Abstract
The natural developmental gradient of light-grown primary leaves of barley (Hordeum vulgare L.) was used to analyze the biogenesis of mitochondrial proteins in relation to the age and physiological changes within the leaf. The data indicate that the protein composition of mitochondria changes markedly during leaf development. Three distinct patterns of protein development were noted: group A proteins, consisting of the E1 β-subunit of the pyruvate dehydrogenase complex, ORF156, ORF577, alternative oxidase, RPS12, cytochrome oxidase subunits II and III, malic enzyme, and the α- and β-subunits of F1-ATPase; group B proteins, consisting of the E1 α-subunit of the pyruvate dehydrogenase complex, isocitrate dehydrogenase, HSP70A, cpn60C, and cpn60B; and group C proteins, consisting of the four subunits of the glycine decarboxylase complex (P, H, T, and L proteins), fumarase, and formate dehydrogenase. All of the proteins increased in concentration from the basal meristem to the end of the elongation zone (20.0 mm from the leaf base), whereupon group A proteins decreased, group B proteins increased to a maximum at 50 mm from the leaf base, and group C proteins increased to a maximum at the leaf tip. This study provides evidence of a marked heterogeneity of mitochondrial protein composition, reflecting a changing function as leaf cells develop photosynthetic and photorespiratory capacity.
In eukaryotic cells mitochondria function in the production of ATP via oxidative phosphorylation, which is coupled to substrate oxidation primarily through the TCA cycle. This function is likely to be particularly important in plant cells with immature chloroplasts. In photosynthetic cells there is an additional demand for the mitochondria to oxidize Gly, which is generated as an intermediate of the photorespiratory pathway (Keys et al., 1978). The presence of chloroplasts as a second site of ATP production via photophosphorylation may lessen the demand for mitochondrial ATP synthesis in photosynthetically mature cells.
To determine whether there are functional differences between mitochondria from photosynthetic and nonphotosynthetic cells, we used the natural developmental gradient that exists within barley (Hordeum vulgare L.) primary leaves. This and other species of Gramineae, such as wheat, have been used in a number of previous studies into the development of photosynthesis and photorespiration (Viro and Kloppstech, 1980; Dean and Leech, 1982a, 1982b; Tobin et al., 1985, 1988; Barkardottir et al., 1987; Baumgartner et al., 1989; Tobin and Rogers, 1992). The linear development of cells away from a basal intercalary meristem produces a gradient of increasing cell age from the leaf base to the tip. The basal leaf cells contain small, immature chloroplasts and are nonphotosynthetic and heterotrophic, importing carbon from more mature leaf tissue or from the seed (Dale, 1972). With increasing maturity, the mesophyll cells enlarge, the plastids increase in size, protein content, and number, and there is coordinated development of key photosynthetic and photorespiratory enzymes within chloroplasts, peroxisomes, and mitochondria (Tobin and Rogers, 1992).
There is some evidence that plant mitochondria differ in structure, activity, and protein composition depending on the metabolic activity and function of the cells in which they are localized (Rios et al., 1991; Colas des Francs-Small et al., 1992; Tobin and Rogers, 1992). Mitochondria from nonphotosynthetic or etiolated tissues, for example, oxidize Gly at very low rates (Arron and Edwards, 1980; Gardestrom et al., 1980) and have much lower amounts of GDC proteins than mitochondria in photosynthetic cells (Walker and Oliver, 1986; Tobin et al., 1989). In wheat primary leaves, we previously found a coordinated increase in GDC subunits during leaf cell development, which results in a greater than 3-fold increase in the concentration of GDC protein within the mitochondria (Rogers et al., 1991). This occurs after the development of maximum activities of other mitochondrial enzymes such as Cyt oxidase and glutamate dehydrogenase (Tobin et al., 1988; Tobin and Rogers, 1992).
This study is an extension of our earlier investigations into mitochondrial development, and identifies significant heterogeneity in the biogenesis of major mitochondrial protein complexes. The physiological significance of this heterogeneity will be discussed with particular reference to the changing metabolic function of mitochondria during leaf cell development.
MATERIALS AND METHODS
Growth and Harvest of Plant Material
Barley (Hordeum vulgare L. cv Klaxon) seeds (Nickerson Seeds, Lincoln, UK) were allowed to imbibe in aerated water for 16 h at room temperature, sown on the surface of compost (M2, Levington Horticulture Ltd., Ipswich, UK), and covered with a thin layer of fine vermiculite (Dupre, Hertford, UK). The plants were grown in a 16-h/8-h light/dark cycle (PPFD 400–700 nm; 225 μmol photons m−2 s−1, 20°C/10°C, and 80% humidity). Primary leaves were sampled at 160 h after imbibition at a mean length of 90 to 95 mm.
Determination of Cell Age
Leaf elongation rates, segmental elongation rates, and VD, a measurement of the velocity (in millimeters per hour) at which a point on the growing leaf is displaced vertically, i.e. toward the leaf tip, were determined using a modified method of Schnyder and Nelson (1988). The VD values were then used to calculate cell age in the following way: By taking the first marked segment (2 mm from the leaf base), we calculated the distance traveled every 0.25 h until that segment had moved to halfway between its starting point and that of the next marked segment. At this point the VD for the next segment was used and the progress every 0.25 h was again calculated. This sequence was repeated until an entire progress curve had been generated for a segment moving from the leaf base to the tip. The curve that was generated then gave a direct relationship between the position of a leaf segment and the time in hours taken for that segment to have moved from the point 2.0 mm from the basal meristem. When the time taken for imbibition (16 h) is taken into account, this provides a measurement of leaf cell age.
