Abstract
DNA mismatches represent a novel target in developing diagnostics and therapeutics for cancer, since deficiencies in DNA-mismatch repair (MMR) are implicated in many cancers and cells that are MMR-deficient show a high frequency of mismatches. We use metal complexes with bulky intercalating ligands serve as probes for DNA mismatches. Here, we report the high resolution (0.92 Å) crystal structure of the ruthenium ‘light switch’ complex Δ-Ru(bpy)2dppz2+ (bpy = 2,2′-bipyridine and dppz = dipyridophenazine), known to show luminescence on binding to duplex DNA, bound to both mismatched and well matched sites in the oligonucleotide 5′-(dCGGAAATTACCG)2-3′ (underline denotes AA mismatches). Two crystallographically independent views reveal that the complex binds mismatches through metalloinsertion, where the dppz inserts into the duplex through the minor groove, ejecting both mispaired adenosines. Additional ruthenium complexes are intercalated at well-matched sites, creating an array of complexes in the minor groove stabilized through stacking interactions between bpy ligands and extruded adenosines. This structure attests to the generality of metalloinsertion and metallointercalation as DNA binding modes.
Deficiencies in DNA mismatch repair (MMR) have been linked to increased rate of mutation and several types of cancers.1-7 Detection of MMR deficiency typically relies on assessment of markers for microsatellite instability, promoter hypermethylation and/or immunohistological staining of MMR proteins.8-10 These methods may not be applicable to all cancers, and more than one MMR protein needs to be considered. On the other hand, all forms of MMR deficiency are expected to show elevated levels of DNA mismatches, which by themselves would be a persistent and universal target for diagnostic agents. Luminescent, mismatch-targeting small molecules are thus ideally suited to become a direct, fast, and sensitive detection method for MMR deficiency in biological samples. We have previously discovered that the metal complex Ru(bpy)2dppz2+ (where bpy = 2,2′-bipyridine and dppz = dipyrido[3,2-a:2′,3′-c]phenazine) shows enhanced luminescence in the presence of base mismatches and abasic sites.11 Its augmented luminescence sensitivity to mismatches makes it a promising parent complex for the design of luminescence-based mismatch sensors. Further development necessitates a thorough structural understanding of the interactions between the ruthenium complex and DNA. Here, we report the atomic resolution structure of Δ-Ru(bpy)2dppz2+ (1) cocrystallized with a 12-mer oligonucleotide containing two adenosine-adenosine mismatches, and examine in detail the binding interactions between the metal complex and DNA.

Dppz complexes of ruthenium have been widely studied due to their unique photophysical responses to DNA. Typically, emission from these complexes is extremely weak in water, but their luminescence is significantly enhanced upon binding to double-stranded DNA, hence the “light switch” effect.12 Extensive studies in solution have established that these complexes bind to DNA by intercalation through the planar dppz ligand.13-17 Some possible structures have been put forward through theoretical calculations;18-19 however, due to the lack of site-specificity in DNA binding, solution and crystal structures have largely remained elusive. Although the discovery of the unique photophysical properties of this class of complexes was made over two decades ago, the first crystal structure of a dppz complex bound to DNA was not obtained until very recently, but it did not capture dppz intercalation into a native DNA duplex.20
Besides binding to well-matched DNA, Ru(bpy)2dppz2+ shows further enhanced luminescence in the presence of DNA defects such as base mismatches.11 We have proposed that the binding of Ru(bpy)2dppz2+ to mismatches occurs by metalloinsertion.11 This binding mode has been elucidated in crystal structures of the mismatch-targeting rhodium complex, Δ-Rh(bpy)2chrysi3+ (where chrysi = chrysene-5,6-quinone diimine), bound to an AC or AA mismatch, as well as in solution NMR studies with a CC mismatch.21-24 In this binding mode, the intercalating chrysi ligand inserts into the mismatch site from the minor groove and extrudes the mispaired bases out of the helix, effectively taking their place in the base stack. Here we show that in the ruthenium-DNA crystal structure, just like the rhodium complex, Ru(bpy)2dppz2+ binds to mismatches also through metalloinsertion, the inserting ligand is dppz. The crystal structure described herein, at 0.92-Å resolution, provides several independent views of ruthenium binding to DNA through dppz intercalation and insertion, illustrating the structural basis of the interactions between DNA and the “light switch” molecule as captured by cocrystallization.
