Abstract
Background
Covalent linkage of the ubiquitin-like protein ISG15 interferes with viral infection and USP18 is the major protease which specifically removes ISG15 from target proteins. Thus, boosting ISG15 modification by protease inhibition of USP18 might represent a new strategy to interfere with viral replication. However, so far no heterologous expression system was available to yield sufficient amounts of catalytically active protein for high-throughput based inhibitor screens.
Results
High-level heterologous expression of USP18 was achieved by applying a chaperone-based fusion system in E. coli. Pure protein was obtained in a single-step on IMAC via a His6-tag. The USP18 fusion protein exhibited enzymatic activity towards cell derived ISG15 conjugated substrates and efficiently hydrolyzed ISG15-AMC. Specificity towards ISG15 was shown by covalent adduct formation with ISG15 vinyl sulfone but not with ubiquitin vinyl sulfone.
Conclusion
The results presented here show that a chaperone fusion system can provide high yields of proteins that are difficult to express. The USP18 protein obtained here is suited to setup high-throughput small molecule inhibitor screens and forms the basis for detailed biochemical and structural characterization.
Background
Posttranslational protein modification by ubiquitin and ubiquitin-like proteins (UBLs) serves as a versatile mechanism to control multiple biological functions in the cell [1]. The IFN-stimulated gene 15 (ISG15) is a UBL strongly induced by type I IFN and ISG15 conjugation (ISGylation) is one of the major antiviral effector systems [2-4]. Consequently, mice lacking ISG15 exhibit enhanced susceptibility upon distinct viral pathogens. Analogous to the ubiquitin modification process, conjugation of ISG15 is mediated by a cascade of E1, E2, and E3 ligases and ISG15 linkage is counteracted by the activity of deconjugating enzymes [5,6]. USP18 (UBP43) was shown to be the major ISG15 deconjugating enzyme and belongs to the peptidase C19 family [7]. As USP18 deficient mice and cells show elevated levels of ISG15- conjugated substrates [8], it appears feasible to enhance ISGylation levels by inhibition of the USP18 protease activity. This might also be of therapeutic interest as USP18 deficient animals were shown to be more resistant against certain viruses [3] and exhibit resistance against PML-RAR- [9] and BCR-ABL-induced leukemia [10]. A prerequisite for the identification of chemical compounds suitable to inhibit USP18 is the availability of a fast and sensitive enzymatic assay monitoring ISG15 deconjugation. High through-put screening based on ubiquitin-AMC (Ub-AMC) has been used with success for the identification of small molecules inhibiting USP protease activity. [11]. The assay is based on the release of the fluorophore AMC upon cleavage of the isopeptide bond by the USP. Thus, presumably ISG15 deconjugase inhibitors could be identified using ISG15-AMC in a similar protease inhibitor assay. However, in the case of USP18 the setup of such an assay for high-throughput screening was hampered so far by limited amounts of recombinant enzymatically active USP18. Attempts to express USP18 in Escherichia coli (E. coli) resulted in degraded protein [5]. Expression in Sf9 cells using the baculovirus expression systems was successful but is difficult to scale up and cost intensive [12] compared to bacterial expression systems. Here we report the development of a bacterial expression system based on the fusion of USP18 to a bacterial chaperone (Trigger Factor = TF) that yields high amounts of enzymatically active protein. USP18 was purified to homogeneity as a TF-fusion protein. The recombinant protease is specific for ISG15 as shown by enzymatic deconjugation of ISG15 from ISGylated cellular proteins and by the formation of a covalent adduct with ISG15 vinyl sulfone. Finally, we established assay conditions for USP18 mediated ISG15-AMC cleavage suited for the setup of large scale inhibitor screens.
Methods
Cloning methods
The consensus sequence for ubiqutinyl hydrolases encompasses residues 46–368 of USP18. The 45 N-terminal residues are of unknown function. cDNA encoding residues 46–368 of mouse USP18 in frame with a His6-tag and the recognition site for the 3 C protease was amplified and cloned into pET15b (Novagen) and pGEMEX (Promega) vector. All cloning steps were performed according to standard protocols [13].
For USP18 constructs with codons optimized for expression in E. coli a synthetic cDNA encoding USP18 residues 46–368, as well as a 3 C protease recognition site and a flexible linker at the 5’ end was purchased from a commercial supplier (Mr. Gene). cDNAs for His6-SUMO and His6-SUMO-TFAAA were generated by PCR using vector pSUMO-tigAAA[14,15] as template. The cDNAs were fused and cloned into the vector pACE by sequence and ligation independent cloning (SLIC) [16] yielding vectors pACE-His6-SUMO-TFAAA-USP18 and pACE-His6-SUMO-USP18. NdeI and XhoI restriction sites were inserted to allow further subcloning of the constructs.