Mitotic Index
Mitotic index was determined using a method modified from Ougham et al. (1987).
Interstomatal Distance
Ten primary leaves were placed abaxial side down onto microscope slides coated with clear nail polish. After the polish had dried the leaves were gently pried away and the impression of the epidermis was examined at ×40 magnification using a light microscope (Dialux, Leitz Ltd., Milton Keynes, Bucks, UK). The distance between stomatal guard cells (interstomatal distance) was recorded along three cell columns per leaf in 1.0-mm sections, 9.0 to 24.0 mm from the leaf base.
Mesophyll Cell Number
Mesophyll cell number was determined using a method modified from Dean and Leech (1982a) for six independent samples from each leaf section.
Measurement of Photosynthesis and Respiration in Leaf Sections
For measurement of photosynthesis, transverse, 5.0-mm sections were taken from between the base and tip of three primary leaves and placed in 1 mL of 50 mm Hepes, 10 mm Mes, pH 6.5, 5 mm CaCl2, and 10 mm NaHCO3 in an O2 electrode (Hansatech, Kings Lynn, UK) at 25°C. The sections were illuminated with white light (PPFD 400–700 nm; 2300 μmol photons m−2 s−1), and the linear rate of O2 evolution was recorded for separate measurements from each region of the leaf.
The procedure for measurement of respiration was the same as for photosynthesis except that NaHCO3 was omitted from the medium and eight leaf sections were used. The linear rate of O2 uptake was recorded in the dark for four separate measurements from each region of the leaf.
Protein Extraction and Quantification
Ten primary leaf sections (5 mm transverse) were frozen in liquid N2 and extracted according to the method of Rogers et al. (1991). The extracts were diluted to a final concentration of 160 μg protein mL−1 in either wash buffer (25 mm Tris, 192 mm Gly, pH 8.3), if required for slot-blot analysis, or in loading buffer (62.5 mm Tris, pH 6.8, 10% [v/v] glycerol, 2% [w/v] SDS, 4% [v/v] β-mercaptoethanol) for SDS-PAGE.
Protein was determined using a microassay (Bio-Rad) with thyroglobulin as the standard.
Slot-Blot Analysis of Mitochondrial Proteins
Sample Application
Leaf protein extracts (50 μL per well, each containing 8 μg of protein) were loaded onto PVDF membranes (Amersham) that had been prewashed in 100% methanol, rinsed in water, and equilibrated in wash buffer (see above) in a 48-well slot-blot apparatus (HSL, Hoefer Scientific Instruments, Newcastle-Under-Lyme, UK). Each well was then washed three times under vacuum with 500 μL of wash buffer, and the membrane was removed, air dried, and stored at 4°C before immunodetection.
Immunodetection of Mitochondrial Proteins
After slot blotting, the PVDF membrane was treated according to the ECL protocol of the manufacturer (Amersham). Incubations with both the primary and secondary antibodies (either goat anti-rabbit or goat anti-mouse horseradish peroxidase conjugate [Sigma] diluted 1:25,000 in TBS containing 0.06% [v/v] Tween and 1% [w/v] dried milk powder) were for 1 h at room temperature.
The PVDF membrane was placed directly onto film (type RX, Fuji Photo Film Co. Ltd., Tokyo, Japan) that had been preflashed using a “sensitized” preflash unit (Amersham). Test strips were exposed to establish the optimum exposure time, and the film was developed using an automatic x-ray film processor (model RGII, Fuji).
Image Analysis of Immunodetected Proteins
After ECL detection, the intensity of any bands on the developed film was evaluated using an image analyzer (AnalySiS 2.1, Norfolk Analytical Ltd., Hilgay, UK) connected to a CCD (charge-coupled device) monochrome camera. The image analyzer was first calibrated to obtain the number of pixels per millimeter, and the captured image was then subjected to brightness and contrast correction. A series of profiles was taken along the film negative, and the gray-scale values were calculated (in pixels) for regions corresponding to sample wells. The intensity of the background (in pixels) was subtracted for each profile. For each analysis, 10 sample wells were used for each leaf extract (8 μg of protein per well), and these images were added together to give a single gray-scale value. From established measurements of the total protein content of cells at each 5.0-mm section along the leaf, the gray-scale values were converted to relative values per cell. Finally, gray-scale values for each individual leaf profile were expressed as a percentage of the most intense band, which allowed comparisons to be made between different experiments. The procedure was rigorously optimized to ensure that the gray-scale value was always within a range that was proportional to the amount of mitochondrial protein present on the PVDF membrane. The following variables were optimized: primary and secondary antibody dilution, protein concentration, period of incubation with ECL reagents, and duration of the film and PVDF membrane exposures. In addition, controls included co-extractions of immature and mature leaf sections and mixing of two separate extracts on single wells of the slot blot (see Results).
Primary Antisera
The primary antisera and the dilution and type of antibody (rabbit polyclonal unless stated otherwise) used in the slot-blot analyses were: α-ATPaseA, β-ATPaseA, α-ATPaseC (mouse monoclonals, 1:10; Luethy et al., 1993); Cyt oxidase II and III (mouse monoclonals, 1:1000; Lightowlers et al., 1991; Taanman and Capaldi, 1993); alternative oxidase (mouse monoclonal, 1:200; Elthon et al., 1989); malic enzyme (1:1000; Winning et al., 1994); RPS12 (1:5000; Gualberto et al., 1988); E1 β-subunit of PDC (1:1000; Luethy et al., 1995b); ORF156 (1:5000; Gualberto et al., 1991); E1 α-subunit of PDC (mouse monoclonal, 1:1000; Luethy et al., 1995a); IDH (1:1000; McIntosh and Oliver, 1992); cpn60C and cpn60B (mouse monoclonal, 1:10; Lund and Elthon, 1993); HSP70A (mouse monoclonal, 1:10; Lund and Elthon, 1993); GDC P, H, T, and L proteins (1:1000; Morgan et al., 1993); fumarase (1:1000; Behal and Oliver, 1997); FDH (1:1000; Colas des Francs-Small et al., 1993); and ORF577 (previously termed ORF589 [Gonzalez et al., 1993]; 1:2000; Handa et al., 1996).