RESULTS AND DISCUSSION
Cocrystallization of Δ-Ru(bpy)2dppz2+ with DNA
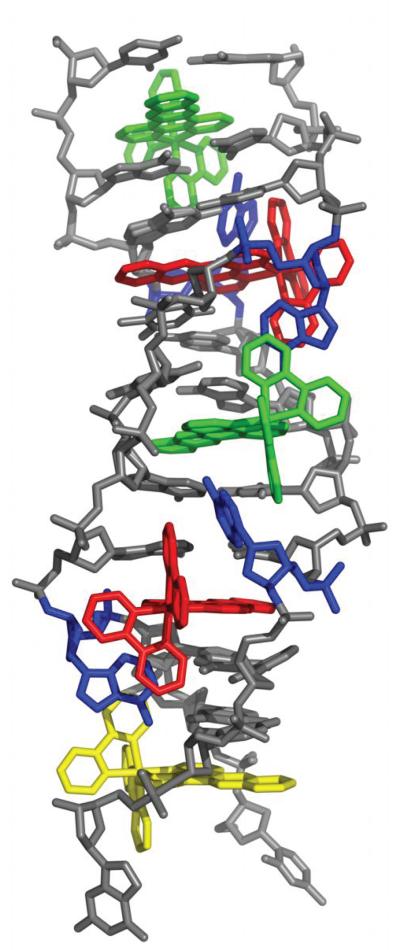
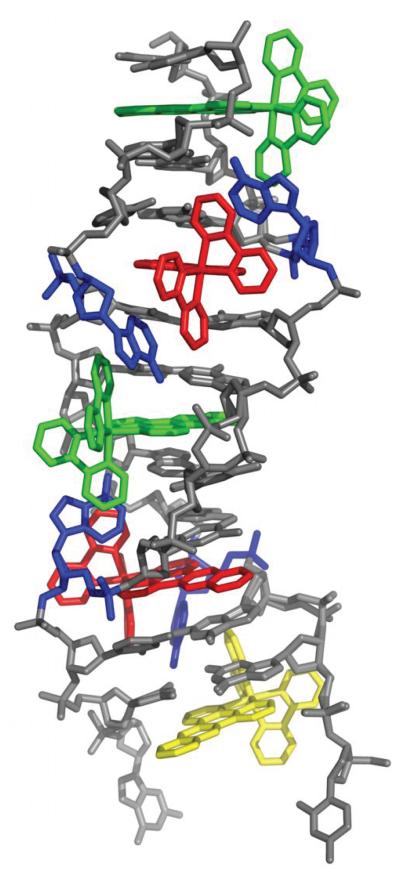
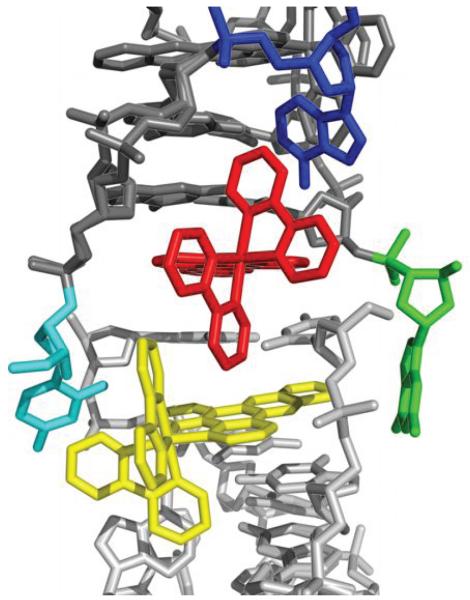
To elucidate the structural basis for Ru(bpy)2dppz2+ interaction with DNA, we cocrystallized Δ-Ru(bpy)2dppz2+ (1) with the 12-mer palindromic DNA sequence, d(C1G2G3A4A5A6T7T8A9C10C11G12)2. This sequence contains two AA mismatches (underlined) and has been previously cocrystallized with Δ-Rh(bpy)2chrysi3+.22 We considered that similar binding of the complex at the mismatched site might yield well defined crystals. Crystals took longer than two months to appear in the crystallization wells, however. The metal complex and the oligonucleotide cocrystallized in space group P1 (see Supplementary Table S1 for data collection and refinement statistics). The asymmetric unit contains one double strand of DNA with five bound ruthenium complexes. The structure, at atomic resolution, revealed three binding modes of the ruthenium complex: (i) metalloinsertion at the mismatched sites with ejection of the mispaired adenosines, (ii) metallointercalation between well-matched base pairs and (iii) end-capping between two (crystallographically-related) duplexes (Figure 1).
Figure 1.
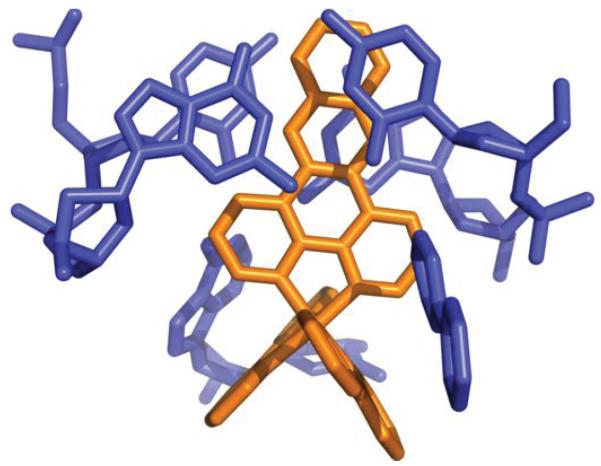
Structure of Δ-Ru(bpy)2dppz2+ (1) bound to the oligonucleotide 5′-C1G2G3A4A5A6T7T8A9C10C11G12-3′ shown in a front view (a) and rotated 90 degrees around the helix axis. Three DNA-binding modes are observed: (i) metalloinsertion, whereby the ruthenium complex (red) inserts the dppz ligand into the DNA duplex (gray) at the mismatched sites through the minor groove, extruding the mispaired adenosines (blue), (ii) metallointercalation, whereby the complex (green) binds between two well matched base pairs, and (iii) end-capping, whereby the complex (yellow) stacks with the terminal Watson-Crick pair of the duplex.
