Abstract
Case-control genetic association studies in admixed populations are known to be susceptible to genetic confounding due to population stratification. The transmission/disequilibrium test (TDT) approach can avoid this problem. However, the TDT is expensive and impractical for late- onset diseases. Case-control study designs, in which cases and controls are matched by admixture, can be an appealing and suitable alternative for genetic association studies in admixed populations. In this study, we applied this matching strategy when recruiting our African American participants in the Study of African American, Asthma, Genes and Environments (SAGE). Group admixture in this cohort consists of 83% African ancestry and 17% European ancestry, which was consistent with reports from other studies. By carrying out several complementary analyses, our results show that there is substructure in the cohort, but that the admixture distributions are almost identical in cases and controls, and also in cases only. We performed association tests for asthma-related traits with ancestry, and only found that FEV1, a measure for baseline pulmonary function, was associated with ancestry after adjusting for socio-economic and environmental risk factors (P = 0.01). We did not observe an excess of type I error rate in our association tests for ancestry informative markers (AIMs) and asthma-related phenotypes when ancestry was not adjusted in the analyses. Furthermore, using the association tests between genetic variants in a known asthma candidate gene, β2 adrenergic receptor (β2AR) and ΔFEF25-75, an asthma-related phenotype, as an example, we demonstrated population stratification was not a confounder in our genetic association. Our present work demonstrates that admixture-matched case-control strategies can efficiently control for population stratification confounding in admixed populations.
Introduction
Population stratification is a potential confounding factor of case-control genetic association studies in admixed populations, such as African Americans (Cardon and Bell 2001; Cardon and Palmer 2003). Population stratification occurs when there are different allele frequencies between cases and controls due to heterogeneity in ancestry, which is unrelated to disease affection status. Ignoring population stratification in association tests may lead to a potential excess of both false positive and false negative results (Burchard et al. 2003b; Lander and Schork 1994; Ziv and Burchard 2003).
The history of African Americans is notable for admixture between Africans, Europeans and Native Americans (Parra et al. 1998). The first attempts to estimate admixture proportions in African Americans were in the 1950s (Glass and Li 1953). Since then, this field has been underdeveloped due to the limited availability of ancestry informative markers (AIMs) and data from ancestral populations. Recent studies have provided fruitful results on AIMs discovery and the development of methodologies for estimating individual and group admixture (Akey et al. 2002; Pfaff et al. 2001; Shriver et al. 2005; Shriver et al. 2003).
Although the transmission/disequilibrium test (TDT) approach is robust against population stratification and has been proposed for finding susceptibility genes in complex traits (Allison 1997; Spielman et al. 1993), the TDT approach is often expensive and impractical for late-onset disorders. One promising solution to control for population stratification is to match cases and controls carefully based on their genetic background. Well matched case-control designs may avoid the confounding effect due to population stratification (Wacholder et al. 2002; Zondervan et al. 2002). To control population stratification confounding, a previous study reported several analytical strategies for matching cases and controls as part of association tests in admixed populations (Hinds et al. 2004). In addition, several groups have proposed to detect and control population stratification confounding in case-control association tests by using two powerful approaches: 1) identifying and including ancestry in the analysis; 2) using genomic control to adjust for potential existing population stratification (Bacanu et al. 2000; Devlin et al. 2001; Freedman et al. 2004; Hoggart et al. 2003; Parra et al. 2004; Pritchard and Donnelly 2001).
Significant worldwide variations in asthma prevalence have been reported by the International Study of Asthma and Allergies in Childhood (ISAAC) and the European Community Respiratory Health Survey (ECRHS) (1998; Pearce et al. 2000). In the U.S., it is well known that there are racial and ethnic differences in asthma prevalence, morbidity, and mortality. Specifically, asthma prevalence and mortality among African Americans is greater than among European Americans (Akinbami et al. 2005; Akinbami and Schoendorf 2002; Mannino et al. 2002). It is important to investigate genetic, environmental and socio-economic factors, which may lead to the racial and ethnic variations.
The β2 adrenergic receptor (β2AR) is one of the candidate genes most consistently identified as being associated with asthma-related phenotypes (Choudhry et al. 2005; Evans et al. 2001; Holloway et al. 2000; Silverman et al. 2003). The Gly16 polymorphism has been associated with asthma severity and lower bronchodilator responsiveness, while the Arg16 allele has been shown to be associated with increased bronchodilator responsiveness (Martinez et al. 1997). It may be because of the difficulties of controlling for population stratification, the effect of the β2AR genetic variants on asthma-related phenotypes among African American asthmatics is unclear.
In this study, we have recruited African American subjects participating in the Study of African American, Asthma, Genes and Environments (SAGE) through well-matched case-control strategies. We have detected population substructure and recent admixture. We have also evaluated group admixture and individual admixture using two programs — ADMIXMAP and Structure2.1. We have examined the relationship between asthma-related phenotypes and ancestry. Moreover, we have demonstrated that there is no evidence of confounding due to population stratification in our genetic association tests of asthma-related traits with AIMs, and with the β2AR genetic variants. Our results have indicated that the inflation of type I error rate in association tests can be efficiently controlled in an admixture-matched case-control study of asthma in African Americans.
Subjects and methods
Study Participants
One hundred and seventy-six African American asthmatics were recruited from three clinics as part of the ongoing Study of African American, Asthma, Genes and Environments (SAGE). One clinic is the San Francisco General Hospital, and the remaining two clinics are located less than two miles away from each other in Oakland, California. Eligible cases were between the ages of 8 and 40 years, had physician-diagnosed asthma, and had experienced two or more asthma symptoms (wheezing, coughing, and/or shortness of breath) in the previous two years. We recruited 176 matched controls whose ages were between 8 and 40. Controls were eligible only if they reported no history of asthma or allergies, no history or report of having experienced symptoms of coughing, wheezing or shortness of breath in the past 2 years, no other history of lung diseases or chronic illness or medications, less than10-pack-per-year smoking history, and no smoking in the last year. All subjects were enrolled into the study only if subjects self-identified as African Americans, and both biological parents and all biological grandparents were identified as African Americans.
Phenotype measurement
Asthma is characterized by recurrent episodes of wheeze, cough and airway obstruction. Airway obstruction is an indicator of asthma severity and can be measured using spirometry. Standard measures of the severity of airway obstruction are FEV1, FEV1/FVC and FEF25-75, all expressed as a percentage of normal predicted values. The lower value, the more severe the airway obstruction. Airway obstruction is reversible with the inhalation of medications such as albuterol, the most commonly prescribed asthma medication in the world. The reversibility of airway obstruction is a measure of drug responsiveness. Reversibility can be measured by performing spirometry before and after the administration of albuterol and measuring the difference (ΔFEV1, ΔFEV1/FVC, and ΔFEF25-75).
