Abstract
The P1B-type ATPases are a ubiquitous family of P-type ATPases involved in the transport of transition metal ions. Divided into subclasses on the basis of sequence characteristics and substrate specificity, these integral membrane transporters play key roles in metal homeostasis, metal tolerance, and the biosynthesis of metalloproteins. The P1B-4-ATPases have the simplest architecture of the five P1B-ATPase families and have been suggested to play a role in Co2+ transport. A P1B-4-ATPase from Sulfitobacter sp. NAS-14.1, designated sCoaT, has been cloned, expressed, and purified. Activity assays indicate that sCoaT is specific for Co2+. A single Co2+ binding site is present, and optical, electron paramagnetic resonance (EPR), and X-ray absorption (XAS) spectroscopic data are consistent with tetrahedral coordination by oxygen and nitrogen ligands, including a histidine and likely a water. Surprisingly, there is no evidence for coordination by sulfur. Mutation of a conserved cysteine residue, Cys 327, in the signature transmembrane SPC metal binding motif does not abolish ATP hydrolysis activity or affect the spectroscopic analysis, establishing that this residue is not involved in the initial Co2+ binding by sCoaT. In contrast, replacements of conserved transmembrane residues Ser 325, His 657, Glu 658, and Thr 661 with alanine abolish ATP hydrolysis activity and Co2+ binding, indicating that these residues are necessary for Co2+ transport. These data represent the first in vitro characterization of a P1B-4-ATPase and its Co2+ binding site.
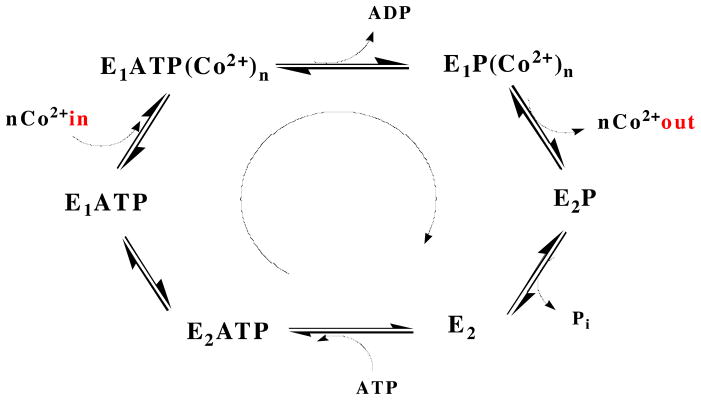
The P-type ATPases, found in a variety of species from bacteria to humans, are a family of integral transmembrane proteins that transport charged substrates across cell membranes using the energy provided by ATP hydrolysis (1, 2). P-type ATPases are divided into subclasses on the basis of substrate specificity. These subclasses include the P1A-ATPases (K+), the P1B-ATPases (heavy metals), the P2-ATPases (Ca2+, Na+/K+, H+/K+), the P3-ATPases (H+), the P4-ATPases (phospholipids), and the P5-ATPases (unknown substrate) (3–5). All P-type ATPases share a common architecture consisting of multiple transmembrane (TM) helices, soluble nucleotide binding and phosphorylation domains (N- and P-domains, referred to collectively as the ATP binding domain or ATPBD), and a soluble actuator domain (A-domain). All P-type ATPAses are believed to follow a Post-Albers mechanism in which catalytic phosphorylation of a conserved aspartic acid residue within an invariant DKTGT sequence in the P-domain causes conformational switching between an E1 high affinity cation binding state and an E2 lower affinity state (6–9) (Figure 1). Crystal structures are available for multiple conformational states of the sarcoplasmic reticulum Ca2+-ATPase (SERCA) (10), and some structures have also been determined of the Na+/K+-ATPase (11, 12), the H+-ATPase (13), and the Cu+-ATPase (14).
Figure 1.
The Post-Albers catalytic cycle, represented for a Co2+ ATPase. The cycle is defined by two conformational states (E1 and E2) that are interconverted by the covalent attachment and release of phosphate to and from a conserved aspartic acid residue. In the E1 state, which has high affinity for the substrate being transported, the binding site is accessible to the cytoplasm. Upon hydrolysis of ATP, a conformational change in the TM helices converts the binding site to being accessible to the periplasm (or other cellular compartment) and lowers the substrate affinity of E2. After the substrate is released, dephosphorylation occurs and the protein reverts to the E1 state.
The P1B-type ATPases transport transition metal ions, including Zn2+/Cd2+/Pb2+ (15–17), Cu2+ (18), Cu+/Ag+ (19, 20), and Co2+ (21). Members of the P1B subgroup are widely distributed across all domains of life, conferring heavy metal tolerance and playing an essential role in the distribution of metal micronutrients and the biosynthesis of metalloproteins (20, 22). In humans, mutations in the Cu+-ATPases ATP7A and ATP7B lead to Menkes syndrome and Wilson disease, respectively (23). The core architectural features of the P1B-type ATPases include at least six TM helices and two large cytoplasmic loops housing the ATPBD (containing the N- and P-domains) and the A-domain (Figure 2). The C-terminal TM helices 4–6 contain residues that are proposed to form the metal binding site(s), and the identities and positions of these residues are believed to determine metal specificity (24). Mutagenesis data indicate that a three-residue cysteine-containing “signature” sequence motif in TM4 (CPC, CPH, SPC, PCP) is of particular importance for metal transport activity (25–29). On the basis of these signature sequence motifs and some experimental data, the P1B-ATPases have been grouped into five substrate-specific subfamilies designated P1B-1 through P1B-5 (24, 30). The P1B-1, P1B-2, and P1B-3 subfamilies contain two additional TM helices at the N-terminus. The subfamilies are further differentiated according to the presence or absence of N- and/or C-terminal soluble metal binding domains (MBDs), which likely regulate ATPase function (31–34).
Figure 2.
Architecture and key residues of P1B-4-ATPases. Left: Overall topology with conserved residues in TM helices shown in approximate locations. Right: Noncontinuous alignment of TM helices 4 and 6 of sCoaT and LpCopA. The phosphorylation site in the P-domain is highlighted in light purple. Site I proposed metal-binding residues of LpCopA are colored in orange, with the corresponding proposed sCoaT metal-binding residues colored in blue. Site II residues of LpCopA are colored in purple along with conserved sCoaT residues, and nonconserved sCoaT residues are colored in red.
The P1B-1-ATPases are specific for Cu+/Ag+, the P1B-2-ATPases are Zn2+/Cd2+/Pb2+ transporters, and the P1B-3 ATPases are specific for Cu2+. Representative members of these three families have been purified and characterized biochemically (17–19, 25, 35, 36). In addition, a crystal structure of the Legionella pneumophila CopA P1B-1-ATPase (LpCopA) has been determined recently (14). Less is known about the P1B-4- and P1B-5-ATPases. While the P1B-4-ATPases have been suggested to play a role in Co2+ transport (21), their substrate specificity has not been established. The substrate for the P1B-5-ATPases is unknown (37). The P1B-4-ATPases have the simplest architecture, with only six TM helices and no N- or C-terminal MBDs. A conserved SPC motif is located in TM4 (Figure 2). Genetic data obtained more than 10 years ago for the Synechocystis PCC6803 P1B-4-ATPase CoaT indicate that it is linked to Co2+ tolerance (21). In Cupriavidus metallidurans CH34, the P1B-4-ATPase CzcP is proposed to play a role in resistance to Zn2+, Cd2+, and Co2+ (38). The plant P1B-4-ATPase HMA1 transports Ca2+ and expression of HMA1 in yeast confers tolerance to Zn2+, Cd2+, Co2+ and Cu+ (39, 40). Thus, the metal substrate of the P1B-4-ATPases has not yet been clearly defined, and no biochemical characterization of a purified P1B-4-ATPase has been reported. To further understand this subfamily of P1B-ATPases, we have expressed, purified, and characterized a P1B-4-ATPase from Sulfitobacter sp. NAS-14.1, designated sCoaT. Isolated sCoaT binds Co2+ and exhibits Co2+-induced ATPase activity. The Co2+ binding site has been probed by spectroscopy and the roles of conserved residues have been assessed by mutagenesis. These data provide new insight into the function and properties of the P1B-4-ATPase subfamily.