Metalloinsertion at mismatched sites
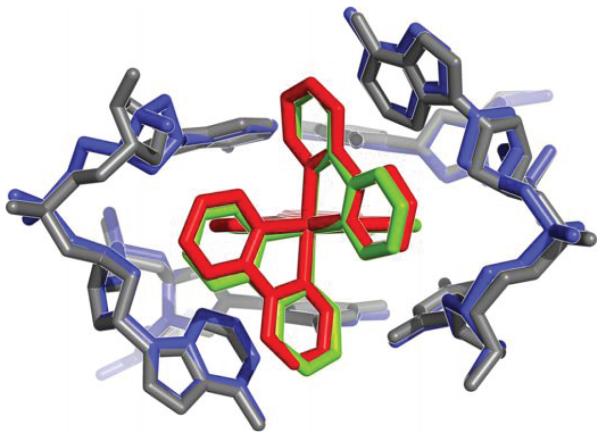
At the two destabilized AA mismatches, the metal complex inserts deeply from the minor groove and fully ejects the mispaired adenosines (Figure 1 and Supplementary Figure S1). We had proposed that Ru(bpy)2dppz2+ binds to DNA mismatches through insertion from the minor groove, and that metalloinsertion may be a general binding mode for octahedral metal complexes bearing planar ligands.11 Increased luminescence is found with a range of DNA mismatches, and, as with the Rh complexes, correlates with thermodynamic instability of the mismatch. Here, consistent with our proposal, the ruthenium complex indeed binds tightly to both mismatched sites through metalloinsertion, with the dppz ligand stacked between the two flanking base pairs, effectively replacing the mispaired adenosines in the base stack. All four ejected adenosines are folded back in the minor groove. They adopt the syn conformation and stack with the very ruthenium that is inserted at their respective mismatched site. An overlay of the two separate metalloinsertion sites shows that the local geometry of the DNA and the relative orientation of the ruthenium complex are highly conserved between the two sites (Figure 2). The dppz is inserted in a head-on fashion, positioned halfway between the phosphate backbones, both phenazine nitrogens are well protected from solvent access. On the other hand, if we only consider the relative orientation of the dppz with respect to the base pairs above and below the insertion site, the dppz is inserted at an angle with respect to the dyad axis of either flanking base pair. Compared to the chrysi ligand, dppz is narrower and symmetric about its long axis. As a result, while the DNA minor groove widens significantly in the rhodium structures to accommodate the sterically expansive and asymmetric chrysi complex,21,22 it does so to a lesser extent in the case of the ruthenium complex (Supplementary Figure S2).
Figure 2.
Two independent views of metalloinsertion at the mismatched sites. a, Superposition of the two independent views of metalloinsertion by the ruthenium complex at the mismatched sites, as viewed from the minor groove (A4-A9 site: ruthenium complex in red and DNA in gray, A9-A4 site: ruthenium complex in green and DNA in blue). The ruthenium complex inserts the dppz ligand from the minor groove and extrudes the mismatched adenosines, which are folded back into the minor groove. The two binding sites were superimposed using only the DNA backbone atoms (rmsd of 42 atoms = 0.607 Å). b, Superimposed metalloinsertion sites as viewed down the helical axis.
Intercalation at matched sites
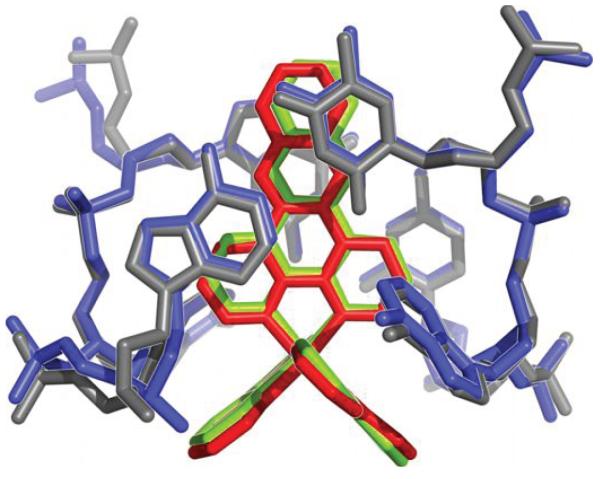
Of the three remaining ruthenium complexes, two are intercalated between well-matched base pairs, also through the minor groove. This mode of intercalation, namely from the minor groove, contrasts what was inferred from NMR and competitive fluorescence studies, in which we had deduced a major groove preference for metallointercalation;14,17,25 linear dichroism studies did, however, suggest minor groove association15,16. We note that in this structure, intercalation occurs in conjunction with stacking interactions between an ancillary bpy ligand of the intercalating complex and either an extruded adenosine or a bpy ligand from a neighboring complex. These stacking interactions serve to stabilize the intercalated complex in the minor groove. These contrasting results indicate that the energetic difference between intercalation from the major groove versus the minor groove must be small. Computational studies of dppz complexes intercalated into a dinucleotide step also support the notion of very small differences in intercalation energetics from the two grooves.18 In this intercalative binding mode, the dppz ligand is positioned inside the base stack also in a head-on fashion. The intercalation of the dppz ligand is so deep that the end most distal from the ruthenium center protrudes into the major groove. Comparison between the two independent intercalation sites reveals subtle differences in the relative position of the complex (Figure 3). In the 5′-A6T7-3′ step, the dppz is right in the middle of the two strands and intercalated more deeply, with most of the stacking interactions formed between the central ring of the phenazine portion and the bases. For the 5′-C1G2-3′ step, the dppz is closer to one strand than the other, and both the distal and the central rings of the phenazine are involved in stacking.