Before the antibodies were used in slot-blot assays their specificity was tested by western-blot analysis of total protein extracts from barley leaves and of isolated barley chloroplasts. In all cases the antibodies were highly specific for polypeptides of the predicted molecular mass, and there was no significant cross-reactivity with chloroplast proteins (data not shown).
RESULTS
Growth and Development of the Barley Primary Leaf
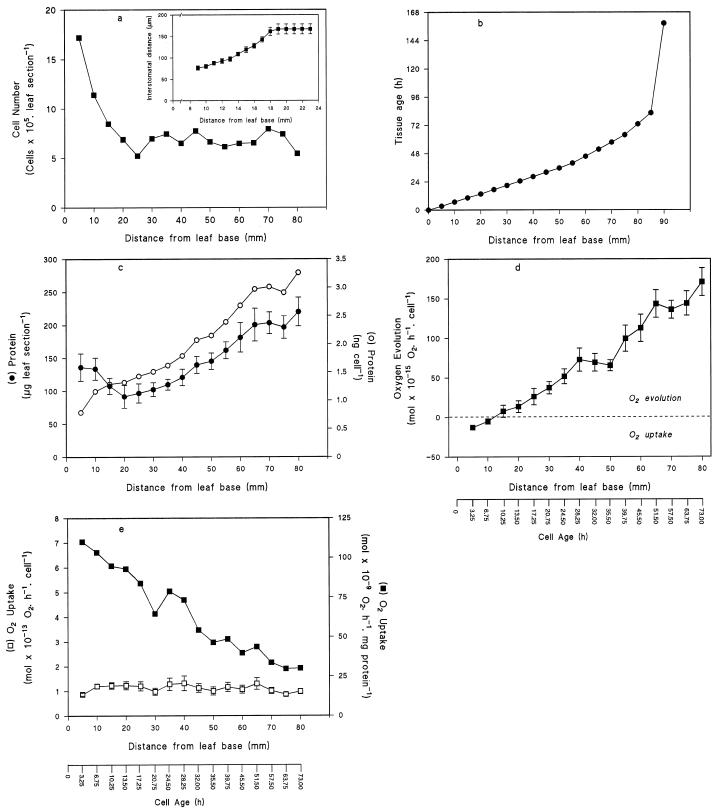
The zone of cell elongation extended to a region approximately 20 mm distal to the base of the primary leaf of 160-h-old barley seedlings. This was apparent from two measurements: the number of mesophyll cells in each 5.0-mm transverse leaf section, which declined to reach a minimum at 20 mm from the leaf base, indicating that mesophyll cell elongation was complete at this point; and the interstomatal distance, which relates to the length of epidermal cells and was maximal at 18 to 20 mm from the leaf base (Fig. 1a, inset), indicating that epidermal cell elongation was also complete.
Figure 1.
a, Change in the number of mesophyll cells during primary leaf development in barley. Transverse 5.0-mm sections were taken at intervals along the length of 160-h-old primary leaves. Each data point represents the mean ± se of six determinations of mesophyll cell number in sections from four individual primary leaves. Inset, Change in the interstomatal distance during primary leaf development in barley. Interstomatal distance was measured at 1.0-mm intervals as shown and each data point represents the mean ± se of three measurements of 10 individual primary leaves. b, Relationship between cell age and the position of a cell within the barley primary leaf. Cell age was calculated from the VD of cells within 160-h-old primary leaves, as described in Methods. c, Soluble protein content in developing cells of barley primary leaves. Soluble protein was extracted from serial transverse sections of 10 individual 160-h-old primary leaves. Each data point represents the mean ± se of three separate protein extractions. d, Change in photosynthetic activity during primary leaf development in barley. Photosynthetic activity (CO2-dependent O2 evolution) was measured in 5.0-mm sections of 160-h-old primary leaves. Each data point represents the mean ± se of four independent measurements. e, Change in respiratory activity during primary leaf development in barley. Respiration (dark O2 uptake) was measured in 5.0-mm sections of 160-h-old primary leaves. Each data point represents the mean ± se of five independent measurements. Rates are expressed relative to the number of mesophyll cells or to the amount of soluble protein within comparable leaf sections (using data from a and c, respectively).
Cell division was confined to the basal 5 mm of the leaf, since no mitotically active cells were detected beyond this region (data not shown).
With the displacement of cells away from the basal meristematic region, there was a gradient of increasing cell age toward the leaf tip (Fig. 1b). The final increase in age of cells between 80 and 90 mm was attributable to the time between imbibition and emergence of the shoot tissue from the seed (see Methods).
The amount of soluble protein within transverse (5.0-mm) leaf sections decreased from the leaf base to the end of the elongation zone (20 mm from the leaf base) and then increased toward the leaf tip (Fig. 1c). The soluble protein content per cell increased linearly from the leaf base to the tip, where it was 4-fold more concentrated (Fig. 1c).
Photosynthetic activity, measured as CO2-dependent O2 evolution, increased along the length of the primary leaf (Fig. 1d). The transition between net O2 uptake and net O2 evolution occurred between 15.0 and 20.0 mm from the basal meristem when cells were approximately 13 h old. This coincided with the end of the zone of cell elongation (compare Fig. 1a).