The following primers were used for vector and insert amplification and ligation: XhoI-pACE-for: 5’-CTCGAGAGATCCGGCTGCTAACAAAG-3’, NdeI-pACE-rev: 5’-CATATGTATATCTCCTTCTTAAAGTTAAAC-3’, XhoI-USP18-rev: 5’-CTTTGTTAGCAGCCGGATCTCTCGAGTTAGGAGCCGGTTTTCG-3’, SUMO-3C-for: 5’-CACAGAGAACAGATTGGTGGTCTGGAAGTTCTGTTCCAGGGTCCG-3’, TF-for: 5’-CACTTTCAACGAGCTGATGAACCAGCAGGC-3’, TF-rev: 5’-GCCTGCTGGTTCATCAGCTCGTTGAAAGTG-3’, SUMO-3C-rev: 5’-CGGACCCTGGAACAGAACTTCCAGACCACCAATCTGTTCTCTGTG-3’, NdeI-His-for: 5’-GTTTAACTTTAAGAAGGAGATATACATATGATGGGTCATCACCATCATC-3’.
For cloning into the pSUMO backbone [14,15], the pACE expression vectors were digested with NdeI and XhoI restriction enzymes and the inserts were ligated into pSUMO vector digested with the same enzymes. A catalytic inactive mutant with substitution of the catalytic cysteine 61 to alanine was generated using the QuikChange II kit (Stratagene).
Expression and purification
The following strains were transformed with the different vectors and tested for expression: E. coli BL21(DE3), E. coli BL21(DE3)pLysS, E. coli Rosetta(DE3) , E. coli Tuner(DE3) and E .coli Tuner(DE3)pLysS (Novagen). Expression was performed in shaking cultures in DYT medium supplemented with appropriate antibiotics, trace elements (Studier) and 0.2% (w/v) glucose. For E. coli strains transformed with pET15b, pGEMEX or pACE 100 μg/ml ampicillin was added to the medium; for pSUMO 50 μg/ml kanamycin was added to the medium. In case of E. coli strains E. coli Rosetta(DE3), E. coli BL21(DE3)pLysS and E. coli Tuner(DE3)pLysS additionally 17 μg/ml choramphenicol was added to the medium.
5 ml DYT medium was inoculated with a single colony and incubated on a shaker at 37°C overnight. For inoculation of expression cultures the overnight culture was diluted 1:100 in the same medium. Test expression cultures had a volume of 20 ml in 200 ml Erlenmeyer flasks at different temperatures (15°C - 37°C). The culture was grown until an OD600 nm of 0.6 was reached and expression was induced by addition of IPTG (Applichem) to a final concentration of 0.1 - 1 mM. Large scale expression was performed in 500 ml in baffled 2 l Erlenmeyer flasks at 15°C for 16 h with a final IPTG concentration of 0.1 mM.
Cells from expression cultures were harvested by centrifugation. Cell pellets were suspended in ice-cold buffer A (20 mM Tris-Cl, 500 mM NaCl, pH 7.9). Cell pellets from small scale expression were disrupted by ultrasonic treatment whereas cells from large scale expression were broken by 2 passages through a French pressure cell at 137 Mpa. Typically, 8 g wet weight cells were used per batch of protein purification.
Crude extracts from test expressions were centrifuged at 4°C at 16,000 g for 60 minutes. Supernatant and pellet fraction were mixed with sample buffer and analyzed on SDS-PAGE. Crude extracts from large scale expression were centrifuged at 100,000 g for 60 min. All purification steps were performed at 4°C using a FPLC system (GE-Healthcare). The supernatant was applied to a Cobalt-IMAC column (1 ml column volume, Novagen) equilibrated in buffer A. Absorption at 280 nm was monitored and column was washed with the same buffer until absorption reached baseline level again. Three washing steps were performed with buffer A supplemented with 10 mM, 20 mM and 30 mM imidazole, respectively. The bound protein was eluted with buffer A containing 1 M imidazole. The pure protein was dialyzed against 5 l buffer A overnight, concentrated to 8 mg/ml and analyzed by SDS-PAGE and subsequent Coomassie staining.