Figure 3.
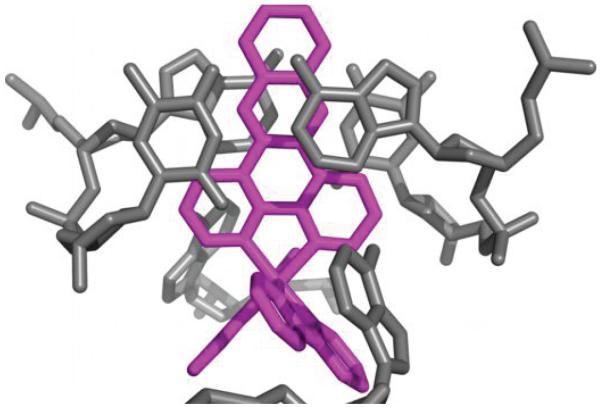
Two independent views of metallointercalation at well-matched sites. a, The ruthenium complex intercalates at the 5′-C1G2-3′ step through the dppz ligand (Ru in orange, DNA and bpy from a neighboring ruthenium in blue). b, Metallointercalation at the 5′-A6T7 -3′ step (Ru in magenta, DNA in gray).
Crystal packing
The fifth ruthenium complex is sandwiched between two crystallographically related duplexes (Figure 4). The last GC base pair becomes a frayed end, with the cytidine and the guanosine pointing down into the minor and major groove of the next duplex, respectively. The dppz ligand of the end-capping ruthenium complex effectively replaces this terminal base pair in the helix, providing an anchor for the next duplex to stack upon. Crystallographically related duplexes are stacked head to tail, forming parallel long rods in the crystal lattice (Supplementary Figure S3). Aside from the end-stacking interactions between consecutive duplexes, mediated by the end-capping metal complex, neighboring parallel rods do not have detectable contacts with one another.
Figure 4.
The end-capping complex. The duplex (dark gray) is end-capped by the ruthenium complex (red), which stacks between an extruded adenosine (blue) and the first complex (yellow) in a crystallographically related duplex (light gray). The last GC base pair (cytidine in cyan and guanosine in green) forms a frayed end.
Local distortions of the oligonucleotide duplex
Throughout the DNA helix, the minor groove is densely populated with alternating metal complexes and extruded adenosines. The five metal complexes are evenly spaced, binding to DNA at every two base steps. The four mismatched adenosines are sandwiched between the five metal complexes, and the last metal complex makes contact with the first one in the next repeating unit. The DNA maintains its B-form, albeit with some local deviations from ideal B-form geometry (Table 1 and Supplementary Table S2). The overall structure is slightly bent toward the major groove, similar to what we have observed in the rhodium structure with the identical DNA sequence.22 All base pairs show some degree of unwinding to accommodate inserted and intercalated complexes (Table 1), as expected, since unwinding of DNA is usually associated with intercalators.26-28 Most sugar puckers are C2′-endo or the closely related C1′-exo (or C3′-exo, Supplementary Table S2), suggesting that alternating C2′-endo/C3′-endo conformations26 are not required for intercalation. The rise is approximately doubled at each ruthenium binding site as the metal complex serves as an additional base pair in the helix, but interestingly, the rise between native consecutive base pairs are less than 3.3 Å (Table 1). This compression of the vertical space between consecutive base pairs may be an indication that stacking between the bpy ligands and extruded adenosines is a dominating interaction, such that the base pairs between the intercalation sites have to shorten their rise in order for the adenosines and bpy ligands to reach each other. The average distance between a bpy and an adenosine is in fact 3.3 Å. This is consistent with our notion that the adenosine-bpy stacking may be driving ruthenium intercalation from the minor groove. But importantly, the adenosines are extruded in the first place because ruthenium complexes are inserted at the mismatched sites, and metalloinsertion has been generally apparent through the minor groove.21,22
Table 1.
