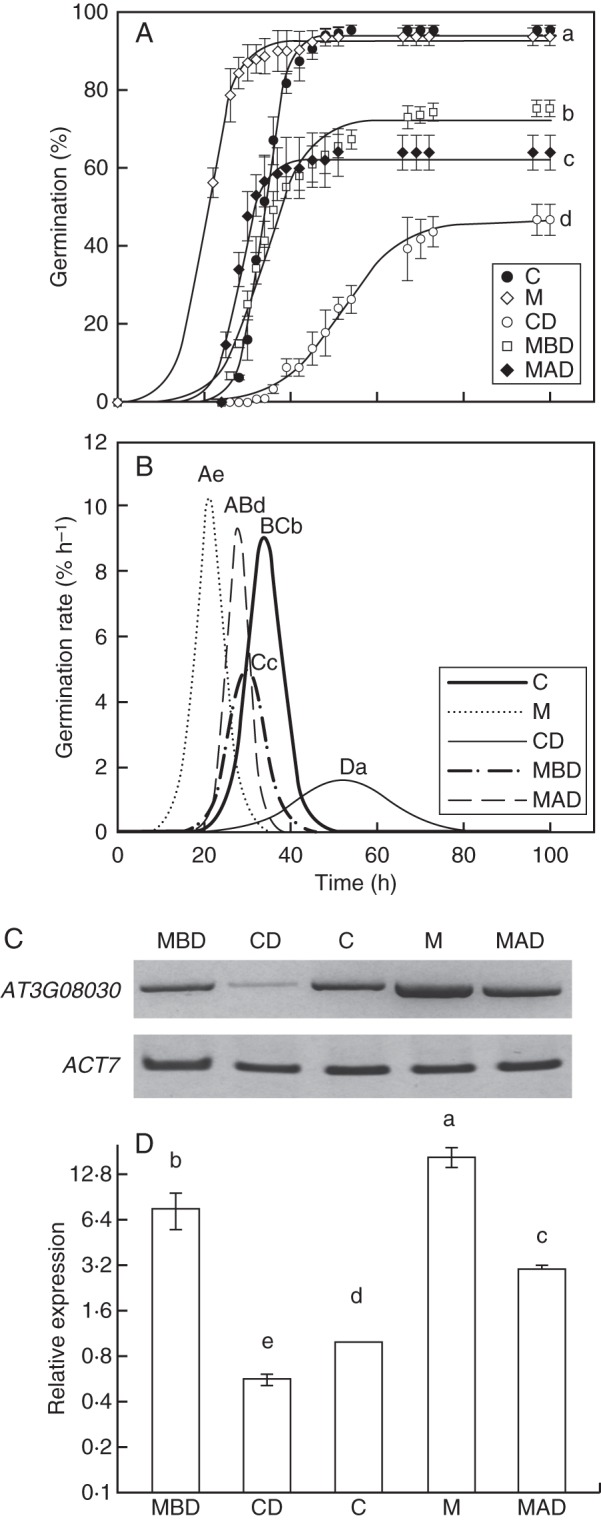
Germination curves and At3g08030 expression: control seeds (C), matrix primed control seeds (M), controlled deteriorated seeds (CD), matrix priming treatment before controlled deterioration treatment seeds (MBD) and matrix priming treatment after controlled deterioration treatment seeds (MAD). (A) Cumulative germination curves. Letters indicate statistical differences between the germination curves. (B) Maximal germination rate. Upper-case letters indicate statistical differences between germination rate and lower-case letters indicate statistical differences between mean germination time. (C) RT-PCR analysis was performed using dry or treated seeds. ACT7 expression was used as an internal control. (D) Relative expression levels of At3g08030 were determined in seeds with different vigour by qRT-PCR using At3g08030-specific primers (Table 1). First-strand cDNA was synthesized and used as template for qRT-PCR amplification as described in the Materials and Methods. Expression levels were normalized against ACT7 values. Note logarithmic scale. Values represent the mean and s.d. of results from duplicate experiments. Letters indicate statistical differences between the germination curves.
At3g08030 expression is increased in matrix primed seeds
At3g08030 was highly expressed in control and matrix primed control seeds (Fig. 3C, D). Low levels of transcript were found in CD seeds and matrix priming treatment significantly increased At3g08030 levels in both C and CD seeds before and after deterioration treatment (F4,15 =101·20, P < 0·05; Fig. 3D).
C. aesculifolia and W. urens At3g08030 orthologous genes
The DUF642 protein family is highly conserved in spermatophytes, so it was feasible to design degenerate primers to amplify the orthologues of At3g08030 in other plant species (see Materials and Methods). With these primers, a 300-bp gene fragment was cloned from C. aesculifolia seedlings and a 297-bp gene fragment from W. urens leaves. Blast analysis of these two nucleotide sequences revealed the best score with At3g08030. The level of identity between A. thaliana and C. aesculifolia for the At3g08030 sequence was 76 % compared with 73 % that of W. urens sequence (Fig. 4). Amino acid sequence identity was also high: 84·5 % for C. aesculifolia and 80·41 % for W. urens. The specific At3g08030 orthologous gene primers for the two species were designed using these data.
Ageing affects germination performance and At3g08030 expression of C. aesculifolia and W. urens seeds
Seed storage deteriorated seeds of both C. aesculifolia and W. urens. Deterioration was detected as a delay in the beginning of germination, a reduction in the germination rate and a reduced germination percentage under either optimal conditions or stress conditions (Figs 5 and 6).
Fig. 5.
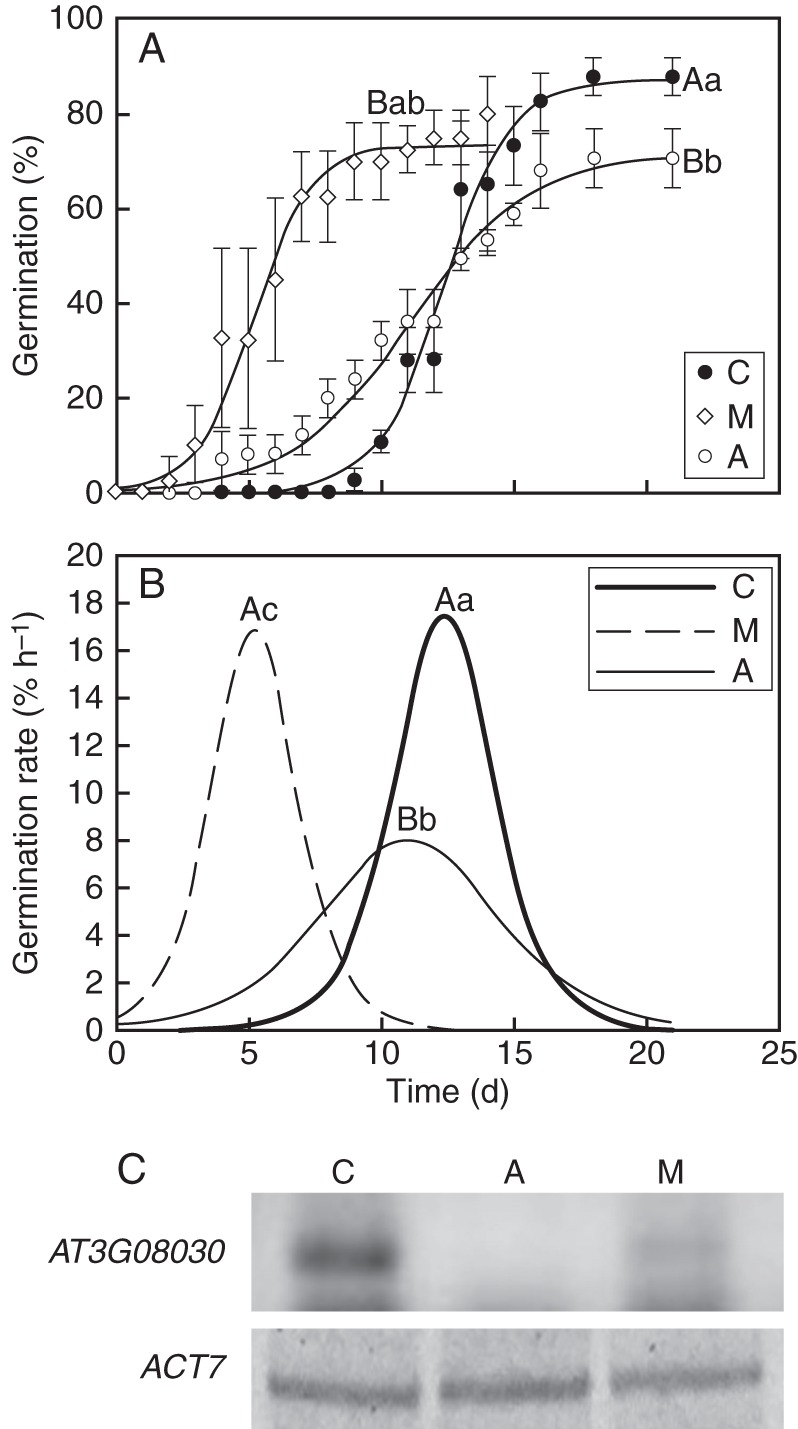
Germination performance and At3g08030 expression of C. aesculifolia seeds. (A) Cumulative germination curves for seeds 6–8 weeks after collection (C), natural priming treatment seeds (M) and 24-month aged seeds (A). (B) Maximal germination rate of C, M and A seeds. (C) RT-PCR analysis of At3g08030 expression was performed in the different seed lots from C. aesculifolia. Expression of ACT7 was analysed simultaneously as an internal standard (lower panel). Letters indicate statistical differences between the germination curves. All the experiments were performed at least three times.
Fig. 6.
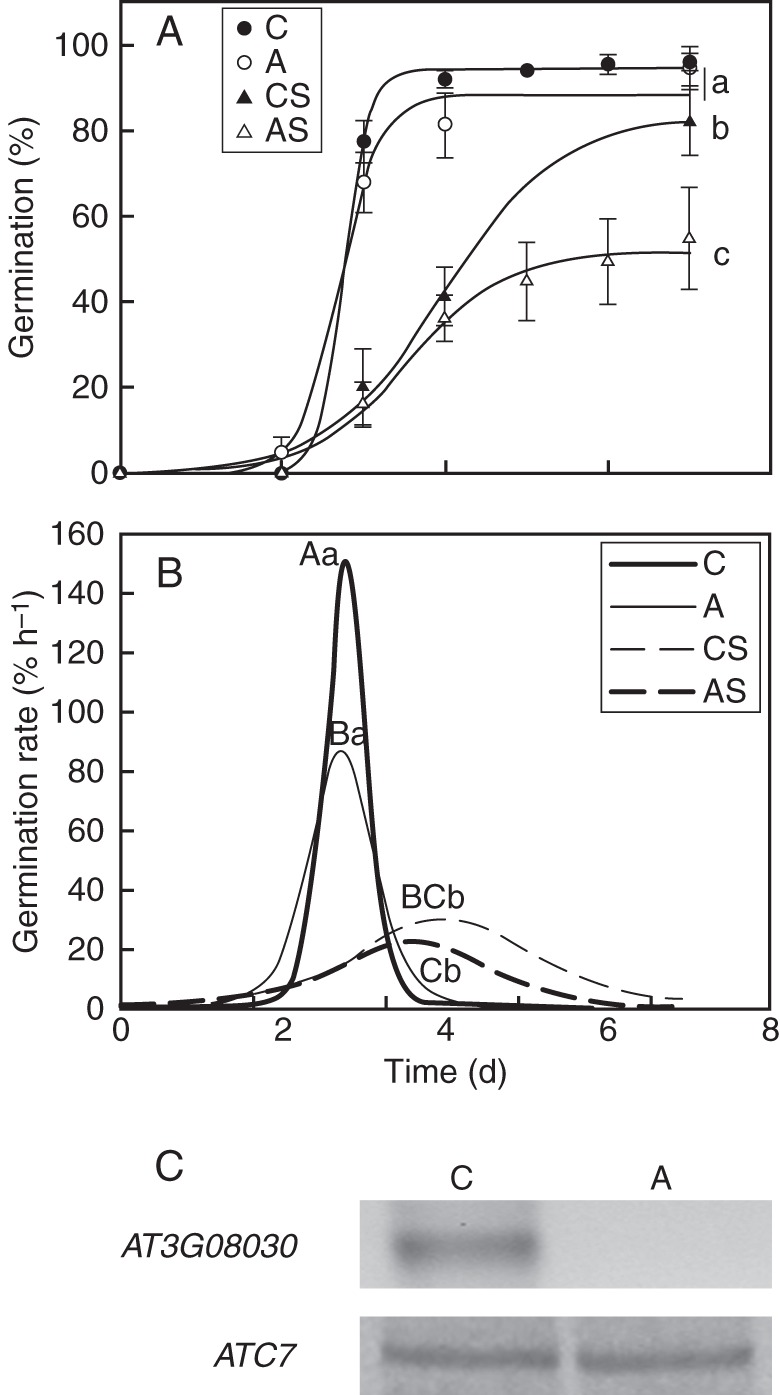
Germination performance and At3g08030 expression of W. urens seeds. (A) Cumulative germination curves of 6–8-week seeds (C) and 12-month aged seeds (A) under optimal conditions and 6–8-week seeds (CS) and 12-month aged-seeds (AS) under stress conditions (50 mm NaCl). (B) Maximal germination rate of C, A, CS and AS seeds. (C) RT-PCR analysis of At3g08030 expression was performed in the different seed lots from W. urens. Expression of ACT7 was analysed simultaneously as an internal standard (lower panel). Letters indicate statistical differences between the germination curves. All the experiments were performed at least three times.
Recently shed seeds of C. aesculifolia exhibited a significant delay in germination compared with buried seeds and the 2-year-old control seeds (F2,6 = 32·69, P = 0·0006) suggesting a physiological dormancy (Fig. 5A, B). Final germination and germination rate were decreased by seed ageing (F2,6 = 39·23, P = 0·0004 and F2,6 = 32·69, P = 0·022, respectively). Burial treatment promoted germination performance of recently shed seeds; in these treated seeds, maximal germination was reached after 7–8 d incubation compared with 15 d for the control and aged seeds. Germination rate and maximal germination of treated seeds were similar to those of recently shed seeds.
To determine whether the At3g08030 orthologous transcript was present in aged seeds of C. aesculifolia and W. urens, RT-PCR analysis was performed using the specific primers designed (Table 1, Figs 5C and 6C). For C. aesculifolia At3g08030 transcript was detected in recently collected seeds and in aged seeds previously treated as described above whereas no transcript was detected in aged seeds (Fig. 5C).
The germination performance of 12-month-old W. urens seeds remained unaltered under optimal conditions, but there was a significant decrease in their germination percentage under salt stress (P = 0·0359), from 82 ± 8·3 to 59 ± 10·6% (Fig. 6A, B).
At3g08030 was not detected in aged seeds of W. urens (Fig. 6C).
DISCUSSION
The DUF642 gene At3g08030 was expressed in all A. thaliana tissues tested, including dry seeds as previously described (Le et al., 2010). Matrix priming, using soil as matrix, increased the rate of germination of A. thaliana seeds (Fig. 2A, B and 3A, B). This matrix priming in soil has been used to increase germination of seeds from different species. Soil characteristics influenced the effect of priming on Zea mays and Avena sterilis (Long et al., 2011; Nicasio-Arzeta et al., 2011). The establishment of maize seedlings from matrix primed seeds was improved when they were grown in saline soil (Nicasio-Arzeta et al., 2011).
An experimental system, using a rapid ageing treatment and matrix priming, produced seeds having different rates of germination and final germination. More rapid germination was detected in the matrix primed aged seeds, as previously described for hydropriming treatment applied to sugarbeet seeds (Fig. 3; Catusse et al., 2011). In this reproducible system with A. thaliana seeds, an association between the rate of germination and final germination and At3g08030 expression levels was established; high expression levels of At3g08030 were associated with primed seeds whereas in CD seeds the transcript was only faintly detected. Matrix priming before and after CD resulted in an important increase of germination performance and At3g08030 expression. Germination parameters and transcript levels of CD seeds were affected differentially by the matrix treatment, supporting the suggestion that the effect of priming is related to seed age (Butler et al., 2009). The At3g08030 protein has not been reported as a priming-associated protein (Gallardo et al., 2001); specific detection of protein in seeds showing different performance is therefore needed. Furthermore, the function of At3g08030 in seed performance remains to be elucidated.
Germination performance studies for different plant species from different ecosystems have been done. A molecular marker for seed ageing will provide an important tool for seed ecophysiology studies because in natural ecosystems seed availability is often a limiting factor for seed collection and use in restoration programmes. Seed storage results in seed ageing that affects seed performance of C. aesculifolia and W. urens. The delay in the initial germination of the recently shed C. aesculifolia seeds indicates that in the seed population some degree of non-deep physiological dormancy is distributed differentially, which is overcome by after-ripening, as in other species (Sánchez and Mella, 1984). During short exposure to the burial treatment, germination is synchronized, indicating homogenization in the physiological stage of the seeds (Fig. 5).