MATERIALS AND METHODS
Homology Modeling of sCoaT Based on LpCopA
Homology modeling of sCoaT and LpCopA was performed using DeepView (http://www.expasy.org/spdbv/) (41). Chain B of the LpCopA structure (PDB accession code 3RFU) was used as the reference sequence onto which the sequence of sCoaT was mapped. Structural alignment was performed on the last four TM helices of both sCoaT and LpCopA (starting from TM helix 3 of sCoaT and TM helix 5 of LpCopA) to focus on the structural region that houses residues believed to determine metal ion specificity (24). The starting position of TM helices was determined using TMHMM 2.0 (42). In the resultant homology model, residues 227–682 of sCoaT are aligned with residues 291–733 of LpCopA.
Cloning and Expression of Wildtype sCoaT (WT-sCoaT)
The gene sequence encoding the P1B-4-ATPase sCoaT was PCR amplified with PfuTurbo DNA polymerase (Stratagene) using Sulfitobacter sp. NAS-14.1 genomic DNA (ATCC) as a template and the following oligonucleotide primers (Integrated DNA Technologies): forward, 5′-CAC-CGT-GCG-GAA-AGT-CGT-TGC-CGA-T-3′; reverse, 5′-CTA-TGC-CGA-TTT-GAC-GCG-ATT-GTC-CTT-GA-3′. The primers were designed on the basis of PubMed entry GI:83956091 with the introduction of an overhang (underlined) for TOPO cloning. The amplified gene (2.04 kbp) was subcloned into the pET151/D-TOPO vector (Invitrogen), which introduces an N-terminal hexahistidine tag with a TEV cleavage site. This vector was transformed into E. coli Top10 chemically competent cells (Invitrogen) following the manufacturer’s protocol and spread on a Luria-Bertani (LB) agar plate containing 100μg/mL ampicillin. DNA sequencing (ACGT Inc.) of individual colonies confirmed the gene sequence.
For expression, E. coli BL21 Star (DE3) chemically competent cells were transformed with the plasmid encoding sCoaT. Overnight cultures were used to inoculate baffled flasks containing 1 L LB media supplemented with 100 mg ampicillin which were then grown at 37 °C with shaking. Protein expression was induced by addition of 1 mM IPTG at an OD600 of 0.6, at which point the growth temperature was lowered to 18 °C. The cells were harvested after 16–18 h by centrifugation at 4800g for 10 min in a Sorvall SLC 4000 rotor at 4 °C, resuspended in 50 mM Tris, pH 7.5, 200 mM NaCl, 500 mM sucrose, and 1 mM PMSF, and stored at −80 °C.
Purification of WT-sCoaT
All purification procedures were carried out at 4 °C. Cells were lysed by sonication for 12 min using 15 s pulses with a 30 s rest period between each pulse. Lysed cells were centrifuged at 8000g for 30 min to remove cell debris. The supernatant was then ultracentrifuged at 163000g for 1 hr. Pelleted membranes were washed once with fresh buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 500 mM sucrose, 1mM PMSF) and homogenized, then pelleted again by ultracentrifugation at 163000g for 45 min. The pelleted, washed membranes were resuspended and homogenized in 25 mM Tris, pH 7.0, 100 mM sucrose, 500mM NaCl, 1 mM PMSF and diluted to 3 mg/mL, with the concentration determined by the detergent compatible Lowry assay (Bio-Rad). For solubilization, n-dodecyl-β-D-maltopyranoside (DDM) was added dropwise to a final concentration of 1% and allowed to incubate for 1 h at 4 °C with gentle stirring. The suspension was cleared by centrifugation at 163000g for 45 min. The supernatant was then applied to a 10 mL Ni2+-loaded HiTrap Chelating HP column (GE Healthcare) and washed with four column volumes of 25 mM Tris, pH 7.5, 100 mM NaCl, 20% glycerol, 0.02% DDM (Buffer A) followed by a wash of six column volumes of Buffer A with 50 mM imidazole. The pure sCoaT protein was eluted with Buffer A containing 300 mM imidazole. Fractions were concentrated in an Amicon 15 mL spin concentrator with a 50 kDa cutoff filter (Millipore), and imidazole was removed by desalting on a HiPrep 26/10 Desalting column (GE Healthcare) equilibrated with Buffer A. The protein concentration was determined by the Pierce 660 nm protein assay, and protein expression and purification were assessed by 15% SDS-PAGE.
To remove the N-terminal hexahistidine tag, purified sCoaT was treated with TEV protease. Hexahistidyl-tagged TEV protease was added to purified sCoaT at a 1:2 molar ratio along with 1 mM DTT and incubated at 4 °C with gentle rocking for 16–18 h. The cleavage reaction mixture was then loaded onto a 10 mL Ni2+-loaded HiTrap Chelating HP column (GE Healthcare) and washed with five column volumes of Buffer A, during which cleaved sCoaT was eluted. Uncleaved sCoaT and hexahistydyl-tagged TEV protease remained bound to the column. Removal of the hexahistidine tag from sCoaT was confirmed by Western blotting followed by immunostaining with anti-hexahistidine antibody (Sigma Aldrich) and by N-terminal sequence analysis (University of Illinois Champaign-Urbana Protein Sciences Facility).
Cloning, Expression, and Purification of sCoaT Mutants
Mutations S325A, C327A, H657A, E658A, and T661A were introduced using the QuikChange II Site-Directed Mutagenesis kit (Stratagene). Plasmid DNA of pET151/WT-sCoaT was used as template DNA for mutagenesis. The entire sequences of the mutants were verified by automated DNA sequencing (ACGT Inc.). The pET151/mutant plasmids were transformed into E. coli BL21 Star (DE3) chemically competent cells, and growth and purification were conducted using the same procedures as for WT-sCoaT.
ATPase Activity Assay
The metal-dependent ATPase activity of purified sCoaT was measured by the pyruvate kinase-lactate dehydrogenase coupled spectroscopic assay (17, 43). In this assay, regeneration of ATP is coupled to the oxidation of NADH, and enzyme activity is determined by monitoring the decrease of the NADH absorption peak at 340 nm. For activity measurements, sCoaT was exchanged into a buffer containing 25 mM Tris, pH 7.5, 50 mM KCl, 10% glycerol, and 0.02% DDM. The ATPase activity assay mixture (1 mL) contained 50 mM HEPES-KOH, pH 7.5, 100 mM KCl, 0.1% asolectin, 0.1% DDM, 5 mM MgSO4, 0.25 mM NADH, 1.25 mM phosphoenolpyruvate, 9 units of pyruvate kinase, 9 units of lactate dehydrogenase, and 10 μg sCoaT, with or without Co2+, Pb2+, Zn2+, Cd2+, Cu2+, Cu+ (generated from Cu2+ in the presence of excess dithiothreitol) and Ni2+. Insolubility of Ag+ under the assay conditions precluded testing Ag+-dependent activity. Assay mixtures were activated by the addition of 5 mM ATP and monitored for 3 min at 25 °C using a Perkin Elmer LAMBDA 1050 UV-Vis spectrophotometer (Keck Biophysics Facility, Northwestern University) at 340 nm.