| Step | Ru binding mode |
Shift (Å) | Slide (Å) | Rise (Å) | Tilt (°) | Roll (°) | Twist (°) |
|---|---|---|---|---|---|---|---|
| C1/G2 | intercalation | 0.1 | 2.2 | 6.3 | 12.5 | 5.5 | 18.1 |
| G2/G3 | - | 0.9 | 0.3 | 2.9 | −3.0 | −5.4 | 30.4 |
| G3/A5 | insertion | 0.6 | 3.5 | 7.0 | 5.4 | 16.6 | 71.7 |
| A5/A6 | - | −0.8 | −0.1 | 3.0 | −0.7 | −0.7 | 23.2 |
| A6/T7 | intercalation | −0.2 | 0.6 | 7.1 | −1.7 | 7.6 | 23.5 |
| T7/T8 | - | 0.4 | −0.4 | 2.8 | −3.4 | 6.6 | 23.6 |
| T8/C10 | insertion | −0.9 | 3.4 | 7.3 | −7.5 | 9.0 | 70.2 |
| C10/C11 | - | −1.0 | −0.3 | 3.0 | 3.2 | −7.5 | 23.4 |
| B-DNA | - | −0.1 | −0.8 | 3.3 | −1.3 | −3.6 | 36 |
Geometrical relationships between consecutive base pairs: shift, translation into the groove, slide, translation toward the phosphodiester backbone, rise, translation along the helix axis, tilt, rotation about the pseudo-twofold axis relating the DNA strands, roll, rotation about a vector between the C1′ atoms, and twist, rotation about the helix axis.
Data were calculated using 3DNA.41
Comparison with other structures
The first crystal structure of a dppz complex bound to DNA is that of the λ-enantiomer of Ru(TAP)2dppz2+ (TAP = 1,4,5,8-tetraazaphenanthrene) bound to a 10-mer oligonucleotide.20 The ruthenium complex is found to bind through semi-intercalation of a TAP ligand between two GC base pairs, as well as intercalation of the dppz ligand between a GC and a reverse Watson-Crick-paired terminal AT pair, with the adenine and thymine coming from symmetry-related strands. Besides having a different ancillary ligand, this complex is of the opposite chirality of our ruthenium complex. We have demonstrated in our structure that the Δ-isomer intercalates through the dppz ligand between natively well-matched base pairs, but this mode of binding is absent in the TAP structure. This is consistent with early solution studies of the binding of intercalative metal complexes to DNA: the ancillary ligands of the λ-isomer would be sterically repelled by the backbone of right-handed B-form DNA, while the Δ-isomer has the correct symmetry to fit in the grooves.29 In the TAP structure, the DNA duplex adopts an overall B-form despite large local distortions, and dppz intercalation occurs only at the interface between two duplexes, which are positioned perpendicular to each other rather than form a continuous helix. At the same time, the semi-intercalation of TAP induces a severe kink in the DNA. For the TAP ligand, full intercalation from the minor groove may be more difficult for the λ–configuration. With bpy ligands metalloinsertion is highly favored for the Δ-isomer, but only small enantiomeric discrimination occurs with intercalation.24 Indeed, the Δ-isomer, as shown in the structure reported herein, binds avidly to the right-handed helix through full intercalation. We may also compare our structure with that of Δ-Rh(Me2trien)phi3+ (Me2trien = 2R,9R-diamino-4,7-diazadecane) intercalated in an 8-mer oligonucleotide.27 The rhodium structure shows only one complex bound per 8-mer duplex, as opposed to five ruthenium molecules per 12-mer duplex in our structure. Functional groups were installed on the rhodium complex to form sequence-specific contacts with the DNA in the major groove, hence the single-site binding at a specific step in the base stack. The complex Ru(bpy)2dppz2+, on the other hand, binds nonspecifically to DNA. Thus, binding at multiple sites along the duplex is observed in the ruthenium structure. The sequence context of the intercalation or insertion sites – pyrimidine-purine (5′-C1G2-3′), pyrimidine-pyrimidine (5′-T8C10-3′), purine-purine (5′-G3A5-3′), and purine-pyrimidine (5′-A6T7-3′) – also speaks to the non-specific nature of DNA binding by the ruthenium complex.
Solution luminescence
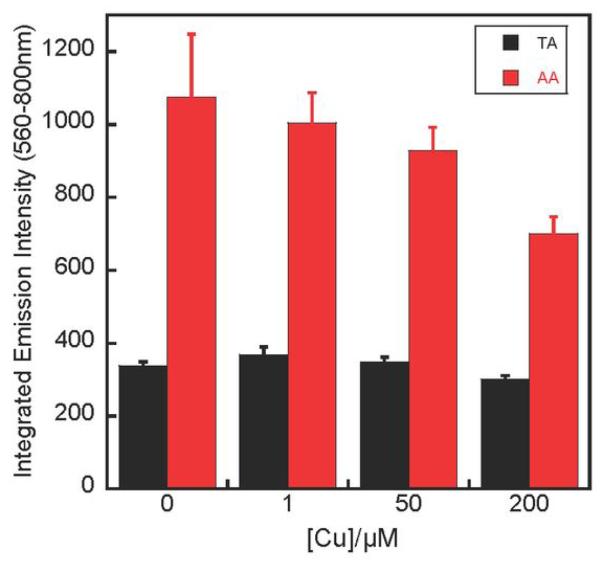
We measured the solution luminescence of Δ-Ru(bpy)2dppz2+ (1) bound to the mismatched oligonucleotide, as well as to a fully complementary 12-mer oligonucleotide d(CGGTAATTACCG)2, in order to determine if the crystal structure reflects binding preferences in solution. As the AA mismatch-containing duplex has a low melting temperature of 22°C, the experiments were conducted at 4°C to ensure all DNA strands are properly hybridized. The luminescence from Δ-Ru(bpy)2dppz2+ (1) bound to mismatched DNA is about three times the luminescence with the equivalent 12-mer well-matched duplex (Figure 5), consistent with previously observed higher sensitivity of Ru(bpy)2dppz2+ luminescence response to mismatched DNA.