Asthmatic subjects were instructed to withhold their bronchodilator medications for at least eight hours before lung function tests. Spirometry was performed according to the American Thoracic Society standards (1995). Pulmonary function test results are expressed as a percentage of the predicted normal value using age-adjusted prediction equations from Hankinson (Hankinson et al. 1999). Baseline pulmonary function results are reported as pre-FEV1, pre-FEV1/FVC and pre-FEF25-75. Albuterol was administered using an extension tube connected to a standard metered dose inhaler (180μg or 2 puffs for subjects < 16 years old and 360μg or 4 puffs for subjects ≥ 16 years old). Fifteen-minutes after albuterol administration, FEV1, FEV1/FVC and FEF25-75 were measured again. Bronchodilator drug responsiveness to albuterol is reported as percent change in FEV1, FEV1/FVC and FEF25-75 between baseline and after albuterol administration (expressed herein as ΔFEV1, ΔFEV1/FVC and ΔFEF25-75, respectively).
Quantitative measures of asthma severity were defined as pre-FEV1, pre-FEV1/FVC, and pre-FEF25-75. Qualitative measures of asthma severity were classified as “mild” or “moderate-severe” asthma based on four “yes/no” questions related to medication use, asthma symptoms, nocturnal awakenings and pre-FEV1 (Burchard et al. 2003a). Total plasma IgE, a measure for determining atopic asthmatic cases, was collected in duplicate for asthmatic subjects using Uni-Cap technology (Pharmacia, Kalamazoo, MI).
Selection of ancestry informative markers (AIMs)
We selected these 31 AIM SNP variants based on their informativeness of ancestry with a large difference of allele frequencies (δ) between Native American, African and European ancestral populations (Bonilla et al. 2004; Parra et al. 1998). For dimorphic variants, δ = |p1 – p2|, where p1 and p2 are defined as the allele frequencies in ancestral populations 1 and 2, respectively. The allele frequencies among these three ancestral populations were obtained by genotyping individuals of the following populations: Irish, English, German and Spanish (Europeans, N = 243); Nigerian, Central African Republic and Sierra Leone (Africans, N = 481); and Mayan, Pima, Cheyenne and Pueblo (Native Americans, N = 148). Detailed information of these 31 AIMs regarding chromosomal location, allele frequencies among different ancestral populations, and difference of allele frequencies between different ancestral populations were provided in Supplementary Table S1. Flanking sequence and other relevant information of these 31 AIMs can be obtained from dbSNP website (http://www.ncbi.nlm.nih.gov/SNP/) and were also described elsewhere (Choudhry et al. 2005, in press).
Genotyping
All thirty-one AIMs and two β2AR SNP variants (SNP-468 in the promoter region and SNP+46 [Arg/Gly 16] within the β2AR coding region) were genotyped using the AcycloPrime-FP™ (PerkinElmer) method (Chen and Kwok 1999). PCR conditions were as follows: 2.4-4.0 ng genomic DNA, 0.1-0.2 μM primers, 50 μM dNTPs, 0.1-0.2 units Platinum Taq (Invitrogen), 6 μl volume with Platinum Taq PCR buffer, 2.5 mM MgCl2 plus 1 μl extra water to counteract evaporation. Cycling conditions were: 95°C for 2 minutes, 35 cycles of 92°C for 10 seconds, 58°C for 20 seconds, 68°C for 30 seconds, and final extension at 68°C for 10 minutes. Enzymatic cleanup and single base extension genotyping reactions were performed with AcycloPrime-FP kits. Plates were read on an EnVision fluorescence polarization plate reader (PerkinElmer) for genotyping calls.
Statistical analyses
Allele frequencies and Hardy-Weinberg Equilibrium
Allele frequencies of each AIM were computed by using genotype data of all individuals, cases and controls, separately. We tested whether there was a significant difference between cases and controls by χ2 test. We tested whether AIMs and β2AR SNPs in our SAGE cohort (N=352) were under Hardy-Weinberg equilibrium (HWE) by using the exact Hardy-Weinberg test, which calculates the probability of the exact number of heterozygotes conditional on the copies of the minor SNP allele. This test has been implemented in the PEDSTATS program (Abecasis et al. 2000). For each AIM, we calculated FST between Africans and Europeans, a measure of ancestral informativeness, as , where δ2 was denoted as variance and was the mean of individual allele frequency (Wright 1969).
Recent admixture
We examined the presence of recent admixture using pair-wise combinations of 30 AIMs in all individuals, cases and controls, respectively. For each marker pair, we first estimated haplotype frequencies using the expectation maximization algorithm (EM) (Excoffier and Slatkin 1995) and computed a likelihood ratio statistic to test the strength of linkage disequilibrium based on the observed genotype data. We then permuted genotype data and computed the same likelihood ratio statistic for 10,000 permutations. Ranks were assigned for the observed and permuted likelihood ratio statistics. The sum of the ranks across all combinations in observed data was compared to the null distribution of the rank sums from 10,000 permutations. This is a global test for evaluating excess linkage disequilibrium across the genome. This statistical approach and original R code were kindly supplied by Dr. Hua Tang.
Population stratification
To detect population stratification, we fit clustering models with K = 1, 2, and 3 clusters, where K is the number of substructures, by using the Structure2.1 program (Falush et al. 2003; Pritchard et al. 2000). We obtained the likelihoods for different K through the MCMC algorithm implemented in the Structure2.1 program. We then selected the most likely K according to the maximum likelihood from the outputs.
Group admixture and individual admixture
The ADMIX 2.0 program based on a coalescent approach was used for estimating group admixture (Bertorelle and Excoffier 1998; Dupanloup and Bertorelle 2001). Admixture proportions are estimated based on the genotype frequencies of the AIMs and their level of divergence – number of generations. Standard deviation of group admixture estimates is calculated according to 10,000 bootstraps.
In order to make sure that we obtained proper estimates of individual admixture, we computed individual admixture estimates (IAEs) using the ADMIXMAP and Structure 2.1 programs, respectively. We then accessed the consistency of IAEs obtained from both programs. A combination of Bayesian and classical approaches has been implemented in the program ADMIXMAP (Hoggart et al. 2003). We input AIMs and trait data from the admixed population and AIMs data from ancestral populations to calculate IAEs by ADMIXMAP with 1,000 burn-in and 20,000 further iterations.