Burial treatment enhanced seed performance as described for other plant species (González-Zertuche et al., 2001). For C. aesculifolia seeds, a decrease in the maximal germination percentage during storage indicated seed ageing. The At3g08030 transcript was not detected in these aged seeds but was still detected in the aged seeds that had been previously buried. At3g08030 expression was not detected in seeds of W. urens that were more sensitive to stress. These results suggest that detection of the At3g08030 transcript might also be used as a molecular marker for seed ageing in other plant species.
The highly conserved gene At3g08030 could be a molecular marker for seed ageing and a tool to determine seed lots from different plant species or habitats. Seed availability is often a limiting factor for reforestation or restoration programmes because it is not possible to predict seed production in nature. Predicting the performance of a particular seed lot requires specific germination tests, and maintaining collections implies re-test intervals (Probert et al., 2009). In this study, low quantities of seeds or tissue are used to determine expression of this gene. In general, the At3g08030 molecular marker could be detected using only 100 mg of tissue, and for plant species with large seeds such as C. aesculifolia, few seeds are needed. The storage compounds mostly found in the endosperm of seeds from different plant species impair RNA extraction or enzyme tests. For detection of At3g08030 transcript, isolated axes can also be used as the mRNA source because it is found in the embryo. In contrast, enzyme assays for catalase or superoxide dismutase and germination curves require many seeds (Evensen and Loy, 1978; Bailly et al., 2000).
Detection of the conserved orthologous At3g08030 transcript exclusively in seeds with high viability but not in seeds with poorer germination performance, as in A. thaliana and C. aesculifolia, or intolerant to stressful saline conditions, as in W. urens, which inhabit different ecosystems suggests that this molecular marker can be widely incorporated into those methods used in revegetation, reforestation and restoration ecology.
ACKNOWLEDGEMENTS
We thank M. and C. Laura Márquez for DNA sequencing, the Ecología Química laboratory for qRT-PCR facilities and Genética Molecular, Desarrollo y Evolución de Plantas laboratory for diverse facilities. This study was supported by a PAPIIT grant IN228109 (Universidad Nacional Autónoma de México). L.E.G.-C. is a PhD student (Posgrado en Ciencias Biológicas, UNAM) and received a CONACyT fellowship.
LITERATURE CITED
- Bailly C, Benamar A, Corbineau F, Côme D. Antioxidant systems in sunflower (Helianthus annuus L.) seeds as affected by priming. Seed Science Research. 2000;10:35–42. [Google Scholar]
- Bradford KJ. Manipulation of seed water relations via osmotic priming to improve germination under stress conditions. HortScience. 1986;21:1105–1112. [Google Scholar]
- Butler LH, Hay FR, Ellis RH, Smith RD, Murray TB. Priming and re-drying improve the survival of mature seeds of Digitalis purpurea during storage. Annals of Botany. 2009;103:1261–1270. doi: 10.1093/aob/mcp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, et al. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. The Plant Journal. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catusse J, Meinhard J, Job C, et al. Proteomics reveals potential biomarkers for seed vigor in sugarbeet. Proteomics. 2011;11:1569–1580. doi: 10.1002/pmic.201000586. [DOI] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiology. 2006;142:1493–1510. doi: 10.1104/pp.106.087452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evensen KJ, Loy JB. Effects of gibberellic acid and gold light on germination, enzyme activities, and amino acid pool size in a dwarf strain of watermelon. Plant Physiology. 1978;62:6–9. doi: 10.1104/pp.62.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, et al. Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiology. 2001;126:835–848. doi: 10.1104/pp.126.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa-deBuen A, Cruz-Ortega R, Martínez-Barajas E, Sánchez-Coronado ME, Orozco-Segovia A. Natural priming as an important metabolic event in the history life of Wigandia urens (hydrophyllaceae) seed. Physiologia Plantarum. 2006;128:520–530. [Google Scholar]
- González-Zertuche L, Vázquez-Yanes C, Gamboa A, Sánchez-Coronado ME, Aguilera P, Orozco-Segovia A. Natural priming of Wigandia urens seeds during burial: effects on germination, growth and protein expression. Seed Science Research. 2001;11:27–34. [Google Scholar]
- Graeber K, Linkies A, Wood ATA, Leubner-Metzger G. A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. The Plant Cell. 2011;23:2045–2063. doi: 10.1105/tpc.111.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydecker W, Higgins J, Gulliver RL. Accelerated germination by osmotic seed treatment. Nature. 1973;246:42–44. [Google Scholar]
- Jamet E, Albenne C, Boudart G, Irshad M, Canut H, Pont-Lezica R. Recent advances in plant cell wall proteomics. Proteomics. 2008;8:893–908. doi: 10.1002/pmic.200700938. [DOI] [PubMed] [Google Scholar]
- Khurana E, Singh JS. Ecology of seed and seedling growth for conservation and restoration of tropical dry forest: a review. Environmental Conservation. 2001;28:39–52. [Google Scholar]
- Le BH, Gheng C, Bui AQ, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proceedings of the National Academy of Sciences. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trick H. Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. BioTechniques. 2005;38:872–876. doi: 10.2144/05386BM05. [DOI] [PubMed] [Google Scholar]
- Long RL, Kranner I, Panetta FD, Birtic S, Adkins SW, Steadman KJ. Wet-dry cycling extends seed persistence by re-instating antioxidant capacity. Plant and Soil. 2011;338:511–519. [Google Scholar]
- Lundano S, Falahati-Anbaran M, Stenoien HK. Seed banks cause elevated generation times and effective population sizes of Arabidopsis thaliana in northern Europe. Molecular Ecology. 2009;18:2798–2811. doi: 10.1111/j.1365-294X.2009.04236.x. [DOI] [PubMed] [Google Scholar]
- Nicasio-Arzeta S, Sánchez-Coronado ME, Orozco Segovia A, Gamboa-deBuen A. Efecto de priming salino y substrato salino durante la germinación y el crecimiento de plántulas de Zea mays var. Chalqueño. Agrociencia. 2011;45:195–205. [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes. 2008;1:93. doi: 10.1186/1756-0500-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley D. A. Seed aging: Implications for seed storage and persistence in the soil. New York: Cornell University Press; 1986. [Google Scholar]
- Probert RJ, Daws MI, Hay FR. Ecological correlates of ex situ seed longevity: a comparative study on 195 species. Annals of Botany. 2009;104:57–69. doi: 10.1093/aob/mcp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MV. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Debeaujon I. Seed longevity: survival and maintenance of high germination ability of dry seeds. C.R. BIOL. 2008;331:796–805. doi: 10.1016/j.crvi.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SP, Belghazi M, Job C, Job D. Proteome-wide characterization of seed ageing in Arabidopsis: a comparison between artificial and natural ageing protocols. Plant Physiology. 2008;148:620–641. doi: 10.1104/pp.108.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D. Seed germination and vigor. Annual Review of Plant Biology. 2012;63:3·1–3·27. doi: 10.1146/annurev-arplant-042811-105550. [DOI] [PubMed] [Google Scholar]
- Sánchez RA, Mella RA. The exit from dormancy and the induction of germination; physiological and molecular aspects. In: Benech-Arnold RL, Sánchez RA, editors. Handbook of seed physiology. Applications to agriculture. The Haworth Press; 1984. ANSI Z39·48. [Google Scholar]
- Soeda Y, Konings MCJM, Vorst O, et al. Gene expression programs during Brassica oleracea seed maturation, osmopriming, and germination are indicators of progression of the germination process and the stress tolerance level. Plant Physiology. 2005;137:354–368. doi: 10.1104/pp.104.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesnier K, Strookman-Donkers HM, van Pijlen JG, van der Geest AHM, Bino RJ, Groot SPC. A controlled deterioration test of Arabidopsis thaliana reveals genetic variation in seed quality. Seed Science and Technology. 2002;30:149–165. [Google Scholar]
- Vázquez-Lobo A, Roujol D, Zúñiga-Sánchez E, et al. The highly conserved spermatophyte cell wall DUF642 protein family: phylogeny and first evidence of interaction with cell wall polysaccharides in vitro. Molecular Phylogenetics and Evolution. 2012;63:510–520. doi: 10.1016/j.ympev.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Vázquez-Yanes C, Orozco-Segovia A. Patterns of seed longevity and germination in the tropical rainforest. Annual Review of Ecology and Systematics. 1993;24:69–87. [Google Scholar]
- Wijayratne UC, Pyke DA. Burial increases seed longevity of two Artemisia tridentate (Asteraceae) subspecies. American Journal of Botany. 2012;99:438–447. doi: 10.3732/ajb.1000477. [DOI] [PubMed] [Google Scholar]