Figure 5.
Solution luminescence. Plot of integrated emission intensity (λex = 440 nm) of 1 μM Δ-Ru(bpy)2dppz2+ (1) with increasing concentration of Cu(phen)22+ in the presence of 12-mer mismatched (AA) and well-matched DNA (TA, 1 μM). Error bars indicate standard deviations in the measurements.
To determine the groove preference of ruthenium complex binding, we employed the minor groove-specific quencher, Cu(phen)22+,30,31 to quench the luminescence from the ruthenium complex (Figure 5). Quenching by paramagnetic Cu(phen)22+ is expected rather than a direct competition for binding sites; Cu(phen)22+, a non-specific groove binder, binds several orders of magnitude more weakly to DNA than the intercalative ruthenium complex. The luminescence associated with mismatched DNA is significantly quenched (by 34%) with increasing concentrations of Cu(phen)22+, while luminescence associated with the matched sequence is quenched to a much lesser extent (12%). This differential quenching is consistent with the mismatch-bound ruthenium complexes being located in the minor groove, but those bound to well-matched DNA are mostly in the major groove. Therefore, although the crystal structure provides a very detailed picture of metalloinsertion and metallointercalation in the minor groove, it may not capture intercalation events occurring in the major groove. Perhaps the inherently dynamic nature of ruthenium intercalation from the major groove, as reflected in the fast exchange and multiple binding conformations revealed in NMR studies, hinders the formation of well-packed crystals. Nonetheless, the crystal structure still provides invaluable insight into intercalation of Δ-Ru(bpy)2dppz2+ (1) when it is in the minor groove.
CONCLUSIONS
The structure presented here at 0.92 Å-resolution depicts in detail the versatile binding modes attainable for octahedral metal complexes bearing an intercalating ligand. It shows two independent views of metalloinsertion, two of intercalation, and one of end-capping. At destabilized regions of the DNA, the metal complex binds through metalloinsertion in the minor groove, accompanied by extrusion of the mismatched bases. This binding mode was previously observed with a sterically expansive ligand, but this structure clearly demonstrates that a narrower ligand such as dppz is equally capable of recognizing mismatches by metalloinsertion, pointing to the generality of this binding mode. The smaller size of the dppz ligand allows the ruthenium complex also to bind through classical intercalation between two consecutive well-matched base pairs. Curiously, intercalated complexes are located in the minor groove as well, which we hypothesize is stabilized by extensive ancillary interactions; given some major groove intercalation in solution, binding from the major and minor groove must be energetically similar. This discrepancy notwithstanding, the crystal structure attests to the remarkable structural flexibility of DNA upon high-density ligand binding, illustrates the nuanced binding geometries sampled by a non-covalently bound small molecule, and highlight the dominance of metalloinsertion as the preferred binding mode to destabilized regions of DNA. We hope these newly garnered structural understandings will help guide the development of future generations of metal complexes as chemical tools and medicinal agents.
METHODS
Materials
[Ru(bpy)2dppz]Cl2 was synthesized according to previously reported procedures.32 The enantiomers were separated using a CYCLOBOND I 2000 DMP HPLC column (Sigma) on a Hewlett-Packard 1100 HPLC, with an isocratic solvent composition of 60/40 (v/v) CH3CN:100 mM KPF6 (aq). The Δ-enantiomer eluted first, followed by the λ-isomer. The assignment of the two fractions was confirmed by circular dichroism.33 The fractions were lyophilized and washed with water to remove excess KPF6 and exchanged for chloride salt on a QAE anion-exchange column. Oligonucleotides (Integrated DNA Technologies) were purified by reverse-phase HPLC using a C18 reverse-phase column (Varian) on a Hewlett-Packard 1100 HPLC. Quantification was performed on a Beckman DU 7400 spectrophotometer.
Crystallization and data collection
Oligonucleotides were incubated with Δ-[Ru(bpy)2dppz]Cl2 (1) before crystallization. Subsequent manipulations were performed with minimal exposure of the complex to light. The crystal was grown from a solution of 1 mM d(C1G2G3A4A5A6T7T8A9C10C11G12)2, 2 or 3 mM enantiomerically pure Δ-Ru(bpy)2dppz2+ (1), 20 mM sodium cacodylate (pH 7.0), 6 mM sperminetetrahydrochloride, 40 mM NaCl or KCl, 10 mM BaCl2, and 5% 2-methyl-2,4-pentanediol (MPD) equilibrated in sitting drops versus a reservoir of 35% MPD at ambient temperature. The crystals grew in space group P1 and unit cell dimensions: a = 24.039 Å, b = 24.797 Å, c = 37.521 Å, α = 74.669°, β = 84.416°, and γ = 76.208° (Supplementary Table S1).
Data were collected from a flash-cooled crystal at 100 K on an R-axis IV image plate using Cu Kα radiation produced by a Rigaku RU-H3RHB rotating-anode generator with double-focusing mirrors and a Ni filter. High-resolution data were subsequently collected from a different crystal on beamline 12-2 at the Stanford Synchrotron Radiation Laboratory (Menlo Park, CA, λ = 0.7749 Å, 100 K, PILATUS 6M detector). The data were processed with or XDS34, and SCALA from the CCP4 suite of programs.35
Structure determination and refinement
The structure was determined by single anomalous dispersion phasing using the anomalous scattering of ruthenium with the Shelxc/d/e suite of programs.36 Five heavy atoms were located per asymmetric unit. The model was built in COOT37 and refined with PHENIX version 1.7.38 The anomalous contribution of ruthenium was taken into account and alternative conformations of phosphates were included in the refinement. Atomic displacement factors have been refined anisotropically. Figures were drawn with Pymol.39 Alignment was performed with LSQMAN.40 The coordinates and structure factors have been deposited in RCSB Protein Data Bank (PDB ID: 4E1U).
Steady state fluorescence
Luminescence spectra (excitation wavelength = 440 nm) with emission intensities ranged from 560 to 800 nm were measured in 40 mM sodium cacodylate (pH 7.0), 80 mM KCl, 20 mM BaCl2 on an ISS-K2 spectrophotometer at 4°C in aerated solutions. Cu(phen)22+ was formed in situ using 1:3 CuCl2 and phenanthroline. Experiments were performed in triplicate.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. C. Virgil for assistance in the separation of enantiomers, and D. C. Rees and J. A. Hoy for valuable discussions. We are grateful to the National Institutes of Health (NIH GM33309 to J. K. B.) for their financial support and the Tobacco-Related Disease Research Program (TRDRP) for a Dissertation Research Award to H. S. We acknowledge the Gordon and Betty Moore Foundation and Sanofi-Aventis Bioengineering Research Program at Caltech for support of the X-ray Facility at the Caltech Molecular Observatory. The rotation camera facility at Stanford Synchrotron Radiation Laboratory is supported by the U.S. Department of Energy and NIH.
Footnotes
AUTHOR CONTRIBUTIONS J.K.B. and H.S. designed the research. H.S. carried out crystallization and solution luminescence experiments. J.T.K. and H.S. solved the crystal structure. H.S. and J.K.B. wrote the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
REFERENCES
- 1.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- 2.Strauss BS. Frameshift mutation, microsatellites and mismatch repair. Mutat. Res. 1999;437:195–203. doi: 10.1016/s1383-5742(99)00066-6. [DOI] [PubMed] [Google Scholar]
- 3.Papadopoulos N, Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Hum. Mutat. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Peltomaki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 5.Lawes DA, SenGupta S, Boulos PB. The clinical importance and prognostic implications of microsatellite instability in sporadic cancer. Eur. J. Surg. Oncol. 2003;29:201–212. doi: 10.1053/ejso.2002.1399. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharyya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Mutator phenotypes in human colorectal carcinoma cell lines. Proc. Natl. Acad. Sci. USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arzimanoglou II, Gilbert F, Barber HRK. Microsatellite instability in human solid tumors. Cancer. 1998;82:1808–1820. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 8.Boland CR, et al. A national cancer institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 9.de la Chapelle A. Microsatellite instability. N. Engl. J. Med. 2003;349:209–210. doi: 10.1056/NEJMp038099. [DOI] [PubMed] [Google Scholar]
- 10.Rosen DG, Cai KQ, Luthra R, Liu J. Immunohistochemical staining of hMLH1 and hMSH2 reflects microsatellite instability status in ovarian carcinoma. Mod Pathol. 2006;19:1414–1420. doi: 10.1038/modpathol.3800672. [DOI] [PubMed] [Google Scholar]
- 11.Lim MH, Song H, Olmon ED, Dervan EE, Barton JK. Sensitivity of Ru(bpy)2dppz2+ luminescence to DNA defects. Inorg. Chem. 2009;48:5392–5397. doi: 10.1021/ic900407n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. A molecular light switch for DNA: Ru(bpy)2(dppz)2+ J. Am. Chem. Soc. 1990;112:4960–4962. [Google Scholar]