In contrast to photosynthesis, the rate of “dark” respiratory O2 uptake per cell remained constant throughout the length of the primary leaf. When expressed relative to the amount of soluble leaf protein, respiration rates decreased to a minimum at the leaf tip (Fig. 1e). Inhibitor-titration experiments indicated that the rate of salicylhydroxamic acid-sensitive O2 uptake in the presence of KCN remained a constant proportion of the total rate of respiration, approximately 40% in all leaf sections (data not shown).
Analysis of Mitochondrial Proteins in Barley Primary Leaves
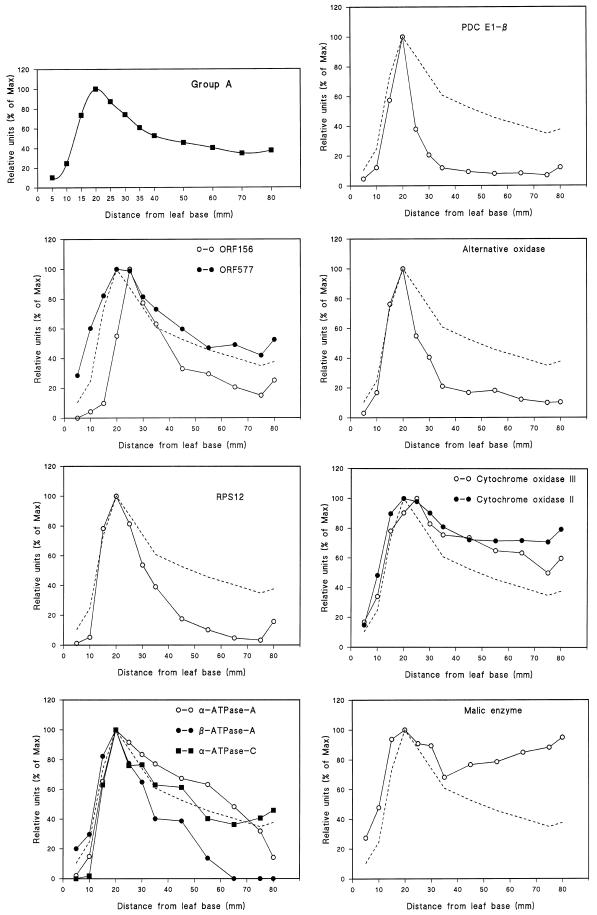
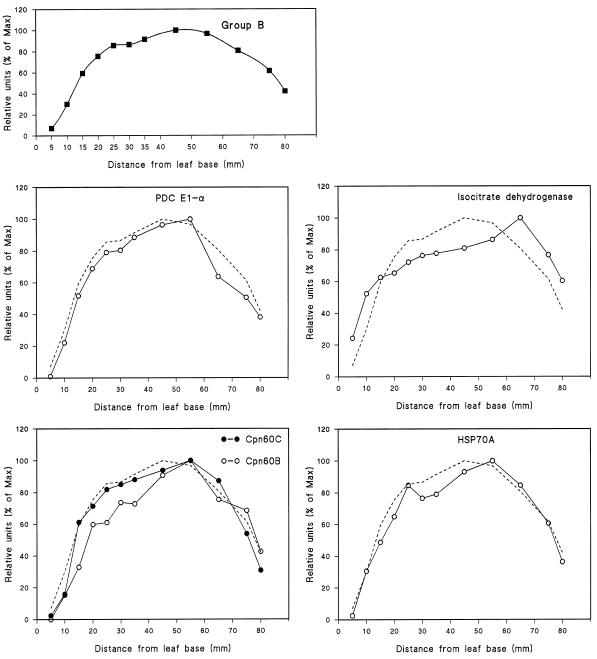
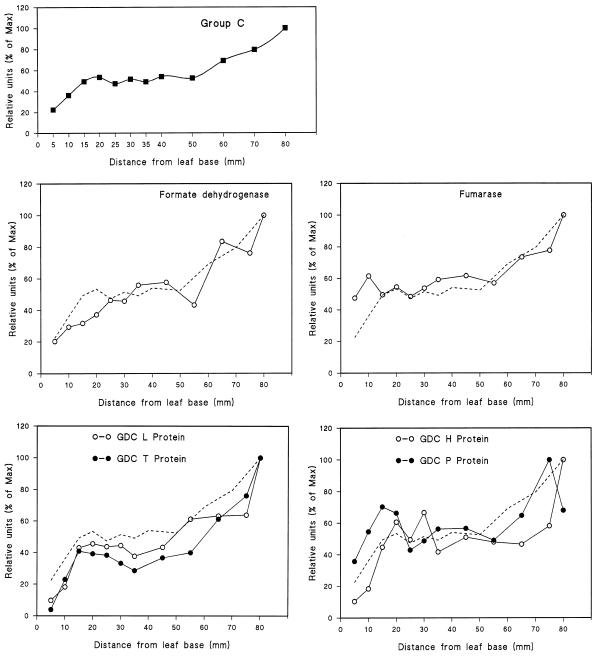
The patterns of development of the 21 different mitochondrial proteins and polypeptides analyzed in this study were one of three distinct types, which we called groups A, B, and C (shown in Figs. 2, 3, and 4, respectively). Note that in each case the data shown in the figures are the mean values of all of the individual proteins considered to be members of that group. To enable comparison between individual protein profiles and the group pattern, the mean values are represented by a dashed line in each panel of the figures.
Figure 2.
Mitochondrial proteins of group A, with maximum intensity of signal occurring 20.0 mm from the basal meristem. The data were calculated from the mean values of each of the mitochondrial proteins categorized as group A. The group A mean is also represented by a dashed line in each of the individual protein profiles. Note that in the case of the α and β ATPase proteins, “A” and “C” refer to different monoclonal antibodies that recognize different epitopes of the same subunit type (Luethy et al., 1993).
Figure 3.
Mitochondrial proteins of group B, with maximum intensity of signal occurring 50.0 mm from the basal meristem. The data were calculated from the mean values of each of the mitochondrial proteins categorized as group B. The group B mean is also represented by a dashed line in each of the individual protein profiles.
Figure 4.
Mitochondrial proteins of group C, with maximum intensity of signal occurring at the leaf tip. The data were calculated from the mean values of each of the mitochondrial proteins categorized as group C. The group C mean is also represented by a dashed line in each of the individual protein profiles.
Eleven of the mitochondrial proteins/polypeptides fell into group A, in which the intensity of the protein band increased to a maximum at 20 mm from the basal meristem and then declined progressively toward the leaf tip (Fig. 2). These proteins were: the E1 β-subunit of PDC, ORF156, ORF577, alternative oxidase, RPS12, Cyt oxidase subunits II and III, malic enzyme, and the α- and β-subunits of ATPase.
The proteins/polypeptides with a group B developmental profile were: the E1 α-subunit of PDC, IDH, HSP70A, cpn60C, and cpn60B (Fig. 3). These increased in labeling intensity to a broad maximum between 50 and 60 mm from the leaf base, where cells were between 35 and 45 h old.
The third category of proteins, group C, showed an initial increase in labeling intensity to reach a “plateau” between 20 and 50 mm from the leaf base, as was the case for proteins of group A. However, in contrast to group A, there was a further increase in the region from 50 mm, where cells were 35 h old, up to the leaf tip (Fig. 4). Proteins in this group were the four subunits of GDC (P, H, T, and L proteins), fumarase, and FDH. This pattern of development was also found when the four GDC subunits were analyzed by western blots (data not shown).
DISCUSSION
Developmental Changes in Leaf Physiology
The barley primary leaves studied here were harvested at a time when their growth rate was at a maximum, which ensured that the age differential between leaf base and tip was also at a maximum. Estimates of mitotic indices (data not shown) indicated that the basal intercalary meristem is located between 0 and 5 mm above the point of attachment of the primary leaf to the seed (i.e. the leaf base). The zone of cell elongation extends to 20 mm from the leaf base, where the number of mesophyll cells per leaf section becomes constant and the interstomatal distance is at its maximum. The former is an indirect estimate of the mesophyll cell length and the latter a direct measurement of epidermal cell length.
The gradient of cell age closely resembled that previously reported for barley primary leaves (Barkardottir et al., 1987). The large increment in cell age within the distal 5 mm of the leaf was attributable to the time taken for this tissue to emerge from the seed after imbibition (Barkardottir et al., 1987).
The change in soluble protein content that we found resembled that found in previous studies (Viro and Kloppstech, 1980; Barkardottir et al., 1987). The initial decrease in protein content coincided with the decrease in cell number per section as the cells were elongating. When protein content was expressed on a per-cell basis, there was an almost linear increase along the leaf. One reason for this is the increased synthesis of Rubisco (Viro and Kloppstech, 1980), with a 20-fold increase in Rubisco protein content per mesophyll cell (Dean and Leech, 1982b). Increases in other photosynthetic proteins, such as the chlorophyll a/b-binding protein (CAB; Mayfield and Taylor, 1984), chloroplast NADP-dependent glyceraldehyde 3-phosphate dehydrogenase (Sibley and Anderson, 1989; Lernmark and Gardestrom, 1994), phosphoglycerate kinase (Shah and Bradbeer, 1991), and Fru-1,6-bisphosphatase (Sibley and Anderson, 1989), have also been reported in developing barley leaves. This increase in photosynthetic protein, together with increases in chloroplast size and number (Baumgartner et al., 1989) and chlorophyll content (Barkardottir et al., 1987; Lernmark and Gardestrom, 1994), contribute to an increase in photosynthetic capacity. We detected photosynthetic activity only in cells located beyond the elongation zone, 20 mm or more from the basal meristem, where the cells were greater than 13 h old. By the time these cells had reached the leaf tip (160 h), their photosynthetic activity had increased approximately 20-fold.
In contrast to the increase in photosynthetic activity, the rate of dark respiratory O2 uptake per cell remained constant along the leaf. Previous studies have reported a decline in respiratory activity in relation to increasing photosynthetic development (Kidd et al., 1921; Azcon-Bieto et al., 1983), and this was also the case when our data were expressed relative to soluble protein content, with a 4-fold decrease in respiration rate between the leaf base and tip. The large increase in photosynthetic protein masks any underlying response of nonphotosynthetic proteins and we therefore recommend the use of cell number as a constant parameter in developmental studies.
Development of Mitochondrial Proteins and Polypeptides
A number of controls were used to minimize the possibility of artifacts that might result from the analysis of slot blots. The amount of protein loaded onto the wells was tested over a wide range to ensure that the signal was proportional to the amount of protein present in the sample. Other variables tested and standardized included the concentration of primary and secondary antibodies, the period of incubation with ECL reagents, and the duration of film and PVDF membrane exposure. In addition, mixing experiments were carried out in which mature leaf sections were co-extracted with immature sections. Separate extracts were also mixed together before loading onto the membrane. All of these controls verified that the signal detected on the image analyzer was proportional to the amount of protein and that there was nothing interfering with the detection of the signal. The data shown here were accumulated from 10 individual scans of slot blots of each of the individual proteins. Pixel values were summed to give pooled data, so se values are not applicable.
The most significant outcome of the present study was our observation that the composition of mitochondrial proteins changes during leaf cell development. All of the proteins that were analyzed increased in the early stages of development to the end of the cell elongation zone (20 mm, 13 h). From this point on there appeared to be three distinct patterns of development: group A proteins decreased and group B and C proteins remained constant up to 50 mm (35 h) from the leaf base, where group B proteins decreased and group C proteins increased up to the leaf tip (160 h).
The initial increase in all mitochondrial proteins occurred within a region of the leaf (0–20 mm) covered by the coleoptile, where the number of mitochondria per mesophyll cell has been found to increase in wheat primary leaves (Tobin and Rogers, 1992). This suggests that there is a general increase in the cellular concentration of mitochondrial proteins throughout the elongation zone. Despite this, there was no marked change in respiratory rate per cell (Fig. 1e), indicating that other factors such as adenylate control or substrate availability exert a stronger control over respiratory flux in vivo (Moore et al., 1992).
The proteins and polypeptides of group A include a number that are mitochondrially encoded in cereals, such as RPS12 (subunit 12 of the small ribosomal subunit), Cyt oxidase subunits II and III, the α- and β-subunits of ATPase (Gualberto et al., 1988; Schuster and Brennicke, 1994), and polypeptides coded by orf156 (Gualberto et al., 1991) and orf577 (Gonzalez et al., 1993; Handa et al., 1996). orf156 is part of the nad3-rps12 transcription unit of wheat mtDNA and encodes an 18-kD mitochondrial membrane protein of unknown function (Gualberto et al., 1991). orf577 is homologous to the ccl1 gene of bacteria and its gene product is thought to be involved in the assembly of c-type Cyts (Gonzalez et al., 1993; Handa et al., 1996). The relative decrease in content of these proteins in cells beyond the elongation zone coincides with the onset of photosynthesis and exposure to light as these cells emerge from beneath the coleoptile.
The remaining three proteins of group A were nuclear encoded: alternative oxidase, malic enzyme, and the E1 β-subunit of PDC. All of these proteins are highly regulated. The alternative oxidase can exist in either a reduced or an oxidized form, with activity associated only with the former (Umbach and Siedow, 1997). A single cross-reacting band at 35 kD, corresponding to the reduced form of the alternative oxidase (Umbach and Siedow, 1993), was detected on western blots of crude leaf extracts from barley; however, we could not detect the 67-kD oxidized homodimer even in the presence of up to 40 mm diamide, a strong oxidizing agent (data not shown). This suggests that either there was no homodimeric form of the alternative oxidase in barley, or that the monoclonal antibody failed to cross-react with this form. The intensity of the 35-kD band increased in the presence of 10% (v/v) mercaptoethanol (data not shown), indicating that a significant proportion of the alternative oxidase was in the oxidized, homodimeric form before mercaptoethanol addition. We conclude from this that the monoclonal antibody does not recognize the oxidized form of alternative oxidase in barley.
Our preliminary analyses of western blots of crude extracts from serial, transverse leaf sections indicated that the 35-kD (reduced) form of alternative oxidase increased to a maximum at the leaf tip (data not shown). Our failure to detect a 67-kD form permits only the tentative conclusion that there is an increase in alternative oxidase protein during barley leaf development. The discrepancy between western-blot and slot-blot analysis may be attributable to the difference in conditions of the two assays. We cannot exclude the possibility that the slot-blot assay, which was carried out in the absence of mercaptoethanol, underestimated the level of alternative oxidase protein. Further work is needed to clarify this point.
Lennon et al. (1995) reported an increase in the reduced form of alternative oxidase protein during leaf development in pea, which coincided with the onset of alternative oxidase activity and with an increase in Gly oxidation capacity of the mitochondria. Although we found that the rate of salicylhydroxamic acid-sensitive respiration in the presence of KCN remained constant per cell at all stages of leaf development (data not shown), this is not a good indication of alternative pathway activity because electrons may be diverted from the Cyt pathway under these conditions (Day et al., 1996). Further analysis, ideally on purified mitochondrial preparations, is required to clearly elucidate the pattern of alternative oxidase development in barley.
The antiserum raised against NAD-dependent malic enzyme from potato recognizes both the 59- and 62-kD subunits (data not shown) and, although these have been found to be coordinately expressed in vivo (Bourguignon and Leaver, 1994), we have yet to determine if this is also the case in barley. The activity of the enzyme is regulated allosterically by a number of metabolites (Grissom et al., 1983; Day et al., 1984), and is modified by changes in the oligomeric state (Grover and Wedding, 1984). Further analysis is required, therefore, to determine if the increase in total malic enzyme protein is caused by the presence of both subunits and whether this relates directly to the activity of the enzyme.
The E1 β-subunit of mammalian PDC binds to the E1 α-subunit to form a heterotetramer (α2β2) E1 subunit, or pyruvate dehydrogenase (Koike and Koike, 1976). It is not clear why the development of the E1 β-subunit in barley leaves differs from that of the E1 α-subunit, although the stoichiometry of PDC has yet to be determined in plants. Furthermore, the E1 α-subunit is regulated by phosphorylation (Camp and Randall, 1985), so its activity cannot be directly related to the amount of protein. It will be necessary to analyze the development of the whole complex, including both the E2 and E3 proteins, to fully understand the control of PDC synthesis. A previous study of barley (Lernmark and Gardestrom, 1994) found that mitochondrial PDC activity remains constant during leaf development, although the degree of in vivo phosphorylation has yet to be established.
The group B proteins, which begin to decline from 50 mm onward, are two components of the TCA cycle, the E1 α-subunit of PDC and NAD-dependent IDH, and proteins involved in the transport of polypeptides into the mitochondrial matrix, mtHSP70 (Lund and Elthon, 1993; Moore et al., 1993), and cpn60 (Tsugeki et al., 1992). These data indicate that there is differential development not only of the PDC but also of mitochondrial dehydrogenases, with malic enzyme protein content declining before that of NAD-IDH. The later development of fumarase protein provides further evidence of a change in the relative content of TCA cycle proteins. In all cases, these profiles have yet to be compared with measurements of enzyme activity. However, it is interesting to note that the TCA cycle proteins involved in the decarboxylative portion of the cycle, malic enzyme, PDC, and IDH, reach a peak before that of the nondecarboxylative portion, which is represented by fumarase. This change in composition of TCA cycle proteins in barley leaves may mark a transition in the role of the mitochondria from bioenergetic to biosynthetic (Tobin and Rogers, 1992).
The peak in cellular content of the mitochondrial HSP70 (Lund and Elthon, 1993) and cpn60 proteins coincides with the stage of leaf development at which there is a major increase in the amount of GDC protein (Figs. 3 and 4). The antibodies used to detect these proteins were specific for mitochondrial HSP70 and cpn60 and did not cross-react with barley chloroplast proteins (see Methods). The synthesis of GDC would be expected to impose significant demands on the protein import machinery given the major accumulation of this protein in the matrix of leaf mitochondria (Oliver, 1994).
The pattern of development of group C proteins closely resembles the development of photosynthesis, with a rapid increase occurring from 50 mm onward. This coincides with the point at which a number of photosynthetic and photorespiratory proteins also increase, and at which chloroplast division is reaching completion (Tobin et al., 1988). The proteins within this group include all four subunits of GDC, the P, H, T, and L proteins. These increase in parallel throughout the development of the barley primary leaf. This is in contrast to the findings of a previous study from our group, in which the L protein was present in high concentrations in basal cells of wheat primary leaves, whereas the remaining GDC proteins developed only after emergence of the leaf cells from beneath the coleoptile (Rogers et al., 1991). It is well established that the synthesis of the GDC proteins is light regulated (Oliver, 1994). One reason for this difference between species, therefore, might be a difference in the amount of light penetration into the immature cells beneath the coleoptile, because, in contrast to wheat, barley coleoptiles lack chlorophyll (data not shown).
The developmental pattern of GDC proteins is consistent with the function of this complex in oxidizing Gly generated during photorespiration. The reason for the parallel development of FDH is intriguing. FDH has been detected only in small amounts in leaf tissue (Oliver, 1981) and is particularly abundant in mitochondria from potato tubers (Colas des Francs-Small et al., 1993). FDH was once considered to be involved in the oxidation of formate generated by the nonenzymatic oxidation of glyoxylate by H2O2 to yield formate and CO2 during photorespiration (Grodzinski and Butt, 1976; Zelitch, 1992). The presence of high concentrations of catalase in leaf peroxisomes indicates that H2O2 is unlikely to accumulate in sufficient quantity for glyoxylate decarboxylation to occur (Walton, 1982), and there is no evidence of increased rates of CO2 production in catalase-deficient mutants of barley (Kendall et al., 1983). Furthermore, Colas des Francs-Small et al. (1993) found FDH to decrease during greening of etiolated leaves. Nevertheless, recent evidence from transgenic tobacco with altered catalase activity, in which CO2 compensation points were inversely related to catalase content, once again raises the possibility that glycolate peroxidation and formate metabolism may take part in photorespiratory carbon cycling (Brisson et al., 1998).
The outcome of the present study is evidence of mitochondrial heterogeneity, with marked changes in protein composition at different stages of leaf development. Without measurements of enzyme activity or analysis of isolated mitochondria, it is not possible to conclude whether these changes affect mitochondrial activity. It is also necessary to consider the spatial heterogeneity of the leaf, for which there is already evidence of differential expression of mitochondrial proteins such as GDC (Tobin et al., 1989) between different cell types. Further analysis is required to determine the full extent of the spatial and temporal control of mitochondrial form and function in higher plants.
Abbreviations:
- ECL
enhanced chemiluminescence
- FDH
formate dehydrogenase
- GDC
Gly decarboxylase complex
- IDH
isocitrate dehydrogenase
- PDC
pyruvate dehydrogenase complex
- VD
displacement velocity
Footnotes
This work was funded by The Royal Society (University Research Fellowship to A.K.T.; Pickering Research Fellowship to C.G.B.) and by a Biological and Biotechnological Sciences Research Council, Biochemistry of Metabolic Regulation in Plants studentship (to P.T.).
LITERATURE CITED
- Arron GP, Edwards GE. Light-induced development of glycine oxidation by mitochondria from sunflower cotyledons. Plant Sci Lett. 1980;18:229–235. [Google Scholar]
- Azcon-Bieto J, Lambers H, Day DA. Respiratory properties of developing bean and pea leaves. Aust J Plant Physiol. 1983;10:237–245. [Google Scholar]
- Barkardottir RB, Jensen BF, Kreiberg JD, Nielsen PS, Gausing K. Expression of selected nuclear genes during leaf development in barley. Dev Genet. 1987;8:495–511. [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. Plastid transcription activity and DNA copy number increase early in barley chloroplast development. Plant Physiol. 1989;89:1011–1018. doi: 10.1104/pp.89.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal RH, Oliver DJ. Biochemical and molecular characterisation of fumarase from plants. Purification and characterisation of the enzyme: cloning, sequencing, and expression of the gene. Arch Biochem Biophys. 1997;348:65–74. doi: 10.1006/abbi.1997.0359. [DOI] [PubMed] [Google Scholar]
- Bourguignon J, Leaver CJ. Plant mitochondrial NAD-dependent malic enzyme: cDNA cloning, deduced primary structure of the 59-kDa and 62-kDa subunits, import, gene complexity and expression analysis. J Biol Chem. 1994;269:4780–4786. [PubMed] [Google Scholar]
- Brisson LF, Zelitch I, Havir EA. Manipulation of catalase levels produces altered photosynthesis in transgenic tobacco plants. Plant Physiol. 1998;116:259–269. doi: 10.1104/pp.116.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp PJ, Randall DD. Purification and characterisation of the pea chloroplast pyruvate dehydrogenase complex. Plant Physiol. 1985;77:571–577. doi: 10.1104/pp.77.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Remy R (1992) Variation of the polypeptide composition of mitochondria isolated from different potato tissues. Plant Physiol 98: 273–278 [DOI] [PMC free article] [PubMed]
- Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Remy R (1993) Identification of a major soluble protein in mitochondria from non-photosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol 102: 1171–1177 [DOI] [PMC free article] [PubMed]
- Dale JE. Growth and photosynthesis in the first leaf of barley: the effect of time and application of nitrogen. Ann Bot. 1972;36:967–979. [Google Scholar]
- Day DA, Krab K, Lambers H, Moore AL, Siedow JN, Wagner AM, Wiskich JT. The cyanide-resistant oxidase. To inhibit or not to inhibit, that is the question. Plant Physiol. 1996;110:1–2. doi: 10.1104/pp.110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Neuburger M, Douce R. Activation of NAD-linked malic enzyme in intact plant mitochondria by exogenous coenzyme-A. Arch Biochem Biophys. 1984;231:233–242. doi: 10.1016/0003-9861(84)90383-7. [DOI] [PubMed] [Google Scholar]
- Dean C, Leech RM. Genome expression during normal leaf development. 1. Cellular and chloroplast numbers and DNA, RNA and protein levels in tissues at different ages within a 7-day-old wheat leaf. Plant Physiol. 1982a;69:904–910. doi: 10.1104/pp.69.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C, Leech RM. The co-ordinated synthesis of the large and small subunits of ribulose bisphosphate carboxylase during early cellular development within a 7-day-old wheat leaf. FEBS Lett. 1982b;140:113–116. [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 1989;89:1311–1317. doi: 10.1104/pp.89.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardestrom P, Bergman A, Ericson I. Oxidation of glycine via the respiratory chain of mitochondria prepared from different parts of spinach. Plant Physiol. 1980;65:389–391. doi: 10.1104/pp.65.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DH, Bonnard G, Grienenberger JM. A gene involved in the biogenesis of cytochromes is co-transcribed with a ribosomal protein gene in wheat mitochondria. Curr Genet. 1993;24:248–255. doi: 10.1007/BF00351799. [DOI] [PubMed] [Google Scholar]
- Grissom CB, Canellas PF, Wedding RT. Allosteric regulation of the NAD malic enzyme from cauliflower: activation by fumarate and coenzyme A. Arch Biochem Biophys. 1983;220:133–144. doi: 10.1016/0003-9861(83)90394-6. [DOI] [PubMed] [Google Scholar]
- Grodzinski B, Butt VS. Hydrogen peroxide production and the release of carbon dioxide during glycolate oxidation in leaf peroxisomes. Planta. 1976;133:261–266. doi: 10.1007/BF00393233. [DOI] [PubMed] [Google Scholar]
- Grover SD, Wedding RT. Modulation of the activity of NAD malic enzyme from Solanum tuberosum by changes in oligomeric state. Arch Biochem Biophys. 1984;234:418–425. doi: 10.1016/0003-9861(84)90288-1. [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Bonnard JM, Lamattina L, Grienenberger JM. Expression of the wheat mitochondrial nad3-rps12 transcription unit: correlation between editing and messenger RNA maturation. Plant Cell. 1991;3:1109–1120. doi: 10.1105/tpc.3.10.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto JM, Wintz H, Weil J-H, Grienenberger JM. The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet. 1988;215:118–127. doi: 10.1007/BF00331312. [DOI] [PubMed] [Google Scholar]
- Handa H, Bonnard G, Grienenberger JM. The rapeseed mitochondrial gene encoding a homologue of the bacterial protein ccl1 is divided into two independently transcribed reading frames. Mol Gen Genet. 1996;252:292–302. doi: 10.1007/BF02173775. [DOI] [PubMed] [Google Scholar]
- Kendall AC, Keys AJ, Turner JC, Lea PJ, Miflin BJ. ) Planta. 1983;159:505–511. doi: 10.1007/BF00409139. [DOI] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ. Photorespiratory nitrogen cycle. Nature. 1978;275:741–743. [Google Scholar]
- Kidd F, Briggs GE, West C. A quantitative analysis of the growth of Helianthus annuus. I. The respiration of the plant and its parts throughout the life cycle. Proc R Soc Ser B. 1921;92:368–384. [Google Scholar]
- Koike M, Koike K. Structure, assembly and function of mammalian α-keto acid dehydrogenase complexes. Adv Biophys. 1976;9:187–227. [PubMed] [Google Scholar]
- Lennon AM, Pratt J, Leach G, Moore AL. Developmental regulation of respiratory activity in pea leaves. Plant Physiol. 1995;107:925–932. doi: 10.1104/pp.107.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lernmark U, Gardestrom P. Distribution of pyruvate dehydrogenase complex activities between chloroplasts and mitochondria from leaves of different species. Plant Physiol. 1994;106:1633–1638. doi: 10.1104/pp.106.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightowlers R, Chrzanowska-Lightowlers Z, Marusich M, