Abstract
The level of mRNAs derived from the plastid-encoded psbD light-responsive promoter (LRP) is controlled by a circadian clock(s) in wheat (Triticum aestivum). The circadian oscillations in the psbD LRP mRNA level persisted for at least three cycles in continuous light and for one cycle in continuous dark, with maxima in subjective morning and minima in subjective early night. In vitro transcription in chloroplast extracts revealed that the circadian cycles in the psbD LRP mRNA level were dominantly attributed to the circadian-regulated transcription of the psbD LRP. The effects of various mutations introduced into the promoter region on the psbD LRP activity in vitro suggest the existence of two positive elements located between −54 and −36, which generally enhance the transcription activity, and an anomalous core promoter structure lacking the functional “−35” element, which plays a crucial role in the circadian fluctuation and light dependency of psbD LRP transcription activity.
Endogenous oscillators (circadian clocks) control diurnal rhythmic phenomena in prokaryotic cyanobacteria and almost all eukaryotes (Sweeney, 1987; Kondo et al., 1993; Hall, 1995; Dunlap, 1996). In plants several biological processes, including enzyme activities, leaf movements, stomatal opening and closing, and the expression of a large number of genes, exhibit endogenous rhythms (Sweeney, 1987; Edmunds, 1988; McClung and Kay, 1994). Photosynthesis is also one of the representative phenomena regulated by circadian clocks at the metabolic (Sweeney and Haxo, 1961; Hennessey and Field, 1992) and gene-expression levels (Kloppstech, 1985; Giuliano et al., 1988; Nagy et al., 1988).
In the unicellular alga Chlamydomonas reinhardtii, several photosynthesis-related genes encoded by both nuclear and plastidial genomes have been reported to exhibit circadian expression (Salvador et al., 1993; Jacobshagen and Johnson, 1994; Hwang et al., 1996). However, in higher plants all of the genes that have been shown to exhibit circadian expression are nuclear encoded (McClung and Kay, 1994). The possibility of circadian control over the plastid-encoded genes was previously evaluated in pea, but no evidence for the existence of rhythmic gene expression in the chloroplast was observed (Adamska et al., 1991). In tomato minor diurnal fluctuations of several plastid-encoded transcripts were detected but such fluctuations have not been clearly defined as circadian rhythms (Piechulla, 1993).
Plastid-encoded psbD and psbC genes, encoding the PSII subunit proteins D2 and CP43, respectively, form an operon (psbD/C) together with other photosynthesis-related genes in higher plants. In the wheat (Triticum aestivum) psbD/C operon, there are four distinct transcription initiation sites. Transcript levels derived from the third promoter (psbD LRP) are specifically induced in etiolated seedlings transferred to light (Wada et al., 1994). In mature wheat chloroplasts, the accumulation of the mRNAs from the psbD LRP is reversibly increased by illumination and reduced by dark adaptation (Satoh et al., 1997). It has been demonstrated in wheat and other higher plants that the light-dependent accumulation of the psbD LRP mRNA is mediated by the light-responsive activation/inactivation of the psbD LRP (Christopher et al., 1992; Allison and Maliga, 1995; Hoffer and Christopher, 1997; Satoh et al., 1997).
The level of psbD LRP mRNA may be controlled not only by light but also by endogenous circadian rhythms. In rice seedlings some of the mRNA levels transcribed within the psbD/C operon have been reported to exhibit diurnal oscillations under a 12-h dark/12-h light cycle (Chen et al., 1995). When etiolated wheat seedlings were transferred to continuous light, the levels of some transcripts, which appeared to be derived from the psbD LRP, showed a fluctuation with an approximately 24-h periodicity for one cycle (Kawaguchi et al., 1992). However, these data are insufficient to confirm the involvement of circadian regulation over the psbD LRP mRNA level. To clearly identify a circadian rhythm, it is critical to demonstrate that a diurnal rhythm can persist for several cycles under constant environmental conditions known as “free-running conditions.”
We examined the time course of the psbD LRP mRNA levels in wheat seedlings under continuous light or continuous dark conditions after 5 d of 12-h light/12-h dark entrainment. The circadian oscillations in the psbD LRP mRNA persisted for at least 3 d in continuous light and for at least 1 d in continuous dark. Judging from the rhythms under free-running conditions, we concluded that the psbD LRP mRNA level is controlled by a circadian clock(s). In vitro transcription in chloroplast extracts prepared from seedlings at several times revealed that the oscillation in mRNA level is dominantly attributed to the circadian fluctuations of the psbD LRP transcription activity. Furthermore, to identify regulatory sequences involved in the clock-regulated transcription of the psbD LRP, we investigated the effects of various mutations introduced into the promoter region on the psbD LRP activity in vitro.
MATERIALS AND METHODS
Plant and Growth Conditions
Wheat (Triticum aestivum) seeds were planted in moist vermiculite and grown in a growth chamber at 25°C under the 12-h white light (approximately 5000 lux)/12-h dark cycles. After 5 or 5.5 d, wheat seedlings were transferred to continuous light or continuous dark conditions. Seedlings were harvested at the times indicated in the figure legends and subjected to total cellular RNA isolation or preparation of chloroplast extracts.
To investigate the effect of light on transcription activity in vitro, 5-d-old light-grown seedlings were maintained under dark conditions for 24 h and again transferred into light for 2 h, at which time the chloroplast extracts were isolated.
RNA Isolation and Analysis
Primary leaves 5 to 7 cm from the top of seedlings were harvested and quickly frozen in liquid N2. The frozen leaf tissue was ground to a fine powder in the presence of liquid N2. The resulting powder suspended into a buffer (100 mm Tris-HCl, pH 8.0, 100 mm LiCl, 10 mm EDTA-NaOH, pH 8.0, and 1% SDS) was extracted twice with an equivalent volume of a phenol mixture (phenol:chloroform:isoamyl alcohol, 25:24:1 [v/v]), and then total cellular RNA was precipitated with one-third volume of 10 m LiCl. The sample was subjected to two additional cycles of phenol extraction followed by ethanol precipitation.
To analyze the accumulation of plastid-encoded transcripts, S1 nuclease-protection analysis was performed as previously described (Satoh et al., 1997). The DNA probes used for detecting the psbA and psbD LRP transcripts were a 472-bp EcoRI-SacI fragment from pW18ES472A and a 834-bp StyI-EcoRI fragment from pW18E866, respectively. Equal amounts of total cellular RNA (1–5 μg) were hybridized with the desired 5′-end-labeled denatured DNA probe in an 80% formamide hybridization buffer at 37°C. The RNA-DNA hybrids were treated with 400 units/mL S1 nuclease for 60 min at 20°C, and the protected DNA fragments were separated on a denaturing polyacrylamide gel. Quantification of mRNA abundance was performed with a bio-imaging analyzer (BAS2000, Fuji, Tokyo, Japan).
Chloroplast Extract Preparation
Harvested (nonfrozen) seedlings were immediately lysed, and intact chloroplasts were isolated by centrifugation of the cell lysate on a 10% to 80% Percoll gradient. Transcriptionally active chloroplast extracts (high-salt extracts) were prepared from the intact chloroplasts, as previously described (Satoh et al., 1997), and subjected to in vitro transcription and a DNA mobility-shift assay.
Plasmid Constructs
Plasmid psbD/−119 (previously designated as pW73BE434 by Satoh et al. [1997]), containing a sequence of the psbD LRP (−119 to +315 of the transcription initiation site) in pSP73 vector, was utilized for preparation of the DNA constructs used in the promoter analysis of the psbD LRP. A 5′ deletion fragment, which has a 5′ end at −36 and a 3′ end at +315 of the psbD LRP transcription initiation site, was amplified by the PCR method with psbD/−119 as a template and cloned between the BamHI and EcoRI sites of pSP73 vector (psbD/−36). The sequence between −72 and −7 of the psbD LRP initiation site was divided into eight blocks, and mutations were introduced in each block by means of site-directed mutagenesis, as described by Kunkel et al. (1987). Each of the obtained fragments were amplified by PCR and cloned between BamHI and EcoRI sites of pSP73 (psbD/MT−1 to −8). Another mutant of the psbD LRP (psbD/MT−9) having the consensus “−35” motif and the optimal spacing distance (17 bp) between −35 and −10 was constructed by PCR. Plasmid psbA/−38, containing the sequence −38 through +278 of the psbA transcription initiation site in the pSP72 vector was used as a template for the psbA transcription in vitro. The psbA promoter with a mutation in the −35 element (psbA/MT−2) was constructed by PCR from psbA/−38. For in vitro transcription, all plasmid constructs were prepared as supercoiled circular forms.
In Vitro Transcription
In vitro transcription was carried out as previously described (Satoh et al., 1997). Each chloroplast extract was prepared from 1.6 × 108 chloroplasts, and 1.5 μg of a given supercoiled DNA template was incubated in a 40-μL reaction mixture at 30°C for 60 min. In vitro-synthesized RNA was isolated and analyzed by S1 nuclease-protection analysis. A 656-bp BamHI-NdeI fragment from pW73BE549 containing the psbD LRP sequence (−234 to +315 of the psbD LRP transcription initiation site), followed by a 107-bp vector sequence and a 521-bp EcoRI-HindIII fragment from pW72ES472A containing the psbA promoter sequence (−194 to +278 of the psbA transcription initiation site), followed by a 49-bp vector sequence were 32P-end-labeled at the NdeI and HindIII sites of the vector sequences, respectively. These fragments were used as S1 probes for transcripts derived from the psbD LRP and psbA promoter in vitro.
DNA Mobility-Shift Assay
For the DNA mobility-shift assay, a 32P-end-labeled, 23-bp, double-stranded oligonucleotide containing the sequence from −56 to −34 of the psbD LRP site was used as a probe. The DNA-binding reaction was carried out in 20 μL of DEAE buffer (Wada et al., 1994) containing 10 μL of chloroplast extract (1.0 × 108 chloroplasts per reaction), the labeled probe (0.1 ng, 1.0 × 104 cpm per reaction), and 2 μg of poly(dI-dC)·(dI-dC) at 30°C for 30 min. The reaction mixture was run on 5% nondenaturing polyacrylamide gel (mono:bis = 30:0.8) at room temperature in 0.5× TBE buffer (45 mm Tris-borate, 1 mm EDTA) and then autoradiographically analyzed. The nonlabeled DNA fragment of the probe sequence, a 26-bp fragment containing the further upstream sequence of the psbD LRP (−85 to −60 of the psbD LRP transcription initiation site), and a 90-bp fragment of the psbD-coding sequence were used for competition assays.
RESULTS
The Level of mRNAs Derived from the Plastid-Encoded psbD LRP Is Controlled by a Circadian Clock(s) in Wheat
We investigated whether the fluctuation of the level of mRNAs transcribed from the psbD LRP initiation site (psbD LRP mRNA level) exhibits clear patterns of circadian rhythm under free-running conditions. Wheat seedlings were grown for 5 d under a 12-h light/12-h dark regime and then shifted into continuous light or continuous dark conditions. Seedlings were harvested every 3 h and frozen in liquid N2. An equivalent amount of total cellular RNA isolated from each sample was subjected to S1 nuclease-protection analysis to determine the relative amount of the psbD LRP mRNA. Figure 1 shows the fluctuation in the psbD LRP mRNA level under continuous light conditions. Sampling times are expressed as ZT (Zerr et al., 1990); ZT 0 is dawn in the sixth cycle from imbibition. The psbD mRNA level fluctuated with a periodicity of approximately 24 h. Starting from ZT 0, the psbD LRP mRNA level peaked in the first subjective morning (ZT 3), declined to a minimum in subjective early night (ZT 15), and again peaked at ZT 27 in continuous light conditions. The psbD LRP mRNA levels varied by approximately 3- to 5-fold between maximum and minimum. This rhythm persisted for at least three cycles under continuous light conditions.
Figure 1.
S1 nuclease-protection analyses of the plastid-encoded psbD LRP and psbA mRNA in wheat seedlings under continuous light. Wheat seedlings were grown for 5 d under 12-h light/12-h dark conditions and transferred to continuous light conditions. A, S1 nuclease-protection analyses were carried out with equal amounts of total cellular RNA (1 and 5 μg for psbA and psbD LRP analyses, respectively) prepared from seedlings harvested every 3 h in continuous light. Black bars, dark periods; white bars, light periods. The inserted dark bars indicate subjective night. S1 analysis of psbA mRNA was not carried out for RNA samples at ZT 51 through 69. B, Quantitative representation of the results shown in A. For each mRNA the lowest signal is defined as 1.0 (arbitrary units), and other abundances are expressed relative to the minimum.
The circadian cycle in the psbD LRP mRNA level was also detected in continuous dark conditions (Fig. 2). The psbD LRP mRNA accumulation was low in the first subjective early night (approximately ZT 15) and then increased, with a peak in subjective morning (ZT 27), and returned to a minimum in the early night phase (from ZT 39–42) with a 3-fold amplitude. From ZT 42 onward, the accumulation of the psbD LRP mRNA was severely reduced to the basal level, although a small peak occurred at ZT 51. Such profiles as the psbD LRP mRNA oscillation rapidly damped in continuous dark have also been observed in the circadian expression of nuclear-encoded cab genes (Millar and Kay, 1991). That the circadian oscillations in the psbD LRP mRNA level were detected under continuous light and partially under continuous dark conditions indicates that the plastid-encoded psbD LRP mRNA level is controlled by an endogenous clock(s) in wheat.
Figure 2.
S1 nuclease-protection analysis of the plastid-encoded psbD LRP and psbA mRNA in wheat seedlings under continuous dark conditions. Wheat seedlings were grown for 5 d under 12-h light/12-h dark conditions, followed by an additional 12-h light period, and were then transferred to continuous dark conditions. A, Sampling of total cellular RNA and S1 nuclease-protection analyses were performed as described in Figure 1. The inserted white bars indicate subjective day. ZT 0 is the initiation of the last light period so that it is the same time as ZT 0 indicated in Figure 1. B, Quantitative representation of the results shown in A. Relative mRNA abundances are as defined in Figure 1.
We also carried out S1 nuclease-protection analyses for the plastid-encoded psbA transcripts encoding the D1 subunit protein of PSII against the same RNA samples (Figs. 1 and 2). We observed that the psbA mRNA levels appeared to fluctuate diurnally. However, these fluctuations were very weak in both continuous light and continuous dark conditions compared with the psbD LRP mRNA levels.
Circadian Oscillation in the psbD LRP mRNA Level Is Dominantly Attributed to the Clock-Regulated psbD LRP Transcription Activity
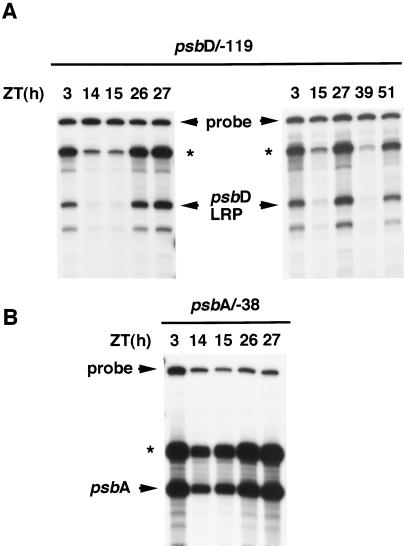
The circadian cycle of the psbD LRP mRNA level can result from changes at the level of transcription, mRNA stability, or both. It has been reported that the psbD LRP mRNA levels are regulated by light through the light-responsive activation/inactivation of the psbD LRP (Christopher et al., 1992; Allison and Maliga, 1995; Hoffer and Christopher, 1997; Satoh et al., 1997). To examine the properties of circadian regulation of the psbD LRP transcription activity, we carried out a series of in vitro transcriptions in chloroplast extracts. Chloroplast extracts were prepared from wheat seedlings in subjective morning (ZT 3, 26, 27, and 51) and subjective early night (ZT 14, 15, and 39) under continuous light conditions (Fig. 3A). In vitro transcription was performed in the extracts prepared from an equal number of chloroplasts with the supercoiled plasmid template containing the sequences between −119 and +315 of the psbD LRP transcription initiation site (psbD/−119). Synthesized transcripts from the psbD LRP initiation site in vitro were specifically detected by S1 nuclease-protection analysis. The in vitro transcription activity of the psbD LRP fluctuated diurnally throughout experimental periods, with about a 10-fold increase in subjective morning (ZT 3, 26, 27, and 51) compared with that in subjective early night (ZT 14, 15, and 39). Although we did not determine the time course of the circadian fluctuation in psbD LRP transcription in detail, this result indicates that the transcription activity of the psbD LRP is under circadian control and that the circadian oscillation in the psbD LRP mRNA level is dominantly attributed to the clock-regulated psbD LRP transcription activity.
Figure 3.
In vitro transcription of the psbD LRP and the psbA promoter in chloroplast extracts prepared from wheat seedlings in continuous light conditions. A, In vitro transcription from the psbD LRP transcription initiation site was carried out in two series with equal amounts of the chloroplast high-salt extracts prepared from seedlings at the indicated times (ZT 3, 14, 15, 26, 27, and ZT 3, 15, 27, 39, and 51, respectively) with the supercoiled circular plasmid pSP73 vector containing the sequences between −119 and +315 of the psbD LRP transcription initiation site (psbD/−119) as the template. Specific in vitro transcripts were detected by S1 nuclease-protection analysis. B, In vitro transcription of the psbA promoter was carried out with one of two series of the chloroplast extracts (ZT 3, 14, 15, 26, and 27) and circular plasmid pSP72 vector containing the sequences between −38 and +278 of the psbA transcription initiation site (psbA/−38) as the template. In vitro transcripts were subjected to S1 nuclease-protection analyses. The arrows indicate the primary transcripts synthesized in vitro. Read-through signals (*) were also seen at the position of the boundary between the vector and the inserted plastid DNA sequences.
We also assayed in vitro transcription activity of the psbA gene with a plasmid template containing the sequence between −38 and +278 of the psbA transcription initiation site (psbA/−38). In vitro transcription of the psbA promoter with the same series of chloroplast extracts showed the diurnal fluctuation of the psbA transcription activity with 2- to 3-fold increases in subjective morning compared with that in subjective early night (Fig. 3B). This result suggests that, in addition to the psbD LRP, the psbA transcription may also be controlled by an endogenous clock(s).
Two cis Elements at the 5′ Upstream Region in the psbD LRP May Act as Positive Elements That Generally Enhance the Transcription Activity Rather Than Circadian Clock-Responsive Elements
The psbD LRP contains a 60-bp region upstream of the transcription initiation site that is highly conserved among several monocotyledonous and dicotyledonous plants (Fig. 4). This region possesses two positive elements involved in the light-dependent transcription of the psbD LRP in barley and tobacco. One is an AAGT repeat located between −54 and −46 of the transcription initiation site (Kim and Mullet, 1995). Another is a GACC/T repeat located between −44 and −36 (Allison and Maliga, 1995). We carried out in vitro transcription in wheat chloroplast extracts from 24-h dark-adapted and 2-h reilluminated seedlings with a series of DNA constructs containing mutations in the upstream region of the psbD LRP initiation site. The results indicated that the same cis elements function as positive regulators in the light-induced transcription of the wheat psbD LRP (data not shown).
Figure 4.
Highly conserved sequences between the wheat psbD LRP region and homologous regions from other higher plants. DNA sequences between −60 and +20 of the wheat psbD LRP transcription initiation site are aligned with barley, rice, tobacco, and Arabidopsis (Christopher et al., 1992; Hoffer and Christopher, 1997). Completely conserved sequences are boxed. The positive regulatory elements, the AAGT and GACC/T repeats, which have been indicated to be involved in light-dependent transcription of the psbD LRP (Allison and Maliga, 1995; Kim and Mullet, 1995), and the putative −35 and −10 elements are marked.
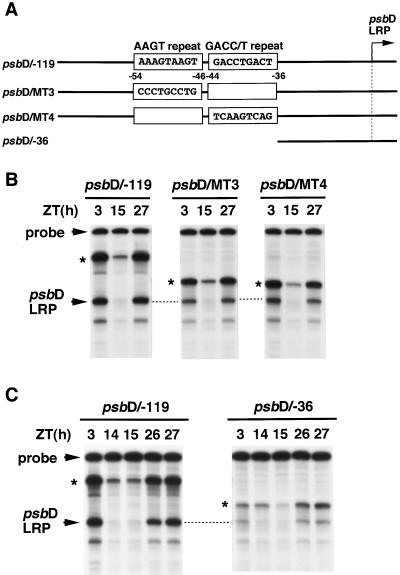
To investigate whether the cis elements would also be involved in circadian regulation of the psbD LRP, we examined the effect of mutations introduced into the two elements on the in vitro transcription activity in chloroplast extracts prepared from seedlings under continuous light conditions. Introduction of mutations in each element (psbD/MT−3 and −4) resulted in a 2-fold reduction of transcription activity, but the transcription levels fluctuated markedly (Fig. 5B). Complete deletion of both elements (psbD/−36) resulted in a severe reduction of the in vitro transcription activity in chloroplast extracts prepared from seedlings in subjective morning (Fig. 5C). However, it should be noted that the psbD/−36 construct still conferred significant circadian fluctuations in the transcription levels initiated from the psbD LRP site.
Figure 5.
Effects on the circadian fluctuation of the in vitro psbD LRP transcription activity with the mutations and deletions of the positive regulatory elements. A, DNA fragments (−119 to +315) containing the indicated base-pair substitutions (psbD/MT−3 and −4) and a DNA fragment deletion containing sequences between −36 and +315 (psbD/−36) were cloned in pSP73 vector and used as templates. B and C, In vitro transcription was carried out in chloroplast extracts prepared from seedlings at indicated times in continuous light conditions. Details are as described in Figure 3.
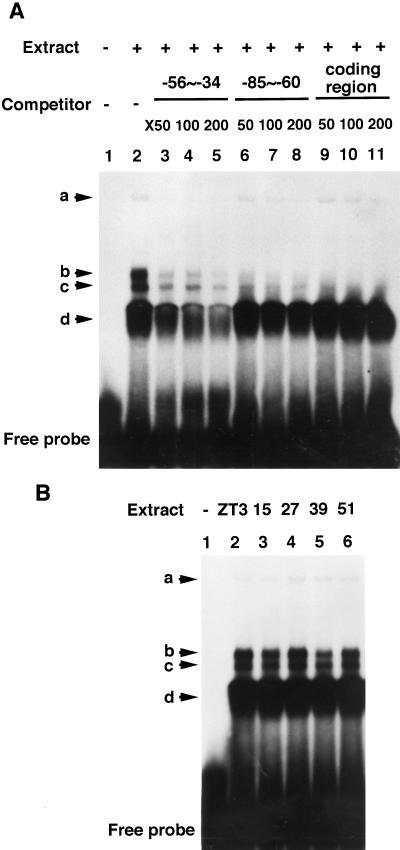
A putative transcription factor that specifically recognizes the AAGT repeat has been suggested in barley chloroplasts (Kim and Mullet, 1995). A DNA-binding protein that recognizes the GACC/T repeat in tobacco chloroplasts has been reported (Allison and Maliga, 1995). To reveal the role of such trans factor(s) on the circadian regulation and the light-dependent behavior of the psbD LRP, we performed a DNA mobility-shift assay using a 23-bp DNA fragment probe containing the sequence from −56 through −34 of the psbD LRP initiation site. Incubation of chloroplast extract prepared from seedlings at ZT 3 in continuous light with the DNA probe resulted in the formation of several DNA/protein complexes (Fig. 6A). Among them, bands “a” and “d” were apparently due to interactions with sequence-specific DNA-binding protein(s), since the nonlabeled DNA fragment of the probe sequence could specifically act as a competitor. However, the relative intensities of the a and d bands revealed neither circadian fluctuations (Fig. 6B) nor light dependency (data not shown). Together with the results shown in Figure 5, the sequence between −54 and −36 is suggested to function as two positive elements that generally enhance the psbD LRP transcription activity rather than acting as essential elements for circadian regulation.
Figure 6.
DNA mobility-shift assay of the upstream positive elements with chloroplast extracts prepared from seedlings in continuous light conditions. A, End-labeled, 23-bp, double-stranded DNA probe (−56 to −34) was incubated in the absence (lane 1) and in the presence (lanes 2–11) of chloroplast extract prepared from seedlings at ZT 3 in continuous light. Binding reactions were performed in the absence (lanes 1 and 2) or in the presence at the indicated times of molecular excess of the unlabeled probe sequence (lanes 3−5), a 26-bp DNA fragment containing another region of psbD LRP (−85 to −60) not involved in transcription activity (lanes 6−8), or a 90-bp psbD-coding region sequence (lanes 9−11) as DNA competitors. Arrows mark detected complexes. B, Binding reactions carried out in chloroplast extracts prepared from seedlings at the indicated times in continuous light conditions (ZT 3, 15, 27, 39, and 51).
The Core Promoter Structure Lacking the Prokaryotic −35 Element May Play a Crucial Role in the Circadian Fluctuation and Light Dependency of the psbD LRP Transcription Activity
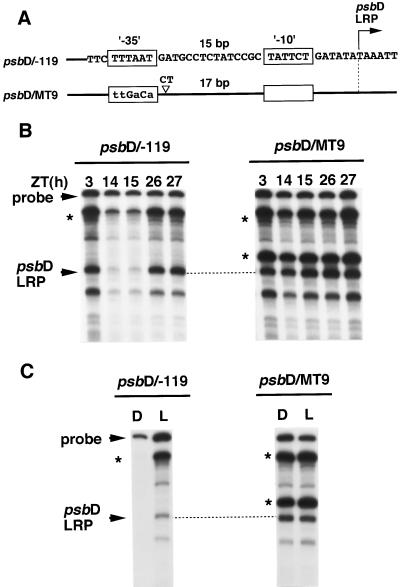
Typical plastid promoters contain sequences resembling prokaryotic −10 (TATAAT) and −35 (TTGACA) elements that are structurally and functionally similar to Escherichia coli ς70-type promoters (Hanley-Bowdoin and Chua, 1987). The importance of the E. coli-like elements for transcription initiation of plastid genes has been demonstrated by in vitro mutational analyses (Link, 1984; Bradley and Gatenby, 1985; Gruissem and Zurawski, 1985). The psbD LRP also contains similar elements (Fig. 4), but the putative −35 element (TTTAAT) only weakly resembles the prokaryotic consensus motif, and the spacing between the −35 and −10 elements is anomalous (15 bp instead of the optimal 17–19 bp).
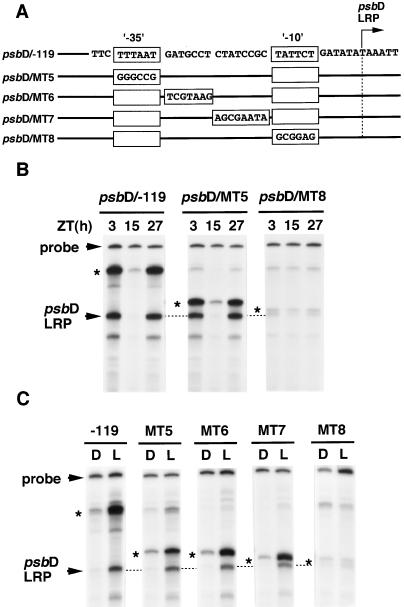
To investigate the relationship between the psbD LRP core promoter sequence and the circadian fluctuation of the psbD LRP transcription, we constructed several DNA constructs containing mutations in the core promoter region of the psbD LRP. Complete destruction of the putative −35 element (−32 to −28 of the psbD LRP transcription initiation site, psbD/MT−5; Fig. 7B) or the spacing sequences between the −35 and −10 elements (−27 to −13, psbD/MT−6 and −7; data not shown) caused no change in the psbD LRP transcription activity in continuous light conditions. Conversely, destruction of the −10 element (−12 to −7, psbD/MT−8) resulted in an almost disappearance of the psbD LRP activity (Fig. 7B). Furthermore, we carried out in vitro transcription with the same set of DNA constructs in chloroplast extracts from 24-h dark-adapted and 2-h reilluminated wheat seedlings (“D” and “L,” respectively), and the results were similar to those for circadian regulation (Fig. 7C). These results indicate that the −10 element is essential for the basal transcription activity, but neither the sequence corresponding to the putative −35 element nor the spacing sequence between two elements plays any positive role in psbD LRP transcription behavior.
Figure 7.
Effects of the generated mutations in the core promoter region of the psbD LRP on its circadian- and light-regulated in vitro transcription. A, DNA fragments (−119 to +315) containing the indicated base-pair substitutions at each block were cloned in pSP73 vector (psbD/MT−5 to −8) and used as templates. B, In vitro transcription was carried out in chloroplast extracts prepared from seedlings at the indicated times in continuous light conditions (ZT 3, 15, and 27). Results for templates psbD/MT−6 and −7 are not shown. C, In vitro transcription with the same set of templates was carried out in chloroplast extracts prepared from 24-h dark-adapted and then 2-h reilluminated seedlings (D and L extracts, respectively). Read-through signals (*) were also seen at the position of the boundary between the vector and the inserted plastid DNA sequences.
However, it might be possible that the anomalous structure of the psbD LRP lacking the functional −35 elements is responsible for the circadian-fluctuation dependency and/or the light dependency of the psbD LRP transcription. To examine this possibility we changed the core promoter structure of the psbD LRP to resemble a typical prokaryotic promoter by substituting the pseudo −35 elements with consensus −35 motif and simultaneously converting the spacing distance between −35 and −10 to 17 bp (psbD/MT−9). This construct abolished the circadian fluctuation of in vitro transcription activity from the psbD LRP initiation site, leaving the promoter to function almost constitutively in continuous light (Fig. 8B). A similar profile was observed in the light dependency of the psbD LRP transcription activity (Fig. 8C). These results suggest that the promoter architecture lacking the −35 element plays a crucial role in the circadian fluctuation and light dependency of the psbD LRP transcription activity.
Figure 8.
Effects on the in vitro psbD LRP transcription activity after modification of the psbD LRP to generating a consensus −35 element and canonical spacing (17 bp). A, DNA fragment (−119 to +315) containing the indicated base-pair substitutions and insertion was cloned in pSP73 vector (psbD/MT−9) and used as templates. B, In vitro transcription carried out in chloroplast extracts prepared from seedlings at the indicated times in continuous light conditions (ZT 3, 14, 15, 26, and 27). C, In vitro transcription performed in chloroplast extracts prepared from 24-h dark-adapted (D) and 2-h reilluminated (L) seedlings, as described in Figure 7C. Read-through signals (*) were also seen at the position of the boundary between the vector and the inserted plastid DNA sequences.
The psbA promoter contains typical −35 and −10 elements, and the spacing between them is optimal (18 bp). Exchange of the −35 element (TTGACA) of the psbA promoter with the pseudo −35 element (TTTAAT) of the psbD LRP resulted in very strong circadian fluctuation of the psbA transcription activity (Fig. 9B). Severe reduction of the transcription levels was observed specifically in chloroplast extracts prepared from seedlings in subjective early night (ZT 14 and 15), with almost no effect in subjective morning (ZT 3, 26, and 27). Furthermore, light induction of the psbA transcription activity also became stronger after the −35 element exchange (Fig. 9C). These results suggest that the robust circadian fluctuation and light induction of the transcription activity observed in the psbD LRP, but not in the psbA promoter, may be attributed to the difference of their core promoter structures, which lack or possess the functional −35 element, respectively.
Figure 9.
Effects on the in vitro psbA transcription activity after substituting the −35 element of the psbA promoter into the pseudo element of psbD LRP. A, DNA fragment between −38 and +278 containing the indicated base-pair substitutions in the −35 element was cloned in pSP72 vector and used as templates (psbA/MT−2). B, In vitro transcription was carried out in chloroplast extracts prepared from seedlings at indicated times in continuous light conditions (ZT 3, 14, 15, 26, and 27). C, In vitro transcription performed in chloroplast extracts prepared from 24-h dark-adapted (D) and 2-h reilluminated (L) seedlings, as described in Figure 7C. Read-through signals (*) were also seen at the position of the boundary between the vector and the inserted plastid DNA sequences.
DISCUSSION
The psbD LRP Transcription Is under Circadian Control in Chloroplasts of Higher Plants
In this report we have demonstrated that the levels of the plastid-encoded psbD LRP mRNAs, which are derived from the third transcription initiation site (psbD LRP site) in the psbD/C operon, are conspicuously controlled by a circadian clock(s) in wheat. The circadian oscillation in the psbD LRP mRNA level persisted for at least three cycles in continuous light and one cycle in continuous dark, with a maximum in subjective morning and a minimum in subjective early night (Figs. 1 and 2). In vitro transcription activity of the psbD LRP in chloroplast extracts prepared from the wheat seedlings under continuous light diurnally fluctuated throughout experimental periods, with about a 10-fold increase in subjective morning compared with that in subjective early night (Fig. 3A). In vitro chloroplast transcription assays in wheat reproduced the light-dependent psbD LRP transcription pattern in vivo (Wada et al., 1994; Satoh et al., 1997). Therefore, it is likely that the results shown in Figure 3A indicate that the circadian oscillations of the psbD LRP mRNA expression is mostly attributable to the transcriptional regulation of the psbD LRP.
Since the promoter structure of the psbD LRP is highly conserved among certain higher plants (Christopher et al., 1992; Hoffer and Christopher et al., 1997; Fig. 4), circadian oscillation in the psbD LRP transcription activity is likely to be exhibited in other higher plants as well. The psbD LRP has been shown to be activated by high-intensity blue and UV-A light in barley chloroplasts (Gamble and Mullet, 1989; Christopher and Mullet, 1994), and it has been proposed that the high-irradiance activation of the psbD LRP serves to maintain D2 protein levels that are photodamaged under high-intensity light conditions (Christopher and Mullet, 1994). In addition to such a role, the circadian oscillation of the psbD LRP may also control the synthetic rate of D2 protein in a diurnal manner.
Circadian-regulated transcription of the plastid-encoded genes in higher plants may not be restricted to the psbD LRP. Although the mRNA level of the plastid-encoded psbA gene showed only weak oscillation with less than a 2-fold variation (Figs. 1 and 2), in vitro transcription activity of psbA promoter fluctuated diurnally with 2- to 3-fold variation (Fig. 3B). Since the psbA mRNA is highly stable (t1/2 = 10–40 h) in mature leaves of higher plants (Mullet and Klein, 1987; Klaff and Gruissem, 1991; Kim et al., 1993), the psbA mRNA level may not reflect the fluctuation of psbA transcription activity.
Activity of a RNA Polymerase, Which Does Not Require the −35 Element for the psbD LRP and psbA Promoter Recognition, May Be Regulated by a Circadian Clock(s) and Light
In vitro transcription with the mutated psbD LRP sequences revealed the existence of two cis elements and a core promoter structure that are involved in the circadian regulation of the psbD LRP activity. The two cis elements are an AAAGTAAGT sequence located between −54 and −46 of the wheat psbD LRP transcription initiation site and a GACCTGACT sequence between −44 and −36. These elements are identical to those demonstrated to function as positive regulatory elements of the psbD LRP in tobacco and barley (Allison and Maliga, 1995; Kim and Mullet, 1995). Mutations introduced into the two elements resulted in a reduction of the in vitro psbD LRP activity, but significant circadian fluctuations in the psbD LRP transcription levels remained (Fig. 5). The ability of DNA-binding factor(s) recognizing these elements was constitutive in continuous light (Fig. 6). These results suggest that the two cis elements act as positive regulators that generally enhance the psbD LRP transcription activity rather than the circadian clock-responsive elements controlling the oscillation.
In contrast, alterations in the core-promoter sequence of the psbD LRP suggest that the core-promoter architecture, lacking the functional −35 element, plays a crucial role in the circadian fluctuation of the psbD LRP transcription activity (Figs. 7B and 8B). A mutation in the psbA core promoter, in which the functional −35 element of the psbA promoter was replaced by the pseudo −35 element from the psbD LRP, resulted in more robust fluctuation of the in vitro psbA transcription activity than the native psbA promoter (Fig. 9B). This suggests that the core promoter structures of the psbA promoter and the psbD LRP, possessing the functional −35 element or not, respectively, may be responsible for the different robustness of fluctuations of the psbA promoter (2- to 3-fold) and psbD LRP (10-fold). Furthermore, the same core promoter structure of the psbD LRP, lacking the functional −35 element, is suggested to be responsible for the light-responsive behavior of psbD LRP transcription activity. Thus, it is likely that the circadian-clock and light-responsive element(s) in the psbD LRP are common.
As a possible mechanism that enables the psbD LRP lacking the functional −35 element to be under circadian and light regulation, we speculate that the chloroplast contains at least two types of transcription apparatus that either require the functional −35 element for promoter recognition or do not. Although activity of the former type of RNA polymerase may not be clearly regulated by a circadian clock(s) and light, the latter type may be under strong circadian and light regulation. This working hypothesis can easily explain the strong circadian and light regulation of the psbD LRP and the mutated psbA promoter, in which the functional −35 element of the psbA promoter was replaced by the pseudo −35 element from the psbD LRP. However, the native psbA promoter, which possesses the functional −35 element, can be recognized by both types of RNA polymerase; therefore, circadian control and light induction of psbA transcription may not be so clearly observed. Since it was reported that the psbD LRP mRNA disappeared in transgenic tobacco lacking a subunit of the plastid-encoded plastid RNA polymerase (PEP), which is similar to RNA polymerase in eubacteria (Allison et al., 1996), our proposed circadian and light-regulated RNA polymerase activity may be mediated through a circadian- and/or light-dependent regulatory mechanism(s) of the PEP.
The PEP comprises α-, β-, β′-, and β“-subunits, which are encoded by rpoA, rpoB, rpoC1, and rpoC2, respectively, in the plastid genome. These subunits form a core enzyme that is structurally and functionally homologous to the bacterial enzyme. In addition to its core enzyme, a bacterial RNA polymerase contains one of the ς factors that are essential for transcription initiation and promoter selectivity (Helmann and Chamberlin, 1988). In higher plants the chloroplast ς factors are encoded by nuclear genomes (Isono et al., 1997; Tanaka et al., 1997; Kestermann et al., 1998; Tozawa et al., 1998). Recently, we also cloned a putative chloroplast ς factor (SigA) from wheat, whose mRNA level is regulated by light (K. Morikawa, Y. Tsunoyama, T. Shiina, and Y. Toyoshima, unpublished data). Northern analysis with a SigA probe revealed that the SigA mRNA exhibited circadian oscillation with phase patterns similar to that of the psbD LRP mRNA level in continuous light (Y. Nakahira, T. Shiina, and Y. Toyoshima, unpublished data). These results led us to propose a mechanism for the circadian and light regulation of the plastid-encoded psbD LRP transcription with which the psbD LRP transcription activity is controlled by the circadian- and light-dependent expression of a nuclear-encoded ς factor. In fact, the rpoD2 encoding a ς70-like transcription factor in cyanobacteria has been shown to be essential for the circadian-regulated transcription of a subset of genes (Tsinoremas et al., 1996).
Alternatively, a unique model that accounts for light-responsive regulation of PEP activity has been described for mustard plastids (Tiller and Link, 1993). With this model low-phosphorylated PEP can effectively initiate transcription in the chloroplast, whereas the highly phosphorylated PEP forms a tightly bound complex among the PEP core enzyme, ς-like factors, and the promoter sequence that arrests transcription in the etioplast. Recently, the light-dependent regulation of phosphorylation state in the nucleus and cytoplasm was shown to be required for the blue-light-responsive transcription of the barely psbD LRP (Christopher et al., 1997), and it was proposed that the light-dependent phosphorylation/dephosphorylation event(s) outside of the plastids may control the phosphorylation state of PEP, which recognizes the psbD LRP through an interorganelle pathway. The same phosphorylation/dephosphorylation event(s) may also be implicated in the circadian regulation of psbD LRP transcription.
ACKNOWLEDGMENTS
We are grateful to Prof. Y. Isozumi (Kyoto University) for allowing us to use the facilities of the Radioisotope Research Center. We also thank Dr. M.V. Lemas for critical reading of the manuscript.
Abbreviations:
- LRP
light-responsive promoter
- PEP
plastid-encoded plastid RNA polymerase
- ZT
Zeitgeiber time
Footnotes
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (no. 10170218 to Y.T.); by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan (no. 09874170 to Y.T. and nos. 10640629, 10309008 to T.S.); and by a grant from the Research for the Future Program, Japan Society for the Promotion of Science (no. JSPS-RFTF96L00604 to T.S.).
LITERATURE CITED
- Adamska I, Scheel B, Kloppstech K. Circadian oscillations of nuclear-encoded chloroplast proteins in pea (Pisum sativum) Plant Mol Biol. 1991;17:1055–1065. doi: 10.1007/BF00037144. [DOI] [PubMed] [Google Scholar]
- Allison LA, Maliga P. Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J. 1995;14:3721–3730. doi: 10.1002/j.1460-2075.1995.tb00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996;15:2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Gatenby A. Mutational analysis of the maize chloroplast ATPase-subunit gene promoter: the isolation of promoter mutants in E. coli and their characterization in a chloroplast in vitro transcription system. EMBO J. 1985;4:3641–3648. doi: 10.1002/j.1460-2075.1985.tb04129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SCG, Wu SP, Lo PK, Mon DP, Chen LFO. Regulation of plastid photosynthetic psbK-I-D-C gene expression by light in rice plants. Physiol Plant. 1995;93:617–623. [Google Scholar]
- Christopher DA, Kim M, Mullet JE. A novel light-regulated promoter is conserved in cereal and dicot chloroplasts. Plant Cell. 1992;4:785–798. doi: 10.1105/tpc.4.7.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Mullet JE. Separate photosensory pathways co-regulate blue light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 1994;104:1119–1129. doi: 10.1104/pp.104.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher DA, Xinli L, Kim M, Mullet JE. Involvement of protein kinase and extraplastidic serine/threonine protein phosphatases in signaling pathways regulating plastid transcription and the psbD blue light-responsive promoter in barley. Plant Physiol. 1997;113:1273–1282. doi: 10.1104/pp.113.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Genetic and molecular analysis of circadian rhythms. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- Edmunds LN. Cellular and Molecular Bases of Biological Clocks. New York: Springer-Verlag; 1988. [Google Scholar]
- Gamble PE, Mullet JE. Blue light regulates the accumulation of two psbD-psbC transcripts in barley chloroplasts. EMBO J. 1989;8:2785–2794. doi: 10.1002/j.1460-2075.1989.tb08424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Hoffman NE, Ko K, Scolnik PA, Cashmore AR. A light-entrained circadian clock controls transcription of several plant genes. EMBO J. 1988;7:3635–3642. doi: 10.1002/j.1460-2075.1988.tb03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W, Zurawski G. Analysis of promoter regions for the spinach chloroplast rbcL, atpB and psbA genes. EMBO J. 1985;4:3375–3383. doi: 10.1002/j.1460-2075.1985.tb04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JC. Tripping along the trial to the molecular mechanisms of biological clocks. Trends Neurosci. 1995;18:230–240. doi: 10.1016/0166-2236(95)93908-g. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Chua NH. Chloroplast promoters. Trends Biochem Sci. 1987;12:67–70. [Google Scholar]
- Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Hennessey TL, Field CB. Evidence of multiple circadian oscillators in bean plants. J Biol Rhythms. 1992;7:105–113. doi: 10.1177/074873049200700202. [DOI] [PubMed] [Google Scholar]
- Hoffer PH, Christopher DA. Structure and blue-light-responsive transcription of a chloroplast psbD promoter from Arabidopsis thaliana. Plant Physiol. 1997;115:213–222. doi: 10.1104/pp.115.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Kawazoe R, Herrin DL. Transcription of tufA and other chloroplast-encoded genes is controlled by a circadian clock in Chlamydomonas. Proc Natl Acad Sci USA. 1996;93:996–1000. doi: 10.1073/pnas.93.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H. Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of sigma70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1997;94:14948–14953. doi: 10.1073/pnas.94.26.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobshagen S, Johnson CH. Circadian rhythms of gene expression in Chlamydomonas reinhardtii: circadian cycling of mRNA abundances of cabII, and possibly of β-tubulin and cytochrome c. Eur J Cell Biol. 1994;64:142–152. [PubMed] [Google Scholar]
- Kawaguchi H, Fukuda I, Shiina T, Toyoshima Y. Dynamic behavior of psb gene transcripts in greening wheat seedlings. I. Time course of accumulation of the psbA through psbN gene transcripts during light-induced greening. Plant Mol Biol. 1992;20:695–704. doi: 10.1007/BF00046454. [DOI] [PubMed] [Google Scholar]
- Kestermann M, Neukirchen S, Kloppstech K, Link G. Sequence and expression characteristics of a nuclear-encoded chloroplast sigma factor from mustard. Nucleic Acids Res. 1998;26:2747–2754. doi: 10.1093/nar/26.11.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE. Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol. 1993;22:447–463. doi: 10.1007/BF00015975. [DOI] [PubMed] [Google Scholar]
- Kim M, Mullet JE. Identification of a sequence-specific DNA binding factor required for transcription of the barley chloroplast blue light-responsive psbD-psbC promoter. Plant Cell. 1995;7:1445–1457. doi: 10.1105/tpc.7.9.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaff P, Gruissem W. Changes in chloroplast mRNA stability during leaf development. Plant Cell. 1991;3:517–529. doi: 10.1105/tpc.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppstech K. Diurnal and circadian rhythmicity in the expression of light-induced plant nuclear messenger RNAs. Planta. 1985;165:502–506. doi: 10.1007/BF00398095. [DOI] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Link G. DNA sequence requirements for the accurate transcription of a protein-coding plastid gene in a plastid in vitro system from mustard (Sinapis alba L.) EMBO J. 1984;3:1697–1704. doi: 10.1002/j.1460-2075.1984.tb02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR, Kay SA. Circadian rhythms in Arabidopsis thaliana. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 615–637. [Google Scholar]
- Millar AJ, Kay SA. Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell. 1991;3:541–550. doi: 10.1105/tpc.3.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet J, Klein R. Transcription and mRNA stability are important determinants of higher plants chloroplast RNA levels. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Kay SA, Chua NH. A circadian clock regulates transcription of the wheat Cab-1 gene. Genes Dev. 1988;2:376–382. [Google Scholar]
- Piechulla B. ‘Circadian clock’ directs the expression of plant genes. Plant Mol Biol. 1993;22:533–542. doi: 10.1007/BF00015982. [DOI] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L. Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas. Regulation by transcription and RNA degradation. Plant J. 1993;3:213–219. doi: 10.1046/j.1365-313x.1993.t01-13-00999.x. [DOI] [PubMed] [Google Scholar]
- Satoh J, Baba K, Nakahira Y, Shiina T, Toyoshima Y. Characterization of dynamics of the psbD light-induced transcription in mature wheat chloroplasts. Plant Mol Biol. 1997;33:267–278. doi: 10.1023/a:1005799001271. [DOI] [PubMed] [Google Scholar]
- Sweeney BM. Rhythmic Phenomena in Plants. San Diego, CA: Academic Press; 1987. [Google Scholar]
- Sweeney BM, Haxo FT. Persistence of a photosynthetic rhythm in enucleated Acetabularia. Science. 1961;134:1361–1363. doi: 10.1126/science.134.3487.1361. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Tozawa Y, Mochizuki N, Shinozaki K, Nagatani A, Wakasa K, Takahashi H. Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 1997;413:309–313. doi: 10.1016/s0014-5793(97)00906-x. [DOI] [PubMed] [Google Scholar]
- Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L.) EMBO J. 1993;12:1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozawa Y, Tanaka K, Takahashi H, Wakasa K. Nuclear encoding of a plastid ς factor in rice and its tissue- and light-dependent expression. Nucleic Acids Res. 1998;26:415–419. doi: 10.1093/nar/26.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsinoremas NF, Ishiura M, Kondo T, Andersson CR, Tanaka K, Takahashi H, Johnson CH, Golden SS. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;15:2488–2495. [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tunoyama Y, Shiina T, Toyoshima Y. In vitro analysis of light-induced transcription in the wheat psbD/C gene cluster using plastid extracts from dark-grown and short- term-illuminated seedlings. Plant Physiol. 1994;104:1259–1267. doi: 10.1104/pp.104.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]