The admixture model implemented in the program Structure2.1 assumes that each individual inherits some proportion of their ancestry from each ancestral population (Falush et al. 2003; Pritchard et al. 2000). To compute IAEs, we input genotype data from each ancestral population, specified as known populations, and admixed subjects, specified as an unknown population, assumed an admixture model and used default values for other parameters by Structure2.1 with 50,000 burn-in and 50,000 further iterations.
The detailed methodologies of these two programs and their differences were described elsewhere (Tsai et al. 2005, in press).
Admixture-matched evaluation
We first obtained individual admixture for all subjects from ADMIXMAP and Structure2.1. To compare the distribution of admixture background between cases and controls, we generated quantile-quantile (Q-Q) plots and performed the Wilcoxon Rank Sum Test to examine whether the admixture distribution in cases was similar to the distribution in controls. To evaluate the admixture distribution in cases only, we carried out 10,000 permutations. We first randomly assigned cases into two groups, then compared the distribution between these two groups by the Wilcoxon Rank Sum Test, recorded P value for each permutation and calculated the empirical P value for 10,000 permutations based on whether the P value for each permutation was less than 0.05.
Tests of association and evaluation of type I error rate
We first applied regression analyses for association tests for asthma-related traits and ancestry as defined by IAEs (individual African and European ancestry estimates). We only incorporated African ancestry estimates in regression analyses to avoid co-linearity. We also performed regression models to test for association between asthma disease status and AIMs under the additive genetic model assumption. For asthmatics, we applied regression models to test for association between asthma-related traits and AIMs. We assessed the normality of quantitative asthma-related phenotypes (asthma severity as defined by: pre-FEV1, pre-FEV1/FVC and pre-FEF25-75). Since drug response traits – ΔFEV1, ΔFEV1/FVC and ΔFEF25-75, and IgE were not normally distributed, we took logarithm transformation of these traits in our regression models.
To evaluate the inflation of the type I error rate, we first tested the association between asthma-related phenotypes and AIMs with or without including covariates: age, gender, socio-economic status (SES), asthma duration, regular use of asthma medication and body mass index (BMI) in the models. We then performed association tests with adjustment for the same covariates and IAEs, specifically, individual African ancestry estimates. We used a P value less than 0.05 as the significance level and recorded the number of positives from regression analyses based on this corresponding threshold.
One way to detect and control population stratification is incorporating ancestry as defined by IAEs in the analysis and examining the results obtained from the models with and without adjusting ancestry. To demonstrate that population stratification did not confound the genetic association in our cohort, we applied linear regression analysis to test the association between two β2AR SNP variants and ΔFEF25-75, an asthma-related trait with and without including IAEs in the models. Data analyses were carried out using statistical packages R 1.9.0 and STATA 8.0 S/E (College Station, TX).
Results
Demographic, clinical and AIMs characteristics
We have recruited 176 African American asthmatic cases and 176 matched controls in the SAGE Study (Table 1). We carried out a χ2 test to examine whether or not there was a difference in socio-economic status (SES) among the subjects recruited from different clinics. Based on our result, there was no significant difference between SES and clinic sites (P = 0.26). However, there was a significant difference in age between cases and controls (P < 0.001). Hence, we included age as a covariate in all the analyses. We genotyped 31 ancestry informative markers (AIMs). One out of 31 AIMs, rs2816, deviated from Hardy-Weinberg equilibrium in the SAGE cohort (N= 352; Supplementary Table S1). Therefore, we excluded this marker in the analyses. The results based on χ2 tests indicated that there was no difference in allele frequencies of 30 AIMs between cases and controls. The average FST of these thirty AIMs, a measure of ancestral informativeness, between African and European populations was 0.35.
Table 1.
Demographic and clinical characteristics in SAGE subjects
| Controls (n=176) | Cases (n=176) | |
|---|---|---|
| Demographic features | ||
| Age (yr) | 31.3 (24.5:36.1) a | 19.4 (13:31) |
| Gender (% male) | 38.07 % | 35.22 % |
| BMI (kg/m2) | - | 25.8 (22:32) |
| Socio-economic status | ||
| low | 44.38 % | 33.73 % |
| moderate | 31.36 % | 40.36 % |
| middle | 20.71 % | 19.28 % |
| upper | 3.55 % | 6.62 % |
| Asthma onset (yr) | - | 5 (2:9) |
| Asthma duration (yr) | - | 12 (7:18) |
| Asthma severity (%severe) | - | 65.91 % |
| Serum IgE (IU/ml) | - | 164 (47.5:417) |
| Baseline Spirometry | ||
| FEV1 (% predicted) | - | 93.15 (84.44:101.69) |
| FEV1 /FVC (% predicted) | - | 93.79 (85.38:98.86) |
| FEF25-75 (% predicted) | - | 76.28 (59.33: 95.65) |
| Bronchodilator Responsiveness | ||
| FEV1 reversibility (%) | - | 7.45 (4.03:14.04) |
| FEV1 /FVC reversibility (%) | - | 4.04 (1.23: 8.15) |
| FEF25-75 reversibility (%) | - | 22.31 (11.22: 39.44) |
Values expressed as median (25th : 75th percentile)
Recent admixture, population substructure and group admixture
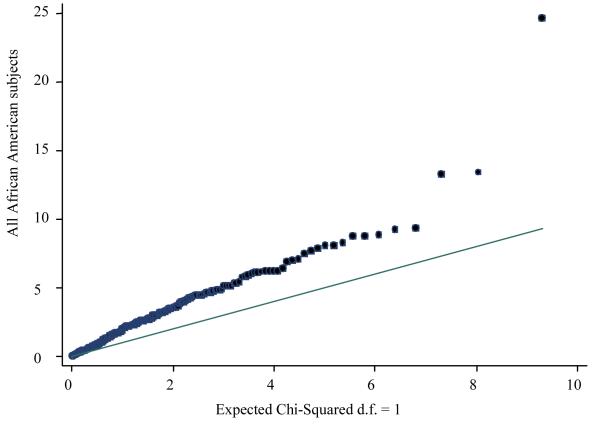
To examine the presence of recent admixture, we applied a global test for evaluating excess genome-wide linkage disequilibrium (LD) by comparing the rank scores of the combination of marker pairs from observed AIMs data and 10,000 permutations. There were 62 (14.3%), 32 (7.4%) and 57 (13.1%) of the 435 marker pairs with significant excess of LD in all subjects, cases and controls, respectively. The results from global rank tests showed significantly higher LD inflation than expected under the null in all subjects, cases and controls, respectively (all three P values < 0.001). The quantile-quantile plot in Figure 1 showed that the global observed LD was higher than the null distribution. These results demonstrated the presence of recent admixture in African Americans.
Figure 1.
Quantile-quantile plot for comparing global LD patterns calculated from observed AIMs data in all African American subjects to the expected null distribution.
We applied Structure2.1 to assess the presence of population substructures within cases, controls and all subjects combined, individually. The results in Table 2 indicated that our African American subjects were most likely descended from two ancestral populations, instead of one or three ancestral populations. We also applied ADMIX to estimate group admixture of the cohort and inferred the cohort descend from either 2 or 3 ancestral populations, respectively. The admixture proportions based on three ancestral populations (Africans, Europeans and Native Americans) were 83.2% ± 1%, 16.5% ± 1% and 0.3% ± 2%, respectively. The admixture proportions based on two ancestral populations (Africans and Europeans) were 83.3% ± 0.8% and 16.7% ± 0.8%, separately. The concordant results from Structure2.1 and ADMIX suggested that our cohort was derived from two ancestral populations (Africans and Europeans).
Table 2.
Number of population substructures within the SAGE cohort estimated by Structure2.1.
| No. of population substructures |
Log likelihood | ||
|---|---|---|---|
| All subjects combined (N = 352) |
Asthmatic cases (N = 176) |
Healthy controls (N = 176) |
|
| 1 | −27332 | −21494 | −21468 |
| 2 | −20276 * | −14610 * | −14570 * |
| 3 | −22314 | −16765 | −16691 |
NOTE:
Number of substructures with the highest log likelihood
Admixture background in cases and controls
We calculated individual ancestry estimates (IAEs) by using ADMIXMAP and Structure2.1, separately. The IAEs obtained from ADMIXMAP were highly correlated with the IAEs computed from Structure2.1 in all subjects (correlation coefficients ρ = 0.99). We observed similar results when evaluating IAEs in asthmatic cases and controls, separately.
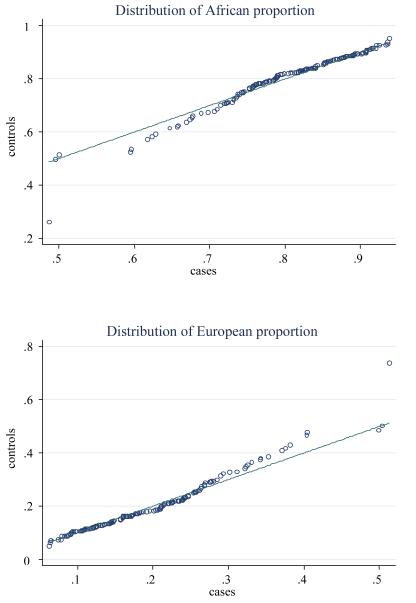
In addition, we examined the distributions of admixture proportions between cases and controls by Q-Q plots (Figure 2) and carried out the Wilcoxon Rank Sum Test for comparing the admixture distributions between cases and controls. The results indicated that there was no difference in the distributions of admixture proportions between African American asthmatic cases and controls (P = 0.49 and 0.48 for IAEs computed by ADMIXMAP and Structure2.1, respectively). We also checked the admixture distribution within cases by carrying out 10,000 permutations (details provided in the ‘Subjects and methods’). The results showed that there was no difference in admixture distribution within cases (P = 0.95 and 0.94 for IAEs calculated by ADMIXMAP and Structure2.1, respectively).
Figure 2.
Quantile-quantile plots of the distribution of admixture proportions in African American cases and controls.
Admixture estimates using different priors
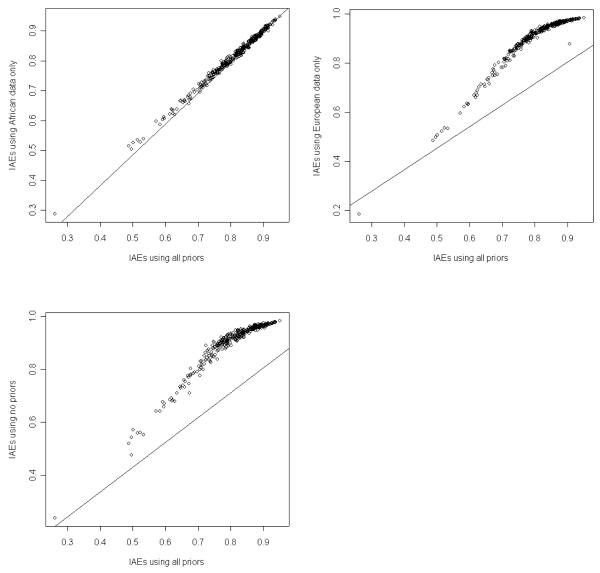
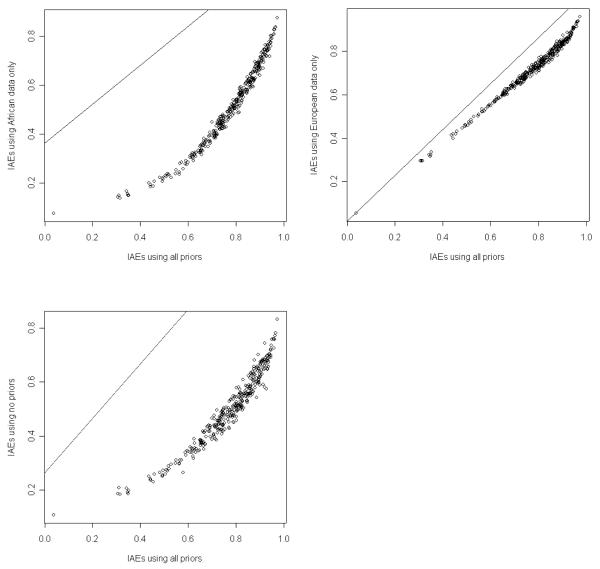
A restriction with respect to studying genetic association in admixed populations is collecting genotyping data of subjects from appropriate ancestral populations. We examined how the ADMIXMAP and Structure2.1 programs performed by either only including the prior data from one ancestral population, or including no prior data from the ancestral populations. We then compared admixture estimates with those obtained by using all prior information from both ancestral populations. The results in Figure 3 showed that admixture estimates obtained by using all priors and only using the data from African subjects were highly concordant when using ADMIXMAP to estimate admixture. In contrast, when using Structure2.1, a high correlation of admixture estimates was observed by using all priors and only using data from European subjects (Figure 4). Both programs provided poor admixture estimates when using no priors.
Figure 3.
Estimates of African proportion based on different prior information using ADMIXMAP.
Figure 4.
Estimates of African proportion based on different prior information using Structure2.1.
Association tests of asthma-related traits with ancestry, and with AIMs
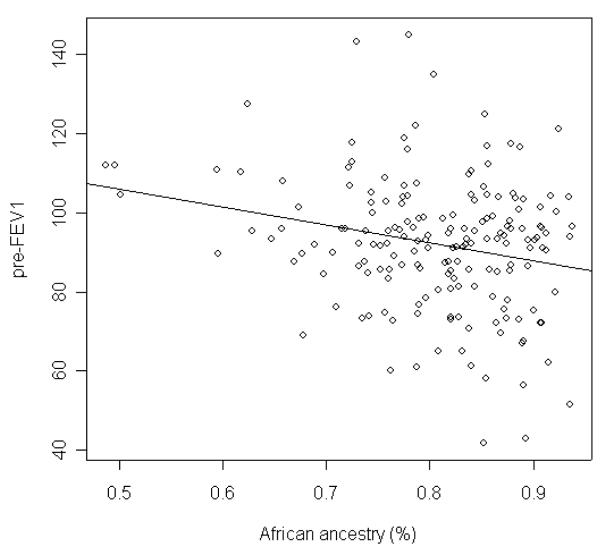
We tested the association of asthma-related traits – affection status, severity (pre-FEV1, pre-FEV1/FVC, pre-FEF25-75), drug response (ΔFEV1, ΔFEV1/FVC, ΔFEF25-75) and IgE with ancestry after adjusting for covariates – age, gender, socio-economic status (SES), asthma duration, regular use of asthma medication and body mass index (BMI). Of note, since individual African and European ancestral proportions were summed to one, we only included individual African proportions as a covariate to account for ancestral information in the analyses. The results in Table 3 showed that a significant association was only observed between pre-FEV1 and ancestry (P < 0.01). Figure 5 presented that individuals with more African background had lower pre-FEV1 values. We also examined the association between asthma-related phenotypes and 30 AIMs with adjustment of admixture background and covariates. We only observed slight inflation of type I error in the association tests between pre-FEV1 and 30 AIMs (Table 4).
Table 3.
Association of asthma-related traits with ancestry in the SAGE subjects.
| Ancestry * | ||||
|---|---|---|---|---|
|
|
||||
| Trait | ADMIXMAP | Structure2.1 | ||
|
|
||||
| Coefficients ** | P value | Coefficients | P value | |
| Disease status | −.303 | 0.818 | −.160 | 0.856 |
| Asthma severity | −.671 | 0.750 | −.515 | 0.716 |
| Pre-FEV1 | −39.322 | 0.008 | −25.980 | 0.009 |
| Pre-FEV1/FVC | −.817 | 0.929 | −.041 | 0.995 |
| Pre-FEF25-75 | −18.596 | 0.442 | −11.935 | 0.461 |
| Δ FEV1 | .472 | 0.226 | .298 | 0.254 |
| Δ FEV1/FVC | −.278 | 0.521 | −.175 | 0.542 |
| Δ FEF25-75 | .301 | 0.455 | .196 | 0.466 |
| IgE | .196 | 0.753 | .131 | 0.752 |
NOTE:
Estimates of ancestry, individual ancestral estimates (IAEs), are obtained either by ADMIXMAP or Structure2.1.
Covariates included in the analyses are age, gender, SES, asthma duration, regular use of asthma medication and BMI.
Figure 5.
Relationship between African ancestry and pre-FEV1 in African American asthmatics.
Table 4.
Association of asthma-related traits with 30 ancestry informative markers in the SAGE subjects.
| Count of positives ‡ (P value < 0.05) * | |||
|---|---|---|---|
|
|
|||
| Trait | No adjustment | w/covariates adjustment, but w/o IAEs adjustment ** |
w/covariates and IAEs adjustment |
| Disease status | 0 | 0 | 0 |
| Asthma severity | 1 | 0 | 1 |
| Pre-FEV1 | 3 | 3 | 2 |
| Pre-FEV1/FVC | 1 | 1 | 1 |
| Pre- FEF25-75 | 1 | 2 | 2 |
| Δ FEV1 | 2 | 2 | 2 |
| Δ FEV1/FVC | 1 | 0 | 0 |
| Δ FEF25-75 | 0 | 0 | 0 |
| IgE | 2 | 2 | 2 |
NOTE:
Under 5% type I error rate (P = 0.05), counts of expected false positives are 1.5 markers.
Analyses were carried out by using multiple regression analysis.
Covariates included in the analyses are age, gender, SES, asthma duration, regular use of asthma medication and BMI.
Genetic association tests of an asthma-related trait with two β2AR SNPs
We have performed comprehensive analyses to examine the association between the β2AR gene and asthma-related traits in our ongoing genetic study (complete results will be presented elsewhere). Here, we presented the results of association tests between an asthma-related phenotype, ΔFEF25-75, and two β2AR SNP variants, SNP-468 and SNP+46 [Arg/Gly 16], to examine whether IAEs, estimates for ancestral background, were a confounder in our cohort. The results of association tests for these two β2AR SNPs remained the same, either with or without ancestry adjustment (Table 5). The significant association between SNP -468 and ΔFEF25-75 did not remain after adjusting for other covariates (Table 5). These results demonstrated that there was no confounding effect due to population stratification in our genetic association tests of the β2AR variants, a well-recognized asthma candidate gene.
Table 5.
Genetic association of Δ FEF25-75 with β2AR SNP variants in African American asthmatics.
| β2AR SNP | No adjustment | With covariates, without IAEs adjustment ** |
With adjustment of covariates and IAEs |
|||
|---|---|---|---|---|---|---|
|
|
||||||
| t scores* | P value | t scores | P value | t scores | P value | |
| −468 C/G | 2.3 | 0.023 | 1.87 | 0.063 | 1.91 | 0.059 |
| +46 A / G | 2.04 | 0.043 | 2.2 | 0.03 | 2.23 | 0.027 |
NOTE:
Analyses were carried out by using regression analysis.
Covariates included in the analyses are age, gender, SES, asthma duration, regular use of asthma medication and BMI.
Discussion
Our results support the notion that a well-matched case-control study design is a feasible solution to overcome population stratification confounding while initiating genetic association studies in admixed populations (Cardon and Palmer 2003). To match admixture background, we have recruited self-identified African American cases and controls from three clinics, two of which are in the same census tract. As expected, we have minimized differences in the degree of admixture by recruiting subjects in such a very specific way. Our results show that subjects share high similarity in genetic background and SES. The minimal degrees of admixture do not lead confounding effect in the genetic association tests. Our results demonstrate that an admixture-matched case-control study design among African Americans can successfully avoid inflation of type I error rate in genetic association tests. The recruitment strategies achieve the goal of matching cases and controls based on admixture background and SES.
In a well-design case-control study, the source population from which cases are ascertained should be that one from which controls are also ascertained (Schlesselman and Stolley 1982). Our strategies for matching admixture are recruiting cases and controls on the basis of geographic proximity, self-reported ancestry and similar SES background. In terms of cost effectiveness, this admixture-matched study design is less expensive, practical for late age-of-onset diseases, and is capable of minimizing the confounding effect due to population stratification. Our findings provide the evidence that it is feasible to control population stratification confounding in the study-design stage.
According to our previous works, three main factors affecting the accuracy of admixture estimates are the number of markers, the informativeness of markers and the number of ancestral subjects. Specifically, the most important factor in determining the accuracy of admixture estimates is the number of AIMs (Tsai et al. 2005, in press). Although we only applied 30 AIMs to obtain admixture estimates, the results of group admixture estimates from our African American cohort agree with the results in previous reports (Hoggart et al. 2003; Parra et al. 1998; Reiner et al. 2005; Shriver et al. 2003). The European admixture proportion in our cohort is approximately 20%, which is consistent to the European admixture proportion in northern or western African American populations from other studies. In addition, we applied two different programs, ADMIXMAP and Structure2.1, for estimating individual admixture proportions. Admixture estimates from both programs showed a very high degree of correlation. Even though admixture estimates here could not be 100% accurate, they should be highly correlated to the underlying individual admixture proportions. Detailed information could be obtained elsewhere (Tsai et al. 2005, in press).
It has been reported that genetic background from different ancestral populations may be associated with socio-economic status (SES) (Burchard et al. 2003b). SES has been considered as an important indicator related to all-cause mortality within and across different racial groups (Lin et al. 2003). We examined the interaction between asthma-related phenotypes, SES and ancestry in our cohort, but did not find any association. One possible explanation as to why we did not observe an association may be due to the fact that admixture proportion and SES were well matched between our African American cases and controls.
We observed the association of pre-FEV1 with ancestry (Table 3 and Figure 5). The results demonstrated that higher African proportions among asthmatics were associated with more severe asthma as defined by lower pre-FEV1 values. Specifically, asthmatics with higher African ancestry had more severe asthma. Because pre-FEV1 is measured at least eight hours after the use of inhaled beta-agonists, this value is presumably an acceptable index of asthma severity. NHLBI guidelines currently use pre-FEV1 as an objective measurement in grading asthma severity. Pre-FEV1 has been validated as a measure of airway obstruction as it closely correlates with pathologic scores of airway diameter (Hogg et al. 1968). Decreased measures of pre-FEV1 were shown to be associated with the risk of future attacks and response to therapy among children with asthma (Enright et al. 1994; Fuhlbrigge et al. 2001). A previous study in Latino Americans showed that asthma severity might be influenced by ancestry in Mexican Americans (Salari et al. 2005). Recognizing that there is no single measure that accurately captures all facets of asthma severity, pre-FEV1 percent predicted has several advantages as a marker of asthma severity, including its objectivity and reproducibility (Enright et al. 1991; Enright et al. 1994; Kitch et al. 2004).
Previous studies based on U.S. vital statistics collected from the Third National Health and Nutrition Examination Survey (NHANES III) have reported that African Americans have higher prevalence of asthma than European Americans (Rodriguez et al. 2002; Romieu et al. 2004). We observed minor excess of false positives in the association tests of pre-FEV1 with AIMs before adjusting ancestry (Table 4). The type I error rate returned back to the expected level after including ancestry in the models. It will be of importance to determine whether the association between ancestry and asthma severity will be reproducible in African American asthmatics across the United States. It will be also important to explore gene-environment interactions of ancestry with SES and/or environmental factors.
A known limitation of genetic association studies in admixed populations is the difficulty in recruiting subjects from appropriate ancestral populations. Recent studies have shown that the principal component approach may be an appealing alternative to account for population stratification confounding, especially when investigators have no data from ancestral populations (Zhang et al. 2003). However, a significant limitation of this approach is how it handles missing data. Since it is common that many study participants do not have complete genotype information for all markers, power on the basis of the principal component approach may be limited. We approached this issue by only incorporating partial prior information from ancestral populations into ADMIXMAP and Structure2.1. The results in Figure 3 indicated that ADMIXMAP provided similar admixture estimates by using all priors and by only using data from African subjects. In contrast, Structure2.1 gave comparable estimates by using all priors and by only using data from European subjects (Figure 4). The difference was likely due to weighting ancestral information differently while inferring admixture proportions in ADMIXMAP and Structure2.1. In future plan, we will assess the difference in performance between ADMIXMAP and Structure2.1 through realistic simulation works. Besides, both programs provided poor admixture estimates when using no priors from ancestral populations. If investigators do not have genotyping data collected from proper ancestral populations, we would recommend investigators using the ‘genome-control’ approach to adjust for population stratification confounding, instead of including poor estimates in the model.
Population stratification occurs when there is an event of nonrandom mating. This permits allele frequencies of markers to vary among segments of the populations, as the results of genetic drift or founder effects (Slatkin 1991). As a consequence, a disease with high prevalence in one subpopulation will be also associated with any alleles that are in high frequency in that subpopulation. Since we detected two subgroups in the cohort, it would be of interest to explore whether the ‘group membership’ was correlated with asthma disease status or asthma-related traits. We grouped the subjects into two clusters based on their IAEs by using k-means cluster analysis (data not shown). We then checked the correlation between group membership and disease status, and between group membership and asthma-related traits. Correlation coefficients were less than ± 0.1 for disease status and asthma-related phenotypes, except for pre-FEV1 (ρ = 0.25). Taken together, substructures observed in our cohort was not correlated with asthma disease status and asthma-related traits.
According to our admixture-matched study design, we did not observe inflation of type I error in our association tests, even without adjustment of ancestry (Table 4). To demonstrate that population stratification can be a potential confounding factor if investigators do not match admixture background during the recruitment stage, we deliberately created a subset from our 352 SAGE subjects. In this subset, we selected asthmatic cases with top 100 African ancestral proportions and healthy controls with bottom 100 African ancestral proportions from the cohort. We then performed association tests of disease status with 30 AIMs in this subset. The results in Table S2 showed that there was an excess of false positives while not adjusting ancestral information in the analysis. The results here strengthened that our recruitment schema— matching admixture background in the study design could efficiently control population stratification confounding in admixed populations.
To adjust and control for potential confounding due to population stratification in the analysis, we applied a two-step approach, in which we estimated IAEs first using ADMIXMAP or Structure2.1. We then included these estimates into a conventional regression model as a covariate. The ADMIXMAP program provides a one-step approach, in which inference of admixture proportions, regression modeling and testing for association are combined in one model simultaneously. We compared the two-step and one-step approaches via comprehensive simulation scenarios that were described elsewhere (Tsai et al. 2005, in press). The findings in this work showed that the most important factor in determining accuracy of IAEs and in minimizing type I error rate was the number of AIMs used to estimate ancestry. For both one-step and two-step approaches, after accounting for precise ancestry information in association tests, the excess of type I error rate was controlled at the 5% level when 100 AIMs were used to calculate IAEs.
In summary, our present study demonstrates that an admixture-matched case-control study design is capable of controlling for confounding due to population stratification in admixed populations. Our results indicate that recruiting admixed subjects in a very specific way such as recruiting from the same clinic or very nearby geographic location can minimize differences in the degree of admixture. Our results show that the minimal differences of admixture in our SAGE cohort do not confound the genetic association tests. Genetic background in our cohort is similar to previously reported genetic background in northern and western African Americans. Ancestry is likely to be associated with asthma severity. We do not observe an excess of false positives in our genetic association tests. Population stratification does not confound the genetic association tests of β2AR SNPs and asthma in our cohort. Our work supports that the admixture-matched case-control study design is a promising strategy for studying genetic association in admixed populations.
Supplementary Material
Acknowledgements
The support for this manuscript came from: National Institutes of Health K23 HL04464, HL07185, GM61390, NCMHD Health Disparities Scholar, Extramural Clinical Research Loan Repayment Program for Individuals from Disadvantaged Backgrounds, 2001-2003, American Lung Association of California and The National Center on Minority Health and Health Disparities to EGB, American Lung Association of California Research Training Fellowship to HJT, Sandler Center for Basic Research in Asthma and the Sandler Family Supporting Foundation. We would like to acknowledge the families and the patients for their participation. We would also like to thank the numerous health care providers for their support and participation in the SAGE Study. We thank Dr. Mark D. Shriver for assistance in development of the AIMs and for providing ancestral DNA. We thank Dr. Hua Tang for providing the R code implemented a global test for estimating recent admixture. We would like to thank Dr. Neil Risch for his support and guidance. Finally, we would like to thank the Sandler Family Foundation, the main sponsor of this project.
Footnotes
Electronic-Database Information
The URLs for data presented herein are as follows: dbSNP website, National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/SNP/
ADMIX web site, Center of Integrative Genomics, University of Lausanne, http://web.unife.it/progetti/genetica/Isabelle/admix2_0.html
ADMIXMAP website, Conway Institute of Biomolecular and Biomedical Research, http://www.ucd.ie/conway/cv1_324.html
Structure2.1 website, Division of Biological Sciences, University of Chicago, http://pritch.bsd.uchicago.edu/
Reference
- American Thoracic Society Standardization of Spirometry, 1994 Update. Am J Respir Crit Care Med. 1995;152:1107–36. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- The International Study of Asthma and Allergies in Childhood (ISAAC) Steering Committee Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. Lancet. 1998;351:1225–32. [PubMed] [Google Scholar]
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–14. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinbami LJ, Rhodes JC, Lara M. Racial and ethnic differences in asthma diagnosis among children who wheeze. Pediatrics. 2005;115:1254–60. doi: 10.1542/peds.2004-0897. [DOI] [PubMed] [Google Scholar]
- Akinbami LJ, Schoendorf KC. Trends in childhood asthma: prevalence, health care utilization, and mortality. Pediatrics. 2002;110:315–22. doi: 10.1542/peds.110.2.315. [DOI] [PubMed] [Google Scholar]
- Allison DB. Transmission-disequilibrium tests for quantitative traits. Am J Hum Genet. 1997;60:676–90. [PMC free article] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66:1933–1944. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Excoffier L. Inferring admixture proportions from molecular data. Mol Biol Evol. 1998;15:1298–311. doi: 10.1093/oxfordjournals.molbev.a025858. [DOI] [PubMed] [Google Scholar]
- Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet. 2004;68:139–53. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, Sylvia JS, Fagan JK, Salas J, Lilly CM, Ziv E, Selman M, Chapela R, Sheppard D, Weiss ST, Ford JG, Boushey HA, Rodriguez-Cintron W, Drazen JM, Silverman EK. Lower Bronchodilator Responsiveness in Puerto Rican Than in Mexican Asthmatic Subjects. Am J Respir Crit Care Med. 2003a doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003b;348:1170–5. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–9. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Chen X, Kwok PY. Homogeneous genotyping assays for single nucleotide polymorphisms with fluorescence resonance energy transfer detection. Genet Anal. 1999;14:157–63. doi: 10.1016/s1050-3862(98)00016-3. [DOI] [PubMed] [Google Scholar]
- Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, Torres A, Gorman JD, Salari K, Rodriguez-Santana JR, Toscano M, Sylvia JS, Alioto M, Castro RA, Salazar M, Gomez I, Fagan JK, Salas J, Clark S, Lilly C, Matallana H, Selman M, Chapela R, Sheppard D, Weiss ST, Ford JG, Boushey HA, Drazen JM, Rodriguez-Cintron W, Silverman EK, Burchard EG. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med. 2005;171:563–70. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, Matallana H, Avila PC, Casal J, Torres A, Nazario S, Castro R, Battle NC, Perez-Stable EJ, Kwok PY, Sheppard D, Shriver MD, Rodriguez-Cintron W, Risch N, Ziv E, Burchard EG. Population Stratification Confounds Genetic Association Studies of Asthma among Latino Americans. Hum Genet. 2005 doi: 10.1007/s00439-005-0071-3. in press. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theoretical Population Biology. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Bertorelle G. Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol. 2001;18:672–5. doi: 10.1093/oxfordjournals.molbev.a003847. [DOI] [PubMed] [Google Scholar]
- Enright PL, Johnson LR, Connett JE, Voelker H, Buist AS. Spirometry in the Lung Health Study. 1. Methods and quality control. Am Rev Respir Dis. 1991;143:1215–23. doi: 10.1164/ajrccm/143.6.1215. [DOI] [PubMed] [Google Scholar]
- Enright PL, Lebowitz MD, Cockroft DW. Physiologic measures: pulmonary function tests. Asthma outcome. Am J Respir Crit Care Med. 1994;149:S9–18. doi: 10.1164/ajrccm/149.2_Pt_2.S9. discussion S19-20. [DOI] [PubMed] [Google Scholar]
- Evans DA, McLeod HL, Pritchard S, Tariq M, Mobarek A. Interethnic variability in human drug responses. Drug Metab Dispos. 2001;29:606–10. [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–7. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–93. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Fuhlbrigge AL, Kitch BT, Paltiel AD, Kuntz KM, Neumann PJ, Dockery DW, Weiss ST. FEV(1) is associated with risk of asthma attacks in a pediatric population. J Allergy Clin Immunol. 2001;107:61–7. doi: 10.1067/mai.2001.111590. [DOI] [PubMed] [Google Scholar]
- Glass B, Li CC. The dynamics of racial intermixture; an analysis based on the American Negro. Am J Hum Genet. 1953;5:1–20. [PMC free article] [PubMed] [Google Scholar]
- Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–87. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stokowski RP, Patil N, Konvicka K, Kershenobich D, Cox DR, Ballinger DG. Matching strategies for genetic association studies in structured populations. Am J Hum Genet. 2004;74:317–25. doi: 10.1086/381716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JC, Macklem PT, Thurlbeck WM. Site and nature of airway obstruction in chronic obstructive lung disease. N Engl J Med. 1968;278:1355–60. doi: 10.1056/NEJM196806202782501. [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JW, Dunbar PR, Riley GA, Sawyer GM, Fitzharris PF, Pearce N, Le Gros GS, Beasley R. Association of beta2-adrenergic receptor polymorphisms with severe asthma. Clin Exp Allergy. 2000;30:1097–103. doi: 10.1046/j.1365-2222.2000.00929.x. [DOI] [PubMed] [Google Scholar]
- Kitch BT, Paltiel AD, Kuntz KM, Dockery DW, Schouten JP, Weiss ST, Fuhlbrigge AL. A single measure of FEV1 is associated with risk of asthma attacks in long-term follow- up. Chest. 2004;126:1875–82. doi: 10.1378/chest.126.6.1875. [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Lin CC, Rogot E, Johnson NJ, Sorlie PD, Arias E. A further study of life expectancy by socioeconomic factors in the National Longitudinal Mortality Study. Ethn Dis. 2003;13:240–7. [PubMed] [Google Scholar]
- Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma--United States, 1980-1999. MMWR Surveill Summ. 2002;51:1–13. [PubMed] [Google Scholar]
- Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100:3184–8. doi: 10.1172/JCI119874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Hoggart CJ, Bonilla C, Dios S, Norris JM, Marshall JA, Hamman RF, Ferrell RE, McKeigue PM, Shriver MD. Relation of type 2 diabetes to individual admixture and candidate gene polymorphisms in the Hispanic American population of San Luis Valley, Colorado. J Med Genet. 2004;41:e116. doi: 10.1136/jmg.2004.018887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce N, Sunyer J, Cheng S, Chinn S, Bjorksten B, Burr M, Keil U, Anderson HR, Burney P. Comparison of asthma prevalence in the ISAAC and the ECRHS. ISAAC Steering Committee and the European Community Respiratory Health Survey. International Study of Asthma and Allergies in Childhood. Eur Respir J. 2000;16:420–6. doi: 10.1183/9031936.00.16337700. [DOI] [PubMed] [Google Scholar]
- Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theor Popul Biol. 2001;60:227–37. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Phong A, Burchard EG, Harris TB, Psaty BM, Kwok PY. Population structure, admixture, and aging- related phenotypes in African American adults: the Cardiovascular Health Study. Am J Hum Genet. 2005;76:463–77. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MA, Winkleby MA, Ahn D, Sundquist J, Kraemer HC. Identification of population subgroups of children and adolescents with high asthma prevalence: findings from the Third National Health and Nutrition Examination Survey. Arch Pediatr Adolesc Med. 2002;156:269–75. doi: 10.1001/archpedi.156.3.269. [DOI] [PubMed] [Google Scholar]
- Romieu I, Mannino DM, Redd SC, McGeehin MA. Dietary intake, physical activity, body mass index, and childhood asthma in the Third National Health And Nutrition Survey (NHANES III) Pediatr Pulmonol. 2004;38:31–42. doi: 10.1002/ppul.20042. [DOI] [PubMed] [Google Scholar]
- Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, Coyle NE, Ung N, Nazario S, Casal J, Torres-Palacios A, Clark S, Phong A, Gomez I, Matallana H, Perez-Stable EJ, Shriver MD, Kwok PY, Sheppard D, Rodriguez-Cintron W, Risch NJ, Burchard EG, Ziv E. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol. 2005;29:76–86. doi: 10.1002/gepi.20079. [DOI] [PubMed] [Google Scholar]
- Schlesselman JJ, Stolley PD. Case control studies: design, conduct, analysis. Oxford University Press; New York: 1982. [Google Scholar]
- Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, Schurr TG, Zhadanov SI, Osipova LP, Brutsaert TD, Friedlaender J, Jorde LB, Watkins WS, Bamshad MJ, Gutierrez G, Loi H, Matsuzaki H, Kittles RA, Argyropoulos G, Fernandez JR, Akey JM, Jones KW. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–99. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- Silverman EK, Kwiatkowski DJ, Sylvia JS, Lazarus R, Drazen JM, Lange C, Laird NM, Weiss ST. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol. 2003;112:870–6. doi: 10.1016/s0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Inbreeding coefficients and coalescence times. Genet Res. 1991;58:167–75. doi: 10.1017/s0016672300029827. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Tsai HJ, Choudhry S, Naqvi M, Rodriguez-Cintron W, Burchard EG, Ziv E. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum Genet. 2005 doi: 10.1007/s00439-005-0067-z. in press. [DOI] [PubMed] [Google Scholar]
- Wacholder S, Rothman N, Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:513–20. [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations. Volume 2: The theory of gene frequencies. University of Chicago Press; 1969. [Google Scholar]
- Zhang S, Zhu X, Zhao H. On a semiparametric test to detect associations between quantitative traits and candidate genes using unrelated individuals. Genet Epidemiol. 2003;24:44–56. doi: 10.1002/gepi.10196. [DOI] [PubMed] [Google Scholar]
- Ziv E, Burchard EG. Human population structure and genetic association studies. Pharmacogenomics. 2003;4:431–41. doi: 10.1517/phgs.4.4.431.22758. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR, Kennedy SH. What makes a good case-control study? Design issues for complex traits such as endometriosis. Hum Reprod. 2002;17:1415–23. doi: 10.1093/humrep/17.6.1415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.