Generation of ISGylated cell lysates and deISGylation assay
USP18 deficient murine embryonic fibroblasts (MEFs) were stimulated with 250 U/ml IFN β (Sigma) for 24 h to induce ISGylation of endogenous proteins or left untreated. MEFs were lysed in 50 mM Tris-Cl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100. The cell lysate was cleared by centrifugation at 16,000 g for 30 min at 4°C. 5 μl of the supernatant (20 μg) were incubated with 2 μl (16 μg) TFAAA-USP18 and 20 μl reaction buffer (50 mM Tris-Cl pH 8.3, 25 mM KCl, 5 mM MgCl2, 1 mM DTT) for 0, 1 and 2 h at 37°C. The reaction was stopped by addition of SDS containing sample buffer. The samples were separated on a 12% SDS-PAGE gel, transferred to a nitrocellulose membrane and analyzed with the following antibodies: ISG15 [17] and β-Actin (I-19, Santa Cruz). For quantification the optical densities of protein bands were obtained using ImageJ [18]. The densitometric values of free and conjugated ISG15 were normalized to β-Actin and depicted relative to the ISG15 values in IFN β-treated cells at 0 h without addition of TFAAA-USP18.
Reaction with ubiquitin and ISG15 vinyl sulfone
HA-Ubiquitin (Ub-VS) and HA-ISG15 vinyl sulfone (ISG15-VS) were purchased from Boston Biochem. 1 μl (8 μg) of TFAAA-USP18 or TFAAA-USP18-C61A was combined with 1 μl (0.5 μg) Ub-VS or ISG15-VS, respectively. Reaction was performed in 50 mM Tris-Cl pH 7.4, 5 mM MgCl2, 250 mM sucrose, 1 mM DTT, 2 mM ATP for 1 h at 37°C [19]. The samples were analysed on a Coomassie-stained 10% SDS-PAGE gel.
Measurement of ISG15-AMC cleavage
ISG15-amidomethyl coumarin (AMC) was purchased from Boston Biochem. Different amounts of TFAAA-USP18 (final concentration 0, 0.36, 0.72, 1.43 μM) were incubated with 600 nM ISG15-AMC in a total volume of 28 μl. For each TFAAA-USP18 concentration duplicates were analyzed. The reaction was performed in 50 mM Hepes-NaOH pH 7.5, 0.01% (v/v) Tween 20, 10 mM DTT. The release of AMC was detected over a period of 30 minutes using a Safire II fluorescence spectrophoto meter with excitation and emission wavelength of 380 nm and 460 nm, respectively.
Results and discussion
High-throughput screening requires large amounts of active protein. Recombinant expression of USP18 in a bacterial system as well as in insect cells has been reported, however with very low yields [5]. Until now, there has been no expression system available for production of sufficient amounts of recombinant USP18. Therefore, we aimed to establish a high-yield and easy-to-apply expression system for catalytically active USP18.
Expression trials using murine cDNA for USP18 cloned into pET15b or a pGEMEX vector were performed in E. coli Rosetta(DE3). However, no expression of His6-tagged USP18 could be observed in Western blots (Table 1). We reasoned that some rare codons in the cDNA of the USP18 clone might obstruct expression and therefore switched to an expression construct with codons optimized for expression in E. coli. In addition, we introduced a SUMO-tag at the N-terminus of USP18 as such a tag was reported to enhance expression levels of this protein in the baculovirus expression system [12]. Sequence and ligation independent cloning (SLIC) was performed to generate the His6-SUMO-USP18 construct in the pACE vector backbone [20,21] (Figure 1A). Subsequently, E. coli BL21(DE3) as well as E. coli BL21(DE3)pLysS were used as host strains for test expressions. In contrast to the clone derived from mouse cDNA, expression of the SUMO-USP18 fusion protein could be detected in both E. coli strains on Western blot with an anti-His6-tag specific antibody. However, expression levels of the fusion protein were too low to be detected on Coomassie-stained SDS-PAGE gel (Table 1).
Table 1.
Constructs tested for expression of different USP18 fusion proteins
| Construct (promoter, antibiotic resistance) | pGEMEX-His-USP18 (T7, ampicillin) | pET15b-His-USP18 (T7, ampicillin) | pACE-His6-SUMO-USP18 (T7, ampicillin) | pACE-His6-SUMO-TFAAA-USP18 (T7, ampicillin) | |||
|---|---|---|---|---|---|---|---|
|
strain |
Rosetta(DE3) |
Rosetta(DE3) |
BL21(DE3) |
BL21(DE3)pLysS |
BL21(DE3) |
BL21(DE3)pLysS |
|
| temperature |
25°C |
25°C |
37°C |
37°C |
25°C |
37°C |
37°C |
| expression |
- |
- |
- |
+ |
+ |
++ |
++ |
| soluble |
n.d. |
n.d. |
n.d. |
n.d. |
n.d. |
n.d. |
n.d. |
|
Construct (promoter, antibiotic resistance) |
pSUMO-His-SUMO-TFAAA-USP18 (T7, kanamycin) |
||||||
|
strain |
BL21(DE3) |
BL21(DE3)pLysS |
Tuner(DE3)pLysS |
Tuner(DE3) |
|||
| temperature |
25°C |
37°C |
15°C |
25°C |
37°C |
15°C |
15°C |
| expression |
+++ |
+++ |
+ |
+++ |
+++ |
++ |
+++ |
| soluble | n.d. | n.d. | + | - | - | ++ | + |
n.d.: not determined, -: no expression observed, +: detectable on Western Blot, but not on Coomassie-stained SDS PAGE gel, ++: detectable on Coomassie-stained gel, but not the dominant band, +++: dominant band on Coomassie-stained gel.
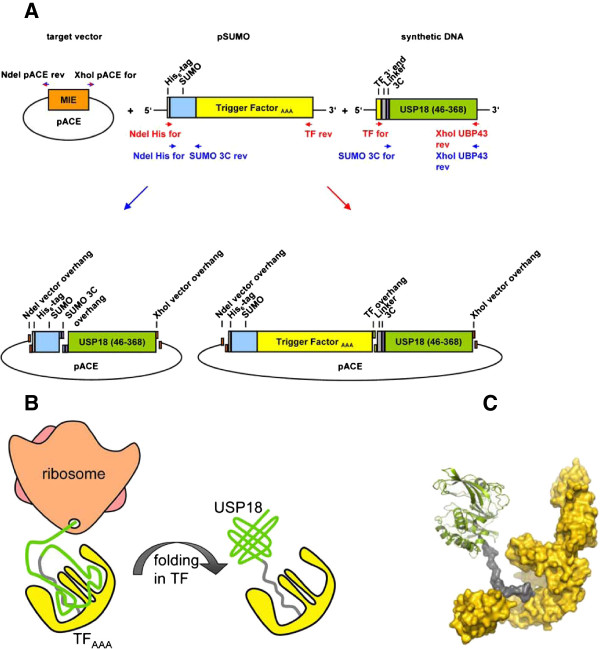
Figure 1.
(A) Generation of SUMO-USP18 and SUMO-TFAAA-USP18 expression vectors using sequence and ligation independent cloning (SLIC). The target vector pACE was linearized using primers NdeI-pACE-rev and XhoI-pACE-for. Recognition sites for the restriction enzymes NdeI and XhoI were introduced during the amplification process. Two vectors served as templates for amplification of the inserts: pSUMO encoding a His6-tag-SUMO-Trigger FactorAAA fusion protein, and a synthetic DNA construct consisting of the 3’ end of the Trigger Factor, a flexible linker, a recognition site for the 3 C protease and a USP18 cDNA for residues 46–368. For both the SUMO-USP18 and the SUMO-TFAAA-USP18 construct, two PCR products with overlapping 5’ and 3’ ends were generated. For the SUMO-USP18 construct primers NdeI-His-for together with SUMO-3C-rev as well as primers SUMO-3C-for together with XhoI-USP18-rev (blue) were used. For the SUMO-TFAAA-USP18 construct, primer NdeI-His-for was combined with TF-rev and primer TF-for with XhoI-USP18-rev (red). Treatment of the PCR products with T4 DNA polymerase in the absence of dNTPs resulted in complementary single stranded overhangs that were subsequently annealed. The resulting vectors encoded for two different USP18 fusion proteins: SUMO-USP18 and SUMO-TFAAA-USP18. Both proteins exhibit a His6-tag for purification and a SUMO-tag to enhance solubility of the protein. In the SUMO-TFAAA-USP18 protein the bacterial chaperone Trigger Factor carrying three exchanges to alanine (F43A, R44A, K45A; =TFAAA) is additionally fused to the N-terminus of USP18 to provide each expressed USP18 molecule a chaperone that facilitates folding. The flexible linker between TFAAA and USP18 was introduced to allow interaction of USP18 with the chaperone. (B) Schematic drawing of TFAAA -USP18. Newly synthesized TFAAA folds and takes up the nascent chain of USP18. The fusion protein dissociates from the ribosome and USP18 can fold in the cradle of TFAAA. (C) Molecular model of TFAAA-USP18. TFAAA is shown as surface representation and USP18 as cartoon with secondary structure elements. TFAAA is shown in yellow, the linker in grey, and USP18 in green, respectively.
Overproduction of soluble recombinant protein in E. coli can be limited by the deprivation of host cell chaperones that are required for correct folding of the respective protein. Co-overexpression of E. coli chaperones was reported previously to enhance solubility and yield of recombinant proteins [22]. Recently, also successful expression of a fusion of the chaperone Trigger Factor with the protein of interest was reported using a cold shock expression system in E. coli[23] (Takara, pCold TF plasmid). This system provides each translated protein its own chaperone. As the chaperone Trigger Factor (TF) is the first chaperone newly translated proteins encounter [24] we fused this chaperone to the N-terminus of USP18. TF interacts with the bacterial ribosome and incorporates nascent polypeptide chains that emerge from the ribosomal exit tunnel. In this way, it provides a protective environment that facilitates folding [24,25]. To assure interaction of USP18 with TF, which forms a large hydrophobic cradle, we introduced a long flexible linker consisting of six GSS repeats between USP18 and the chaperone (Figure 1B, C). Moreover, the long linker ensures that the folded USP18 is accessible for substrates and not sterically blocked by TF.
TF binds to the ribosome via the motif 43-GFRxGxxP-50 [26,27]. Although TF binds with low affinity to the ribosome [28], overexpression of TF might become a serious problem for protein synthesis in the expression host. In order to reduce binding of the TF-USP18 fusion protein to the ribosome and facilitate dissociation, residues G43, F44 and R45 of TF were exchanged to alanine (TFAAA). These residues have been shown previously to be critical for TF-ribosome interaction [27].
The resulting fusion protein consists of an N-terminal His6-tag, SUMO, Trigger FactorAAA, and USP18 (= TFAAA-USP18). TFAAA-USP18 in the pACE vector backbone was tested for expression in E. coli BL21 (DE3) and E. coli BL21 (DE3) pLysS. In contrast to SUMO-USP18, insertion of TF increased expression levels so that the fusion protein could be detected on Coomassie-stained SDS-PAGE gel (Table 1). However, it did not represent the major fraction compared to endogenous bacterial proteins.
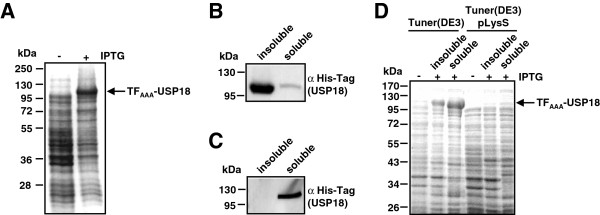
Therefore, we changed the vector backbone from pACE to pSUMO. This boosted expression of the fusion protein which now represented the major band on SDS gel when expression was performed at 37°C (Table 1 and Figure 2A). However, these expression conditions resulted in poor solubility of the protein as demonstrated by Western Blot with a His6-tag specific antibody (Figure 2B). Almost all recombinant protein was detected in the pellet fraction whereas only a weak band was detected in the soluble fraction. Lowering temperature is often reported to increase yield and solubility of expressed proteins [29-31]. Test expressions at 25°C had no observable effect and resulted in insoluble protein (not shown). Decreasing further the expression temperature to 15°C yielded soluble TFAAA-USP18 (Figure 2C). However, the drop in temperature caused also a severe decrease in protein expression (Table 1).
Figure 2.
Expression of TFAAA-USP18 in pSUMO vector backbone under different conditions(A) TFAAA-USP18 was expressed in E. coli BL21(DE3)pLysS at 37°C. Expression was verified by analyzing protein content directly after lysis on SDS-PAGE followed by Coomassie staining. After 3 h of induction TFAAA-USP18 fusion protein made up more than 50% of whole cellular proteins. (B) Soluble and insoluble fractions from (A) were analysed by Western blot with an anti His6-Tag antibody. Almost all fusion protein was present in the insoluble fraction and only a faint band for soluble protein was observed. (C) Expression of TFAAA-USP18 at 15°C in E. coli BL21(DE3)pLysS yielded soluble protein. Western blot analysis using an anti His6-Tag antibody detected TFAAA-USP18 only in the soluble fraction. (D) E. coli Tuner(DE3) and E. coli Tuner(DE3)pLysS were tested for expression of TFAAA-USP18 at 15°C. Soluble and insoluble fractions were analysed by SDS-PAGE and subsequent Coomassie staining. Strong expression was only observed in E. coli Tuner(DE3). The major portion of the fusion protein was observed in the soluble fraction.
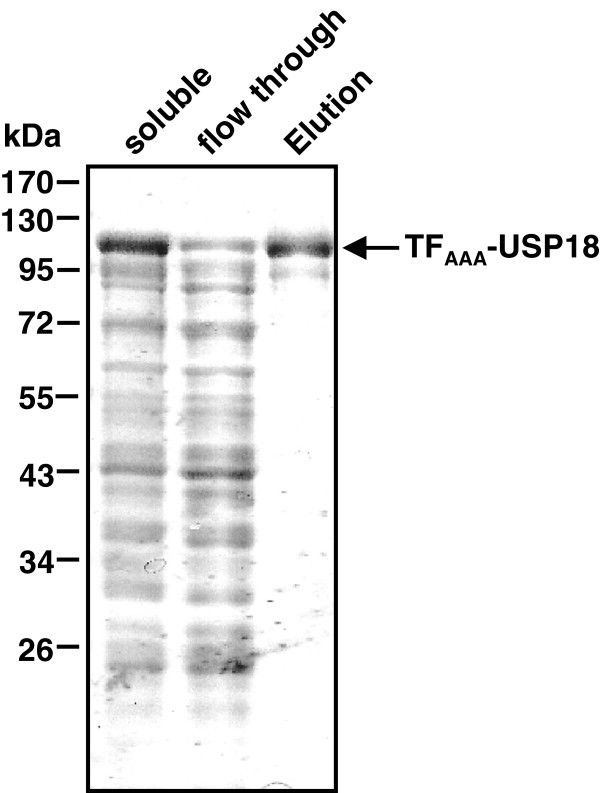
To achieve again high expression levels combined with high solubility of TFAAA-USP18 we changed to the stringent expression host strains E. coli Tuner(DE3) and E. coli Tuner(DE3)pLysS. Tuner strains are deficient in lactose permease (lacY) and thus allow uniform uptake of IPTG via diffusion. Whereas E. coli Tuner(DE3)pLysS only showed a weak expression of TFAAA-USP18, strong expression of soluble fusion protein was observed when the E. coli Tuner(DE3) strain was grown at 15°C (Table 1 and Figure 2D). Therefore, these conditions were applied for large scale expression and purification. 2 liter expression cultures typically yielded 24 g of wet weight pellet. For purification of TFAAA-USP18, different IMAC columns were tested of which a cobalt IMAC column provided the best results. Using this column, pure TFAAA-USP18 was eluted allowing one-step purification without need of further purification steps (Figure 3). A minor band running at lower molecular weight was observed when the sample was boiled only for a short time or without fresh DTT added. This band most likely represents protein containing an intramolecular disulfide bond formed during boiling. Typical yield was 10 mg pure protein out of 8 g wet weight pellet.
Figure 3.
One-step purification of TFAAA-USP18. TFAAA-USP18 was bound to a Co-IMAC column and eluted with imidazole. Purity of the eluted fusion protein was visualized by SDS-PAGE with subsequent Coomassie staining.
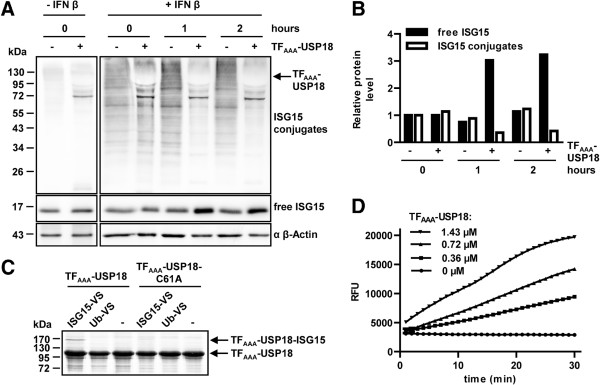
Once expression and purification was established we checked whether the large scale preparations represent also catalytically active enzyme. Therefore, we tested isopeptidase activity of TFAAA-USP18 towards ISG15 modified cellular proteins (Figure 4A). High levels of ISGylated cellular protein were obtained using USP18 deficient mouse embryonic fibroblasts (MEFs) stimulated with IFN β. MEF cell lysates were incubated with and without TFAAA-USP18 and changes in ISGylation levels were monitored by Western blot with an ISG15-specific antibody. Incubation with TFAAA-USP18 drastically decreased the amount of ISGylated proteins, simultaneously the amount of free ISG15 increased demonstrating the ability of the TFAAA-USP18 to recognize and cleave ISG15 from cellular target proteins. To further evaluate enzymatic specificity, TFAAA-USP18 as well as a TFAAA-USP18 variant, where the catalytic cysteine is exchanged to alanine (TFAAA-USP18-C61A), were incubated with the suicide inhibitors ubiquitin vinyl sulfone (Ub-VS) and ISG15 vinyl sulfone (ISG15-VS), respectively. These suicide inhibitors form a covalent adduct upon reaction with the active site cysteine of ubiquitin-specific proteases. The reaction can be visualized as a shift in molecular mass on a Coomassie-stained gel. For TFAAA-USP18, covalent complex formation was detected with ISG15-VS whereas mutation of the catalytic cysteine to alanine resulted in complete loss of the interaction. Neither TFAAA-USP18 nor TFAAA-USP18-C61A showed cross-reactivity towards Ub-VS (Figure 4B). In summary, these experiments show that TFAAA-USP18 is catalytically active and underline its specificity towards ISG15.
Figure 4.
Enzymatic activity of TFAAA-USP18 (A) Cell lysates of USP18 knockout mouse embryonic fibroblasts were stimulated with IFN β resulting in elevated ISGylation or left untreated. Cell lysates were incubated with and without TFAAA-USP18 for the indicated times at 37°C. TFAAA-USP18-mediated ISG15 deconjugation was monitored by Western blot with an ISG15-specific antibody. A decrease of protein ISGylation with a concomitant increase of free ISG15 was observed. (B) Quantification of ISGylation and free ISG15 from (A) revealed a 3-fold decrease of conjugated and a corresponding increase of free ISG15 upon incubation with TFAAA-USP18. Densitometric values of free ISG15 and ISGylated proteins were normalized to β-Actin and to the protein levels in IFN β-treated cells at 0 hours without addition of TFAAA-USP18. (C) TFAAA-USP18 or the catalytically inactive mutant TFAAA-USP18-C61A were incubated with substoichiometric amounts of either ISG15 vinyl sulfone (ISG15-VS) or ubiquitin vinyl sulfone (Ub-VS) for 1 h at 37°C. The covalent adduct of USP18 with ISG15 is shown by a shift to higher molecular mass visualized on a Coomassie-stained SDS PAGE. (D) Catalytic activity of TFAAA-USP18 monitored by cleavage of ISG15-AMC: different amounts of TFAAA-USP18 were incubated with 600 nM ISG15-AMC. Release of AMC was monitored by its specific fluorescence at 460 nm over a period of 30 minutes. RFU: Relative fluorescence units.
Screening for potential USP18 inhibitors requires a method that allows quantification of USP18 activity and is compatible with standard detection instruments. Therefore, we established assay conditions for TFAAA-USP18-mediated ISG15-AMC cleavage. Different amounts of the fusion protein were incubated with ISG15-AMC and cleavage was measured over a period of 30 minutes. The measured rate of ISG15-AMC cleavage was constant for more than 20 minutes and the rate increased linearly with enzyme concentration (Figure 4C). At the highest concentration of TFAAA-USP18, a slight decrease in the rate was observed after 25 minutes that is most likely due to a limitation of substrate and not caused by a decrease in enzyme activity. TF itself has no isopeptidase catalytic activity and does not interfere with the assay. These results demonstrate that TFAAA-USP18 is very well suited for kinetic analysis and the assay presented here can be easily adapted for high-throughput screening for specific inhibitors of USP18.
Conclusion
Today, the analysis of genomic and expressions array data provide a plethora of data on proteins in the regulation of vital cellular processes representing potential therapeutic targets. The entire process of drug development relies on the availability of correctly folded and active target proteins provided by heterologous production in eukaryotic and prokaryotic expression systems. However, for fast, efficient, and easy-to-scale-up expression E. coli is still the expression system of choice. Here, we describe a new and efficient method to express and purify high yields of recombinant catalytically active USP18. Starting from zero expression we could boost the yields of active protein by optimization of codons, vector backbone, and a novel chaperone fusion system. The excellent yields obtained for USP18 put forward that this system is also very well suited for other proteins where recombinant expression failed so far.
Abbreviations
ISG: IFN-stimulated gene; USP: Ubiquitin-specific protease; HA: Hemagglutinin; AMC: 7-amino-4-methylcoumarin; IFN β: Interferon β.
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
AB carried out the cloning of vectors, performed expression, purification, and analysis of proteins, and wrote the manuscript. LK carried out design and cloning of expression vectors. ED designed expression vector and expression experiments. CA and JR have conducted assays with USP18 using ISG-15-AMC as a substrate. KPK designed expression experiments and activity assay, and wrote the manuscript. GF designed expression vectors and experiments, purified protein and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Anja Basters, Email: anja.basters@uniklinik-freiburg.de.
Lars Ketscher, Email: lars.ketscher@uniklinik-freiburg.de.
Elke Deuerling, Email: elke.deuerling@uni-konstanz.de.
Christoph Arkona, Email: arkona@uni-leipzig.de.
Jörg Rademann, Email: rademann@uni-leipzig.de.
Klaus-Peter Knobeloch, Email: klaus-peter.knobeloch@uniklinik-freiburg.de.
Günter Fritz, Email: guenter.fritz@uniklinik-freiburg.de.
Acknowledgements
GF is supported by a Heisenberg Fellowship by the Deutsche Forschungsgemeinschaft (FR 1488/3-2). This work was further supported by grants from the Deutsche Forschungsgemeinschaft KN 590/3-1 and KN 590/1-3 to KPK and DE 783/3-1 DFG to ED.
References
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobeloch KP. In vivo functions of ISGylation. Subcell Biochem. 2011;54:215–227. doi: 10.1007/978-1-4419-6676-6_17. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. IFN-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Chen ZJ. Emerging role of ISG15 in antiviral immunity. Cell. 2010;143(2):187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KI, Zhang DE. UBP43, an ISG15-specific deconjugating enzyme: expression, purification, and enzymatic assays. Methods Enzymol. 2005;398:491–499. doi: 10.1016/S0076-6879(05)98040-3. [DOI] [PubMed] [Google Scholar]
- Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277(12):9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database issue):D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobeloch KP, Utermohlen O, Kisser A, Prinz M, Horak I. Reexamination of the role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol Cell Biol. 2005;25(24):11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SJ, Blumen S, Pitha-Rowe I, Kitareewan S, Freemantle SJ, Feng Q, Dmitrovsky E. UBE1L represses PML/RARα by targeting the PML domain for ISG15ylation. Mol Cancer Ther. 2008;7(4):905–914. doi: 10.1158/1535-7163.MCT-07-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Luo JK, Ritchie KJ, Sakai I, Takeuchi K, Ren R, Zhang DE. Ubp43 regulates BCR-ABL leukemogenesis via the type 1 IFN receptor signaling. Blood. 2007;110(1):305–312. doi: 10.1182/blood-2006-07-033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama K, Aoki S, Nishikawa K, Matsumoto T, Wada K. Identification of novel chemical inhibitors for ubiquitin C-terminal hydrolase-L3 by virtual screening. Bioorg Med Chem. 2007;15(21):6810–6818. doi: 10.1016/j.bmc.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Liu L, Spurrier J, Butt TR, Strickler JE. Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions. Protein Expr Purif. 2008;62(1):21–28. doi: 10.1016/j.pep.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW(ed) Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Eichmann C, Preissler S, Riek R, Deuerling E. Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Proc Natl Acad Sci USA. 2010;107(20):9111–9116. doi: 10.1073/pnas.0914300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaux J, Horst J, Scior A, Preissler S, Koplin A, Bukau B, Deuerling E. Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. J Biol Chem. 2009;285(5):3227–3234. doi: 10.1074/jbc.M109.075804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Nie Y, Frey D, Olieric N, Schaffitzel C, Collinson I, Romier C, Berger P, Richmond TJ, Steinmetz MO. et al. Automated unrestricted multigene recombineering for multiprotein complex production. Nat Methods. 2009;6(6):447–450. doi: 10.1038/nmeth.1326. [DOI] [PubMed] [Google Scholar]
- Osiak A, Utermohlen O, Niendorf S, Horak I, Knobeloch KP. ISG15, an IFN-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol. 2005;25(15):6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9(10):1149–1159. doi: 10.1016/S1074-5521(02)00248-X. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3(12):1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DJ, Schaffitzel C, Berger P, Wellinger R, Bieniossek C, Richmond TJ, Berger I. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure. 2007;15(3):275–279. doi: 10.1016/j.str.2007.01.016. [DOI] [PubMed] [Google Scholar]
- de Marco A, Deuerling E, Mogk A, Tomoyasu T, Bukau B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007;7:32. doi: 10.1186/1472-6750-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki K, Terashima Y, Toda E, Yoshinaga S, Araki N, Matsushima K, Terasawa H. Expression and purification of human FROUNT, a common cytosolic regulator of CCR2 and CCR5. Protein Expr Purif. 2011;77(1):86–91. doi: 10.1016/j.pep.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Merz F, Rutkowska A, Zachmann-Brand B, Deuerling E, Bukau B. Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J Biol Chem. 2006;281(10):6539–6545. doi: 10.1074/jbc.M512345200. [DOI] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B. et al. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J. 2008;27(11):1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431(7008):590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419(6903):171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- Rutkowska A, Mayer MP, Hoffmann A, Merz F, Zachmann-Brand B, Schaffitzel C, Ban N, Deuerling E, Bukau B. Dynamics of trigger factor interaction with translating ribosomes. J Biol Chem. 2008;283(7):4124–4132. doi: 10.1074/jbc.M708294200. [DOI] [PubMed] [Google Scholar]
- Han KG, Lee SS, Kang C. Soluble expression of cloned phage K11 RNA polymerase gene in Escherichia coli at a low temperature. Protein Expr Purif. 1999;16(1):103–108. doi: 10.1006/prep.1999.1061. [DOI] [PubMed] [Google Scholar]
- Shirano Y, Shibata D. Low temperature cultivation of Escherichia coli carrying a rice lipoxygenase L-2 cDNA produces a soluble and active enzyme at a high level. FEBS Lett. 1990;271(1–2):128–130. doi: 10.1016/0014-5793(90)80388-y. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Taiming L, Liu J. Low temperature and glucose enhanced T7 RNA polymerase-based plasmid stability for increasing expression of glucagon-like peptide-2 in Escherichia coli. Protein Expr Purif. 2003;29(1):132–139. doi: 10.1016/S1046-5928(03)00002-0. [DOI] [PubMed] [Google Scholar]