Metal Loading and Analysis
Purified sCoaT was first exchanged into a buffer containing 25 mM Tris, pH 7.5, 50 mM NaCl, 10% glycerol, and 0.02% DDM using repeated dilution and concentration in an Amicon spin concentrator. Cobalt loading was performed by addition of 5 molar equivalents of CoCl2 to the sCoaT solution followed by overnight (16–18 h) incubation at 4 °C with gentle rocking. Excess cobalt was removed using an EconoPac 10DG desalting column (Bio-Rad). Metal content was measured by inductively coupled plasma optical emission spectrometry (ICP-OES) using a Varian Vista MPX ICP-OES (Integrated Molecular Structure Education and Research Center, Northwestern University). All ICP-OES samples were prepared by digestion of protein in 5 mL of 5% TraceSelect nitric acid (Sigma Aldrich) in Chelexed water followed by filtration through a 22 μm sterile filter. Cobalt standards for calibration (Sigma Aldrich) were also prepared in 5% nitric acid. Reported ICP-OES results are the average of three replicate experiments performed for at least two independent protein sample preparations. Optical spectra were measured using a Perkin Elmer LAMBDA 1050 UV-Vis instrument.
Electron Paramagnetic Resonance (EPR) Spectroscopy
Samples for EPR spectroscopy were concentrated to 0.6–1mM Co2+ in 25 mM Tris, pH 7.5, 50 mM NaCl, 0.02% DDM, 50% glycerol. Deuterated samples were prepared in the same buffer using D2O and d8-glycerol (Sigma Aldrich). Samples were transferred to custom Q-band EPR tubes, frozen, and stored in liquid nitrogen. Q-band continuous wave (CW) EPR and Electron Nuclear Double Resonance (ENDOR) spectra were collected on custom-built instruments each equipped with a liquid helium immersion dewar at 2 K using 100 kHz field modulation and dispersion mode detection under rapid passage conditions (44). These spectrometers have been described previously (45). Q-band pulsed ENDOR spectra were collected at 2 K on custom-built instruments previously described (45–47) and employed the Davies (π – T – π/2 – τ – π – τ – echo) microwave pulse sequence, in which the RF pulse is applied during time period T (48). All pulse experiment data acquisition was performed with the SpecMan software package (49) (http://specman4epr.com) in conjunction with a Spin-Core PulseBlaster ESR_PRO 400 MHz word generator and Agilent Technologies Acquiris DP235 500MS/sec digitizer.
X-ray Absorption (XAS) Spectroscopy
Samples for XAS were prepared in 25 mM Tris, pH 7.5, 50 mM NaCl, 0.02% DDM and 50% glycerol with cobalt concentrations in the 1–1.5 mM range. Two independent replicates of cobalt-loaded, WT- and C327A-sCoaT were prepared. Samples were loaded into Lucite cells wrapped with Kapton tape, frozen in liquid nitrogen and stored at −80 °C until data collection. XAS data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) on beamline 7-3 and at the National Synchrotron Light Source (NSLS) on beamline X3-b. The SSRL beamline was equipped with a Si[220] double crystal monochromator and the NSLS beamline was equipped with a Si[111] monochromator; both beamlines were equipped with a harmonic rejection mirrors. During data collection, samples were maintained at 10 K using an Oxford Instruments continuous-flow liquid helium cryostat at SSRL and at 24 K using a He Displex Cryostat at NSLS. Protein fluorescence excitation spectra were collected using 30-element Ge solid-state detectors at both beamlines. At SSRL, iron filters, 0.3 μm in width, were placed between the cryostat and detector to filter background fluorescence scattering not associated with Co signals. XAS spectra at both facilities were measured in 5 eV increments in the pre-edge regions (7542–7702 eV), 0.25 eV increments in the edge regions (7702–7780 eV), and 0.05 Å−1 increments in the extended X-ray absorption fine structure (EXAFS) region (to k = 13.0 Å−1), integrating from 1s to 25s in a k3 weighted manner for a total scan length of approximately 40 min. X-ray energies were individually calibrated by collecting Co foil absorption spectra simultaneously with protein data. The first inflection point of the Co foil spectrum was assigned to 7709.5 eV. Each fluorescence channel of each scan was examined for spectral anomalies prior to averaging and spectra were closely monitored for photoreduction. SSRL data represent the average of 6 to 7 scans, and NSLS data represent the average of 9 to 10 scans.
XAS data were processed using the Macintosh OS X version of the EXAFSPAK program suite (50) integrated with the Feff v8 software (51) for theoretical model generation. Data reduction utilized a Gaussian function in the pre-edge region and a three-region cubic spline throughout the EXAFS region. Data were converted to k-space using a cobalt E0 value of 7745 eV. The k cubed weighted EXAFS was truncated at 1.0 and 12.5 Å−1 for filtering purposes. This k range corresponds to a spectral resolution of ca. 0.14 Å for all cobalt-ligand interactions; therefore only independent scattering environments outside 0.14 Å were considered resolvable in the EXAFS fitting analysis (52). EXAFS fitting analysis was performed on raw/unfiltered data. EXAFS data were fit using both single and multiple scattering amplitude and phase functions calculated with the program Feff v8. Single scattering theoretical models were calculated for carbon, oxygen, sulfur and cobalt coordination to simulate copper-nearest neighbor ligand environments. A multiple scattering Co-imidazole theoretical model was calculated to simulate the numerous scattering interactions observed from the linear ring. A scale factor (Sc) of 0.98% and an E0 value of −3 eV for a Co-O/N bond, utilized during the protein simulations, were determined by fitting a crystallographically characterized hexaquacobalt (II) nitrate (53). Criteria for judging the best-fit simulation utilized both the lowest mean square deviation between data and fit (F′), corrected for the number of degrees of freedom (54), and a reasonable Debye-Waller factor (σ2 < 0.006 Å2).
RESULTS AND DISCUSSION
Homology Modeling
On the basis of sequence alignments, seven conserved residues were proposed to participate in metal binding by P1B-4-ATPases: serine, proline, and cysteine in TM4, and histidine, glutamine, glycine, and threonine in TM6 (24) (Figure 2). Additional insight into specific ligands can be obtained by comparison of sCoaT to LpCopA, the only structurally characterized P1B-ATPase to date (14). Although there are no bound metal ions in the structure, LpCopA is predicted to have two metal binding sites, one composed of Asn 689, Met 717, and Ser 721 (site I), and the second involving the transmembrane cysteines Cys 382 and Cys 384 as well as Tyr 688 (site II). Homology modeling reveals that four of the putative LpCopA metal binding residues align with corresponding conserved residues in sCoaT (Figure 2, right). The residues in the SPC motif, Ser 325 and Cys 327, align with LpCopA Cys 382 and Cys 384, and residues His 657 and Thr 661 align with LpCopA proposed ligands Met 717 and Ser 721. The other two LpCopA binding residues, Asn 689 and Tyr 688, align with nonconserved valine residues. Conserved sCoaT residue Glu 658 corresponds to an alanine in LpCopA, but would be close enough to His 657 and Thr 661 to participate in cobalt binding.
Expression and Purification of WT-sCoaT and Variants
Recombinant expression of WT-sCoaT yielded approximately 2 mg of purified protein/L of cultured media. Overnight induction of cells at 18 °C increased the yield of purified WT-sCoaT compared to a 3 h induction period at 37 °C. DDM-solubilized WT-sCoaT was purified in a single step by Ni2+-affinity chromatography (Figure 3), and cleavage of the hexahistidine tag using TEV protease yielded ~70% recovery of untagged WT-sCoaT. The removal of the hexahistidine tag was verified by Western blot using an anti-hexahistidine antibody (Figure 3) and by N-terminal sequence analysis, which indicated complete cleavage. Variant proteins in which Ser 325, Cys 327, His 657, Glu 658 and Thr 661 were mutated to alanine were also prepared to assess the role of these conserved residues in metal binding and activity. These mutants did not express as well as WT-sCoaT, yielding approximately 0.3–0.5 mg of pure protein/L, but utilization of the same WT-sCoaT purification protocol resulted in >90% pure proteins (Figure S1). The low yields precluded TEV protease cleavage of the sCoaT variants, however.
Figure 3.
SDS-PAGE and Western analysis of purified WT-sCoaT. Lane 1, SDS-PAGE of purified tagged WT-sCoaT; lane 2, SDS-PAGE of purified TEV-cleaved WT-sCoaT; lane 3, anti-6xHis tag Western blot of purified tagged WT-sCoaT; lane 4, anti-6xHis tag Western blot of purified TEV-cleaved WT-sCoaT.
ATPase Activity
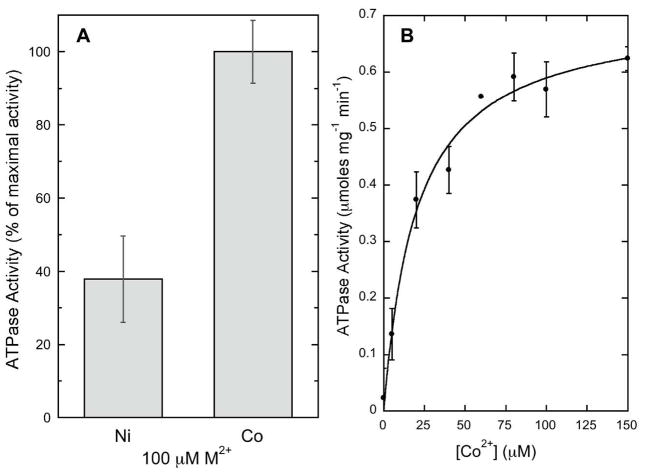
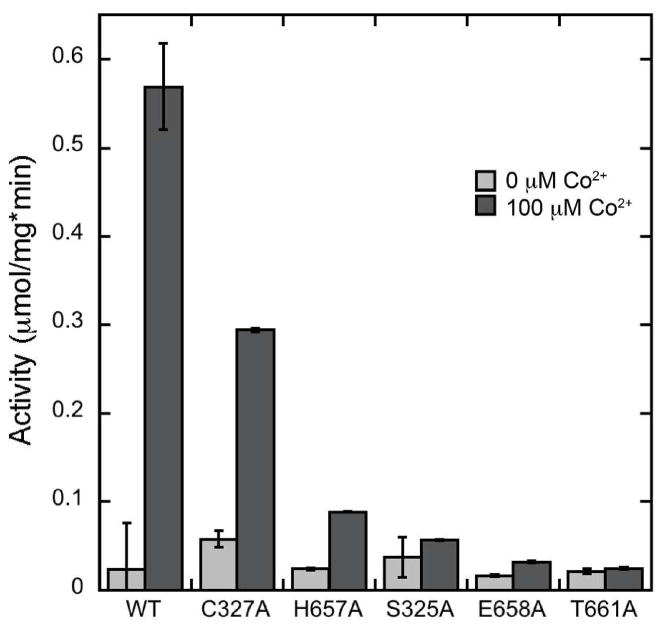
The ATPase activity of WT-sCoaT was specifically stimulated by Co2+, and was also stimulated by Ni2+, but to a much lesser extent than Co2+ (Figure 4A). None of the other metals tested (Zn2+, Cd2+, Cu2+, Cu+, Pb2+) were able to activate the enzyme. The Co2+-dependent activity of WT-sCoaT has a KM value of 20.05 ± 3.6 μM and a Vmax value of 0.71 ± 0.03 μmol mg−1min−1 (Figure 4B). The Vmax value is similar to those reported for the Pb2+-stimulated activity of ZntA (17) and the Ag+-stimulated activity of CopA (19). The activity of the variants was negligible, with the notable exception of the C327A variant (C327A-sCoaT), which has approximately 50% of the WT-sCoaT activity (Figure 5). This result is unexpected and differs from previous studies of P1B-1-, P1B-2- and P1B-3-ATPases. For A. fulgidus CopA, mutation of either or both cysteines in the CPC motif to alanine abolishes ATPase activity (26), and in the case of E. coli CopA, mutation of these cysteines results in loss of copper resistance, transport, and phosphoenzyme formation (25). Replacement of either cysteine with alanine in the E. coli ZntA CPC motif also results in inactive enzyme, but ZntA variants in which the cysteines are replaced with histidine retain some activity (27). For the E. hirae CopB Cu2+-ATPase, alteration of the CPH transmembrane motif to SPH abolished the ability to restore copper resistance in a CopB knockout mutant in vivo, suggesting impaired Cu2+ transport (55).
Figure 4.
Activity of WT-sCoaT. (A) Co2+ stimulates the highest ATP hydrolysis activity. Results are the average of at least three independent replicates. Maximal activity for 100 μM Co2+ is 0.57 μmoles ATP mg−1 min−1. (B) Co2+ stimulated ATPase activity of WT-sCoaT. Fits were performed using Kaleidagraph software with the equation y = (Vmax*x)/(KM+x).
Figure 5.
Comparison of ATP hydrolysis activity of WT-sCoaT, C327A-sCoaT, and variants in which His 657, Ser 325, Glu 658, and Thr 661 are replaced with alanine.
Metal-Binding Stoichiometry
Tagged WT-sCoaT binds 1.55 ± 0.07 Co2+ ions per protein monomer, and cleaved WT-sCoaT binds 1.38 ± 0.21 Co2+ ions per protein monomer, indicating that the hexahistidyl tag does not bind a significant amount of Co2+ during the loading procedure. WT-sCoaT is thus proposed to contain one metal binding site for Co2+. ZntA from E. coli also is reported to contain a single metal binding side (35) whereas A. fulgidus CopA binds two Cu+ ions with high affinity (56). Interestingly, C327A-sCoaT also appears to have a single Co2+ binding site, with a stoichiometry of 1.49 ± 0.53 Co2+ ions per monomer. This result suggests that the cysteine in the TM metal binding SPC motif is not essential for Co2+ binding and is consistent with the observation of ATPase activity for C327A-sCoaT (Figure 5). None of the other mutants were able to bind Co2+, further indication that the hexahistidyl tag does not bind Co2+. This finding is consistent with the lack of ATPase activity of these variants and indicates that Ser 325, His 657A, Glu 658 and Thr 661 are essential for Co2+ binding and activity, and may be ligands as predicted by the homology modeling based on the LpCopA structure.
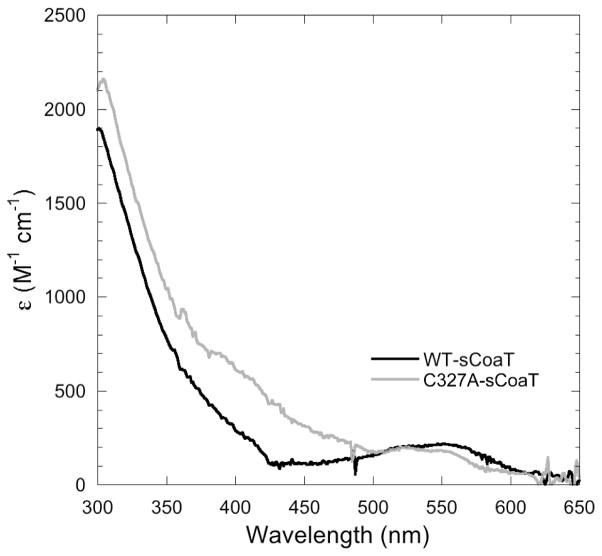
The optical spectra of Co2+-loaded WT-sCoaT and C327A-sCoaT both exhibit a broad shoulder extending from the 280 nm protein absorbance to ~400 nm (Figure 6). Although this region of the spectrum is typically associated with Co2+-thiolate ligand-to-metal charge transfer (LMCT) bands, the lack of distinct peaks observed for these intense transitions (57–59) and the similarity between the spectra of WT-sCoaT and the cysteine-lacking C327A-sCoaT suggest that this absorbance does not arise from interactions with sulfur ligands. This shoulder is more likely attributable to histidine-to-Co2+ LMCT, and resembles features observed in the spectrum of Co2+-substituted copper,zinc superoxide dismutase (60). The geometry of Co2+ sites can be correlated with the position and intensity of d-d transitions in the visible region (59, 61). The broad features in the 500–600 nm region of the sCoaT spectra (ε ~ 300 M−1 cm−1) are most consistent with regular tetrahedral geometry (61, 62). The shift of the peak in C237A-sCoaT towards ~525 nm may indicate some distortion of the Co2+ binding site, perhaps due to perturbation of the secondary coordination environment.
Figure 6.
Difference spectra of the Co2+-bound forms of WT- and C327A-sCoaT, using the respective apo forms as reference spectra.
EPR and ENDOR
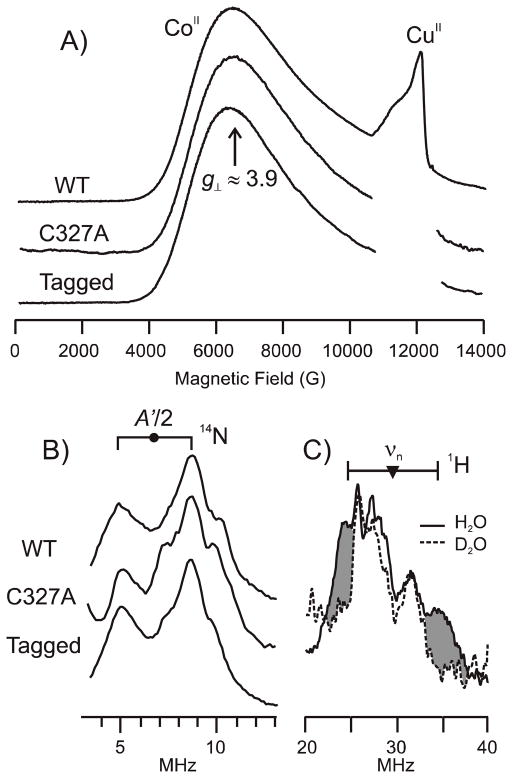
WT-, tagged WT-, and C327A-sCoaT all exhibit an S = 3/2 Co2+ EPR signal with a broad feature at g⊥ = 3.9 and g|| ~ 2, consistent with tetrahedral symmetry (63). Tagged and untagged WT-sCoaT show identical EPR signals, indicating that Co2+ does not bind to the hexahistidyl tag (Figure 7a). The presence of the same feature in the C327A-sCoaT spectrum indicates that the Co2+ coordination sphere is not altered by deletion of the conserved cysteine residue.
Figure 7.
35 GHz EPR and ENDOR spectra. (A) Absorption-display CW Q-band EPR spectra of tagged WT-sCoaT, C327A-sCoaT and WT-sCoaT at 2K. Each sample exhibits a S = 3/2 Co2+ signal as characterized from the broad feature at g⊥ = 3.9. Adventitious Cu2+ in variable amounts is observed at higher fields, but no copper is detectable in the samples by ICP-OES. (B) 14N Davies ENDOR spectra taken near g⊥. Goalpost centered at A′/2, split by 2 × ν(14N). (C) 1H CW ENDOR spectra of WT-CoaT in 1H2O (solid line) and 2H2O buffer (dashed line) taken near g⊥. Shading highlights intensity from exchangeable proton. Goalpost centered at ν(1H), split by A′. CW EPR and ENDOR conditions: microwave frequency, 34.9 GHz; modulation amplitude, 2 G; time constant, 32 ms; EPR scan time, 4 min; ENDOR scan rate, 2 MHz/sec. Pulse ENDOR conditions: microwave frequency, 34.6 GHz; π, 80 ns; τ, 600 ns; TRF, 30 μs; repetition rate, 20 ms.
ENDOR spectra collected at g⊥ = 3.9 from WT-, C327A- and tagged WT-sCoaT (Figure 7b), reveal identical signals from a single, strongly-coupled 14N. The signal is a doublet of broad peaks that is centered at half the hyperfine coupling of A′ = 13.6 MHz (45), with a separation of twice the 14N nuclear Larmor frequency. This coupling treats the Co2+ as having an effective spin, S′ = 1/2; in an S = 3/2 representation the hyperfine coupling is A = 21.0 MHz, comparable to histidine bound to Cu2+, S = 1/2. The observed hyperfine coupling A′, taken at a given g-value is modified from the intrinsic A value. A′ = gA/ge. The higher-frequency (ν+) partner shows poorly resolved structure that indicates the Co2+ site is structurally heterogeneous, with several contributing conformers; an additional contribution from the nuclear quadrupole interaction in at least one conformer may also be present. An attempt to measure this interaction with ESEEM spectroscopy failed because rapid spin dephasing quenched the spin-echo modulation. The strongly-coupled nitrogen can be assigned to a bound histidine residue, based on the near identity of its signal to that of a histidine bound to Co2+ of the Zn-finger in transcription factor IIIA (TFIIIA) from Xenopus laevis (64). This residue is very likely to be His 657, which is found in transmembrane helix 6 of sCoaT (Figure 2), is conserved in the P1B-4 subgroup, and has been proposed to be a metal binding ligand (24). The equivalence of the 14N signals from untagged and tagged WT-sCoaT shows that there is no coordination to Co2+ by a residue from the hexahistidine tag. The equivalence of the 14N signals from WT-sCoaT and C327A-sCoaT, like the invariance of the EPR signal, indicates that the C327A mutation does not alter the Co2+ coordination sphere. Lastly, 1H ENDOR spectra taken near g⊥ exhibit a 1H doublet, hyperfine coupling A′ = 13 MHz, that is lost when the sample is prepared with 2H2O buffer. As this coupling is of the magnitude expected from the proton(s) of an HxO coordinated along the g|| direction, these observations indicate that Co2+ binds a single aqua species.
XAS Spectroscopy
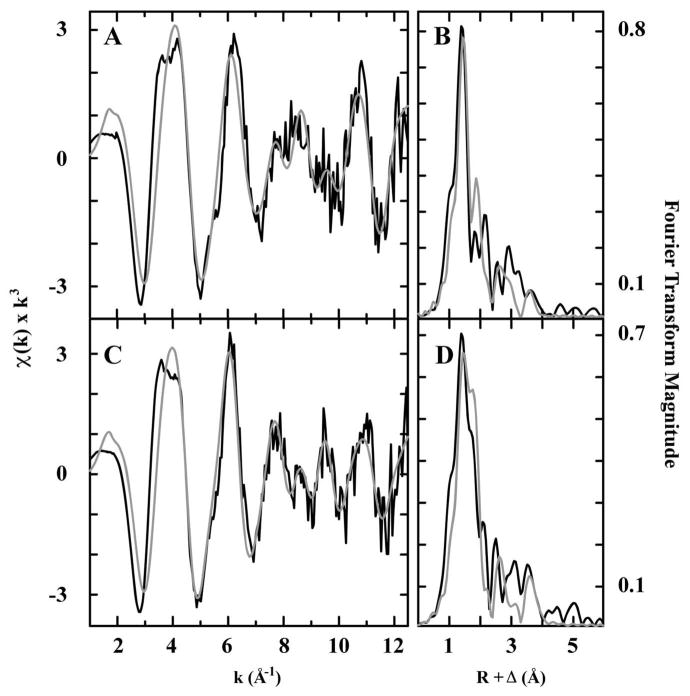
The XANES spectra of WT- and C327A-sCoaT resemble that of Co(NO3)2•6H2O (Figure 8), indicating cobalt is stable in the divalent redox state and most likely dominated by coordination of oxygen/nitrogen based ligands. The Fourier transforms of the EXAFS spectrum for Co2+ bound to WT- and to C327A-sCoaT suggest complex patterns of Co-nearest neighbor ligand environments at R < 3 Å, with additional scattering patterns of ligands at R > 3 Å (Figure 9). Best-fit simulations of the WT- and C327A-sCoaT Co EXAFS indicate a nearest neighbor environment constructed completely of oxygen and/or nitrogen based ligands. Nearest neighbor metrical parameters for include a short Co-O/N distance at 1.95 and 1.96 Å, and a longer distance set of Co-O/N ligands at 2.14 and 2.12 Å for WT- and C327A-sCoaT, respectively. The shorter ~1.95 Å distances are highly reminiscent of the values observed for four-coordinate Co-O small molecule compounds in the Cambridge structural database, whereas the longer ~2.13 Å distance would be consistent with Co-imidazole nitrogen distances (65). Long-range carbon scattering environments at ca. 3.1, 3.5, and 4.1 Å are also consistent with imidazole multiple scattering interactions expected for histidines coordinated to Co (66) (Table 1). There is no evidence for a Cu-S interaction in the EXAFS, consistent with the activity assays, metal binding studies, UV-Vis, and EPR data, indicating Cys 327 of the SPC motif is not involved in Co2+ binding. Moreover, the presence of only oxygen/nitrogen ligands is compatible with the predicted metal binding residues that are conserved in the P1B-4 subclass, including the serine in TM4, and the histidine, glutamate, and threonine residues in TM6 (Figure 2). The observed imidazole multiple scattering interactions support the EPR data and are consistent with His 657 in TM4 being a ligand.
Figure 8.
Normalized XANES spectra of Co bound WT- (solid) and C327A-sCoaT (dashed line), displayed with the spectrum of solid Co(NO3)2 (gray triangles).
Figure 9.
EXAFS and Fourier transform for WT- and C327A-sCoaT. Raw unfiltered EXAFS data (black) and simulations (gray) for cobalt bound to WT- (A) and C327A-sCoaT (C). Fourier transforms of the raw EXAFS (black) and best fit simulations (gray) for cobalt bound to WT- (B) and C327A-sCoaT (D).
Table 1.
Summary of best fit EXAFS simulations for cobalt bound to WT- and C327A-sCoaT. Data fit over a k range of 1 to 12.5 Å−1.
| Sample | Nearest Neighbor Ligand Environmenta | Long Range Ligand Environmentb | F′g | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Atomc | R (Å)d | CNe | σ2f | Atomc | R (Å)d | CNe | σ2f | ||
| WT | O/N | 1.95 | 2 | 3.62 | C | 3.04 | 1 | 5.21 | 0.49 |
| O/N | 2.14 | 2 | 3.42 | C | 3.33 | 1 | 3.62 | ||
| C | 4.09 | 1 | 3.31 | ||||||
| C327A | O/N | 1.96 | 1 | 1.97 | C | 3.03 | 1 | 4.85 | 0.49 |
| O/N | 2.12 | 3 | 5.69 | C | 3.39 | 0.5 | 3.15 | ||
| C | 4.08 | 2 | 5.06 | ||||||
Independent metal-ligand scattering environment at R < 3.0 Å
Independent metal-ligand scattering environment at R > 3.0 Å
Scattering atoms: O (oxygen), N (nitrogen), C (carbon)
Average metal-ligand bond length for multiple independent samples
Average metal-ligand coordination number for multiple independent samples
Average Debye-Waller factor in Å2 × 103 for multiple independent samples
Number of degrees of freedom weighted mean square deviation between data and fit
Implications for the sCoaT metal binding site
The combined data on WT-sCoaT and the sCoaT variants indicate the presence of a single Co2+ binding site that is coordinated by nitrogen and oxygen ligands, including a histidine and a water molecule, in a tetrahedral geometry. Surprisingly, the data indicate that the cysteine residue in the conserved SPC transmembrane metal binding motif is not directly involved in Co2+ binding in this state of the enzyme. In several P1B-ATPases that contain cysteines in this motif (vide supra), such as E. coli CopA, A. fulgidus CopA, E. hirae CopB, and E. coli ZntA, the transmembrane cysteines appear to play essential and equivalent roles in the catalytic cycle (25–27, 55). However, in other P1B-ATPases, the cysteine residues in this motif are believed to be nonequivalent and function at different stages of the transport mechanism. For example, mutation of both cysteines in S. cerevisiae Ccc2 yields no in vivo or ATPase activity, but phosphorylation by ATP is still possible for a CPC to SPC variant, although not for a CPC to CPS variant. These findings indicate that the second cysteine is necessary for metal binding at the transport site whereas the first cysteine plays a role in metal dissociation and/or enzyme dephosphorylation (67). Similarly, mutation of either cysteine in the CPC motif of CadA, a Zn2+/Cd2+/Pb2+-ATPase from Listeria monocytogenes, inhibits in vivo Cd2+ transport, but only the CPC to CPA variant retains ATP hydrolysis activity. In addition, the CPA variant can be phosphorylated by ATP, suggesting that the first cysteine is more directly involved in Cd2+ binding and the second cysteine may participate in occlusion and release (29). The data for C327A-sCoaT are consistent with this type of scenario in which Cys 327 does not play a role in initial Co2+ binding and catalytic steps involving the E1 conformation (Figure 1), but may play a role in the E2 conformation, perhaps participating in metal release or dephosphorylation. Taken together, our data indicate that Co2+ can bind in the absence of the TM4 cysteine and that histidine, probably His 657, and water are coordinated. Besides His 657, Ser 325, Glu 658, and Thr 661 are necessary for Co2+ binding and transport and thus likely comprise the additional nitrogen and oxygen ligands. Further studies to elucidate the molecular details of Co2+ coordination are underway.
Supplementary Material
Acknowledgments
Portions of this research were carried out at both the Stanford Synchrotron Radiation Laboratory (SSRL) and at the National Synchrotron Light Source (NSLS). SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program. NSLS, located at Brookhaven National Laboratory, is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, under Contract No. DE-AC02-98CH10886.
Abbreviations
- sCoaT
Sulfitobacter sp. NAS-14.1 P1B-4-ATPase
- WT-sCoaT
wild type sCoaT
- C327A-sCoaT
variant sCoaT in which Cys 327 is replaced with alanine
- LpCopA
Legionella pneumophila Cu+ P1B-1-ATPase
- ATPBD
ATP binding domain
- N-domain
nucleotide binding domain
- P-domain
phosphorylation domain
- A-domain
actuator domain
Footnotes
This work was supported by NIH grants GM58518 (A. C. R.), DK068139 (T. L. S.), and HL13531 (B. M. H). E. L. Z. was supported in part by NIH grant 5T32 GM008382. G.E.C. was supported by NSF fellowship DGE-0824162.
SDS-PAGE analysis of purified sCoaT mutants. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bublitz M, Morth JP, Nissen P. P-type ATPases at a glance. J Cell Sci. 2011;124:2515–2519. doi: 10.1242/jcs.088716. [DOI] [PubMed] [Google Scholar]
- 2.Palmgren MG, Nissen P. P-Type ATPases. Ann Rev Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 3.Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- 4.Bublitz M, Poulsen H, Morth JP, Nissen P. In and out of the cation pumps: P-Type ATPase structure revisited. Curr Op Struct Biol. 2010;20:431–439. doi: 10.1016/j.sbi.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Lutsenko S, Kaplan JH. Organization of P-type ATPases: significance of structural diversity. Biochemistry. 1995;34:15607–15613. doi: 10.1021/bi00048a001. [DOI] [PubMed] [Google Scholar]
- 6.Albers RW. Biochemical aspects of active transport. Annu Rev Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 7.Post RL, Hegyvary C, Kume S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J Biol Chem. 1972;247:6530–6540. [PubMed] [Google Scholar]
- 8.Solioz M, Camakaris J. Acylphosphate formation by the Menkes copper ATPase. FEBS Lett. 1997;412:165–168. doi: 10.1016/s0014-5793(97)00770-9. [DOI] [PubMed] [Google Scholar]
- 9.Voskoboinik I, Mar J, Strausak D, Camakaris J. The regulation of catalytic activity of the Menkes copper-translocating P-type ATPase. J Biol Chem. 2001;276:28620–28627. doi: 10.1074/jbc.M103532200. [DOI] [PubMed] [Google Scholar]
- 10.Møller JV, Olesen C, Winther AML, Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Quart Rev Biophys. 2010;43:501–566. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 11.Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TLM, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 12.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 angstrom resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- 14.Gourdon P, Liu XY, Skjørringe T, Morth JP, Møller LB, Pedersen BP, Nissen P. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475:59–74. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 15.Rensing C, Mitra B, Rosen BP. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rensing C, Sun Y, Mitra B, Rosen BP. Pb(II)-translocating P-type ATPases. J Biol Chem. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- 17.Sharma R, Rensing C, Rosen BP, Mitra B. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J Biol Chem. 2000;275:3873–3878. doi: 10.1074/jbc.275.6.3873. [DOI] [PubMed] [Google Scholar]
- 18.Mana-Capelli S, Mandal AK, Argüello JM. Archaeoglobus fulgidus CopB is a thermophilic Cu2+-ATPase. J Biol Chem. 2003;278:40534–40541. doi: 10.1074/jbc.M306907200. [DOI] [PubMed] [Google Scholar]
- 19.Mandal AK, Cheung WD, Argüello JM. Characterization of a thermophilic P-type Ag+/Cu+-ATPase from the extremophile Archaeglobus fulgidus. J Biol Chem. 2002;277:7201–7208. doi: 10.1074/jbc.M109964200. [DOI] [PubMed] [Google Scholar]
- 20.Raimunda D, González-Guerrero M, Leeber BW, Argüello JM. The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals. 2011;24:467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutherford JC, Cavet JS, Robinson NJ. Cobalt-dependent transcriptional switching by a dual-effector MerR-like protein regulates a cobalt-exporting variant CPx-type ATPase. J Biol Chem. 1999;274:25827–25832. doi: 10.1074/jbc.274.36.25827. [DOI] [PubMed] [Google Scholar]
- 22.Lutsenko S, Gupta A, Burkhead JL, Zuzel V. Cellular multitasking: The dual role of human Cu-ATPases in cofactor delivery and intracellular copper balance. Arch Biochem Biophys. 2008;476:22–32. doi: 10.1016/j.abb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bull PC, Cox DW. Wilson disease and Menkes disease: new handles on heavy-metal transport. Trends Genet. 1994;10:246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 24.Argüello JM. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J Membr Biochem. 2003;195:93–108. doi: 10.1007/s00232-003-2048-2. [DOI] [PubMed] [Google Scholar]
- 25.Fan B, Rosen BP. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J Biol Chem. 2002;277:46987–46992. doi: 10.1074/jbc.M208490200. [DOI] [PubMed] [Google Scholar]
- 26.Mandal AK, Argüello JM. Functional roles of metal binding domains of the Archaeoglobus fulgidus Cu+-ATPase CopA. Biochemistry. 2003;42:11040–11047. doi: 10.1021/bi034806y. [DOI] [PubMed] [Google Scholar]
- 27.Dutta SJ, Liu JB, Stemmler AJ, Mitra B. Conservative and nonconservative mutations of the transmembrane CPC motif in ZntA: Effect on metal selectivity and activity. Biochemistry. 2007;46:3692–3703. doi: 10.1021/bi0616394. [DOI] [PubMed] [Google Scholar]
- 28.Okkeri J, Haltia T. The metal-binding sites of the zinc-transporting P-type ATPase of Escherichia coli. Lys(693) and Asp(714) in the seventh and eighth transmembrane segments of ZntA contribute to the coupling of metal binding and ATPase activity. Biochim Biophys Acta. 2006;1757:1485–1495. doi: 10.1016/j.bbabio.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Wu CC, Gardarin A, Martel A, Mintz E, Guillain F, Catty P. The cadmium transport sites of CadA, the Cd2+-ATPase from Listeria monocytogenes. J Biol Chem. 2006;281:29533–29541. doi: 10.1074/jbc.M604658200. [DOI] [PubMed] [Google Scholar]
- 30.Argüello JM, Eren E, González-Guerrero M. The structure and function of heavy metal transport P1B-type ATPases. Biometals. 2007;20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 31.González-Guerrero M, Hong D, Argüello JM. Chaperone-mediated Cu+ delivery to Cu+ transport ATPases. Requirement of nucleotide binding. J Biol Chem. 2009;284:20804–20811. doi: 10.1074/jbc.M109.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatori Y, Majima E, Tsuda T, Toyoshima C. Domain organization and movements in heavy metal ion pumps - papain digestion of CopA, a Cu+-transporting ATPase. J Biol Chem. 2007;282:25213–25221. doi: 10.1074/jbc.M703520200. [DOI] [PubMed] [Google Scholar]
- 33.Eren E, Kennedy DC, Maroney MJ, Argüello JM. A novel regulatory metal binding domain is present in the C terminus of Arabidopsis Zn2+-ATPase HMA2. J Biol Chem. 2006;281:33881–33891. doi: 10.1074/jbc.M605218200. [DOI] [PubMed] [Google Scholar]
- 34.Barry AN, Shinde U, Lutsenko S. Structural organization of human Cu-transporting ATPases: learning from building blocks. J Biol Inorg Chem. 2010;15:47–59. doi: 10.1007/s00775-009-0595-4. [DOI] [PubMed] [Google Scholar]
- 35.Liu JB, Dutta SJ, Stemmler AJ, Mitra B. Metal-binding affinity of the transmembrane site in ZntA: Implications for metal selectivity. Biochemistry. 2006;45:763–772. doi: 10.1021/bi051836n. [DOI] [PubMed] [Google Scholar]
- 36.Okkeri J, Haltia T. Expression and mutagenesis of ZntA, a zinc-transporting P-type ATPase from Escherichia coli. Biochemistry. 1999;38:14109–14116. doi: 10.1021/bi9913956. [DOI] [PubMed] [Google Scholar]
- 37.Traverso ME, Subramanian P, Davydov R, Hoffman BM, Stemmler TL, Rosenzweig AC. Identification of a hemerythrin-like domain in a P1B-type transport ATPase. Biochemistry. 2010;49:7060–7068. doi: 10.1021/bi100866b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scherer J, Nies DH. CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Molec Microbiol. 2009;73:601–621. doi: 10.1111/j.1365-2958.2009.06792.x. [DOI] [PubMed] [Google Scholar]
- 39.Moreno I, Norambuena L, Maturana D, Toro M, Vergara C, Orellana A, Zurita-Silva A, Ordenes VR. AtHMA1 is a thapsigargin-sensitive Ca2+ heavy metal pump. J Biol Chem. 2008;283:9633–9641. doi: 10.1074/jbc.M800736200. [DOI] [PubMed] [Google Scholar]
- 40.Seigneurin-Berny D, Gravot A, Auroy P, Mazard C, Kraut A, Finazzi G, Grunwald D, Rappaport F, Vavasseur A, Joyard J, Richaud P, Rolland N. HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J Biol Chem. 2006;281:2882–2892. doi: 10.1074/jbc.M508333200. [DOI] [PubMed] [Google Scholar]
- 41.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 42.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 43.Vogel G, Steinhart R. ATPase of Escherichia coli: purification, dissociation, and reconstitution of the active complex from the isolated subunits. Biochemistry. 1976;15:208–216. doi: 10.1021/bi00646a032. [DOI] [PubMed] [Google Scholar]
- 44.Mailer C, Hoffman BM. Tumbling of an adsorbed nitroxide using rapid adiabatic passage. J Phys Chem. 1976;80:842–846. [Google Scholar]
- 45.Werst MM, Davoust CE, Hoffman BM. Ligand spin densities in blue copper proteins by Q-band 1H and 14N ENDOR spectroscopy. J Am Chem Soc. 1991;113:1533–1538. [Google Scholar]
- 46.Davoust CE, Doan PE, Hoffman BM. Q-band pulsed electron spin-echo spectrometer and its application to ENDOR and ESEEM. J Magn Reson. 1996;119:38–44. [Google Scholar]
- 47.Zipse H, Artin E, Wnuk S, Lohman GJS, Martino D, Griffin RG, Kacprzak S, Kaupp M, Hoffman B, Bennati M, Stubbe J, Lees N. Structure of the nucleotide radical formed during reaction of CDP/TTP with the E441Q-α2β2 of E. coli ribonucleotide reductase. J Am Chem Soc. 2009;131:200–211. doi: 10.1021/ja806693s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schweiger A, Jeschke G. Principles of pulse electron paramagnetic resonance. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- 49.Epel B, Gromov I, Stoll S, Schweiger A, Goldfarb D. Spectrometer manager: A versatile control software for pulse EPR spectrometers. Concepts in Magnetic Resonance Part B-Magnetic Resonance Engineering. 2005;26B:36–45. [Google Scholar]
- 50.George GN, George SJ, Pickering IJ. EXAFSPAK. 2001 http://wwwssrl.slac.stanford.edu/~george/exafspak/exafs.htm.
- 51.Ankudinov AL, Rehr JJ. Relativistic calculations of spin-dependent X-ray absorption spectra. Phys Rev B. 1997;56:R1712–R1715. [Google Scholar]
- 52.Lee PA, Citrin PH, Eisenberger P, Kincaid BM. Extended x-ray absorption fine structure - its strengths and limitations as a structural tool. Rev Mod Phys. 1981;53:769–806. [Google Scholar]
- 53.Prelesnik BV, Gabela F, Ribar B, Krstanovic IR. Hexaquacobalt(II) nitrate, Co[OH2]6[NO3]2. Cryst Struct Commun. 1973;2:581–583. [Google Scholar]
- 54.Bencze KZ, Kondapalli KC, Stemmler TL. X-ray absorption spectroscopy. In: Scott RA, Lukehart CM, editors. Applications of Physical Methods to Inorganic and Bioinorganic Chemistry: Handbook, Encyolopedia of Inorganic Chemistry. 2. John Wiley & Sons, Ltd; Chichester, UK: 2007. pp. 513–528. [Google Scholar]
- 55.Bissig KD, Wunderli-Ye H, Duda PW, Solioz M. Structure-function analysis of purified Enterococcus hirae CopB copper ATPase: Effect of Menkes/Wilson disease mutation homologues. Biochem J. 2001;357:217–223. doi: 10.1042/0264-6021:3570217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.González-Guerrero M, Eren E, Rawat S, Stemmler TL, Argüello JM. Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. J Biol Chem. 2008;283:29753–29759. doi: 10.1074/jbc.M803248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Regan L, Clarke ND. A tetrahedral zinc(II)-binding site introduced into a designed protein. Biochemistry. 1990;29:10878–10883. doi: 10.1021/bi00501a003. [DOI] [PubMed] [Google Scholar]
- 58.May SW, Kuo JY. Preparation and properties of cobalt(II) rubredoxin. Biochemistry. 1978;17:3333–3338. doi: 10.1021/bi00609a025. [DOI] [PubMed] [Google Scholar]
- 59.Bertini I, Luchinat C. High spin cobalt(II) as a probe for the investigation of metalloproteins. Adv Inorg Biochem. 1984;6:71–111. [PubMed] [Google Scholar]
- 60.Salvato B, Beltramini M, Ricchelli F, Tallandini L. Cobalt substitution studies on bovine erythrocyte superoxide dismutase: evidence for a novel cobalt-superoxide dismutase derivative. Biochim Biophys Acta. 1989;998:14–20. doi: 10.1016/0167-4838(89)90112-x. [DOI] [PubMed] [Google Scholar]
- 61.Lever ABP. Inorganic electronic spectroscopy. 2. Elsevier Science Publishing Company, Inc; New York: 1984. [Google Scholar]
- 62.Brader ML, Kaarsholm NC, Harnung SE, Dunn MF. Ligand perturbation effects on a pseudotetrahedral Co(II)(His)3-ligand site. J Biol Chem. 1997;272:1088–1094. doi: 10.1074/jbc.272.2.1088. [DOI] [PubMed] [Google Scholar]
- 63.Makinen MW, Kuo LC, Yim MB, Wells GB, Fukuyama JM, Kim JE. Ground term splitting of high-spin Co2+ as a probe of coordination structure. 1 Dependence of the splitting on coordination geometry. J Am Chem Soc. 1985;107:5245–5255. [Google Scholar]
- 64.Walsby CJ, Krepkiy D, Petering DH, Hoffman BM. Cobalt-substituted zinc finger 3 of transcription factor IIIA: Interactions with cognate DNA detected by P-31 ENDOR spectroscopy. J Am Chem Soc. 2003;125:7502–7503. doi: 10.1021/ja034589o. [DOI] [PubMed] [Google Scholar]
- 65.Allen FH. The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Cryst. 2002;B58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 66.Feiters MC, Navaratnam S, Alhakim M, Allen JC, Spek AL, Veldink GA, Vliegenthart JFG. EXAFS of poly[•-hexakis(2-methylimidazolato-N,N′)triiron(II)]: implications for metalloprotein studies. J Am Chem Soc. 1988;110:7746–7750. [Google Scholar]
- 67.Lowe J, Vieyra A, Catty P, Guillain F, Mintz E, Cuille M. A mutational study in the transmembrane domain of Ccc2p, the yeast Cu(I)-ATPase, shows different roles for each Cys-Pro-Cys cysteine. J Biol Chem. 2004;279:25986–25994. doi: 10.1074/jbc.M308736200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.