- 13.Hiort C, Lincoln P, Norden B. DNA binding of Δ- and λ-[Ru(phen)2DPPZ]2+ J. Am. Chem. Soc. 1993;115:3448–3454. [Google Scholar]
- 14.Dupureur CM, Barton JK. Use of selective deuteration and 1H NMR in demonstrating major groove binding of Δ-[Ru(phen)2dppz]2+ to d(GTCGAC)2. J. Am. Chem. Soc. 1994;116:10286–10287. [Google Scholar]
- 15.Haq I, et al. Interaction of Δ- and λ-[Ru(phen)2DPPZ]2+ with DNA: a calorimetric and equilibrium binding study. J. Am. Chem. Soc. 1995;117:4788–4796. [Google Scholar]
- 16.Lincoln P, Broo A, Norden B. Diastereomeric DNA-binding geometries of intercalated ruthenium(II) trischelates probed by linear dichroism: [Ru(phen)2DPPZ]2+ and [Ru(phen)2BDPPZ]2+ J. Am. Chem. Soc. 1996;118:2644–2653. [Google Scholar]
- 17.Dupureur CM, Barton JK. Structural studies of λ- and Δ-[Ru(phen)2dppz]2+ bound to d(GTCGAC)2: characterization of enantioselective intercalation. Inorg. Chem. 1997;36:33–43. [Google Scholar]
- 18.Ambrosek D, Loos P-F, Assfeld X, Daniel C. A theoretical study of Ru(II) polypyridyl DNA intercalators: structure and electronic absorption spectroscopy of [Ru(phen)2(dppz)]2+ and [Ru(tap)2(dppz)]2+ complexes intercalated in guanine– cytosine base pairs. J. Inorg. Biochem. 2010;104:893–901. doi: 10.1016/j.jinorgbio.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Vargiu AV, Magistrato A. Detecting DNA mismatches with metallo-insertors: a molecular simulation study. Inorg. Chem. 2012;51:2046–2057. doi: 10.1021/ic201659v. [DOI] [PubMed] [Google Scholar]
- 20.Hall JP, et al. Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand. Proc. Natl Acad. Sci. USA. 2011;108:17610–17614. doi: 10.1073/pnas.1108685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierre VC, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl Acad. Sci. USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeglis BM, Pierre VC, Kaiser JT, Barton JK. A bulky rhodium complex bound to an adenosine-adenosine DNA mismatch: general architecture of the metalloinsertion binding mode. Biochemistry. 2009;48:4247–4253. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordier C, Pierre VC, Barton JK. Insertion of a bulky rhodium complex into a DNA cytosine-cytosine mismatch: an NMR solution study. J. Am. Chem. Soc. 2007;129:12287–12295. doi: 10.1021/ja0739436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeglis BM, Pierre VC, Barton JK. Metallo-intercalators and metallo-insertors. Chem. Commun. (Camb.) 2007;44:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmlin RE, Stemp EDA, Barton JK. Ru(phen)2dppz2+ luminescence: dependence on DNA sequences and groove-binding agents. Inorg. Chem. 1998;37:29–34. doi: 10.1021/ic970869r. [DOI] [PubMed] [Google Scholar]
- 26.Sobell HM, Tsai C-C, Jain SC, Gilbert SG. Visualization of drug-nucleic acid interactions at atomic resolution: III. Unifying structural concepts in understanding drug-DNA interactions and their broader implications in understanding protein-DNA interactions. J. Mol. Biol. 1977;114:333–365. doi: 10.1016/0022-2836(77)90254-6. [DOI] [PubMed] [Google Scholar]
- 27.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Structure of a photoactive rhodium complex intercalated into DNA. Nat. Struct. Mol. Biol. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 28.Adams A, Guss JM, Denny WA, Wakelin LPG. Crystal structure of 9- amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide bound to d(CGTACG)2: implications for structure–activity relationships of acridinecarboxamide topoisomerase poisons. Nucleic Acids Res. 2002;30:719–725. doi: 10.1093/nar/30.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton JK. Metals and DNA: molecular left-handed complements. Science. 1986;233:727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- 30.Sigman DS, Chen C-HB. Chemical nucleases: new reagents in molecular biology. Annu. Rev. Biochem. 1990;59:207–236. doi: 10.1146/annurev.bi.59.070190.001231. [DOI] [PubMed] [Google Scholar]
- 31.Lim MH, Lau IH, Barton JK. DNA strand cleavage near a CC mismatch directed by a metalloinsertor. Inorg. Chem. 2007;46:9528–9530. doi: 10.1021/ic701598k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amouyal E, Homsi A, Chambron J-C, Sauvage J-P. Synthesis and study of a mixed-ligand ruthenium(II) complex in its ground and excited states: bis(2,2′- bipyridine)(dipyrido[3,2-a:2′,3′-c]phenazine-N4N5)ruthenium(II) J. Chem. Soc., Dalton Trans. 1990:1841–1845. [Google Scholar]
- 33.Liu J-G, et al. Enantiomeric ruthenium(II) complexes binding to DNA: binding modes and enantioselectivity. J. Biol. Inorg. Chem. 2000;5:119–128. doi: 10.1007/s007750050015. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. XDS. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project Number 4 The ccp4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrodinger LLC. The PyMOL Molecular Graphics System. 2010.
- 40.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 41.Lu X, Olson WK. DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.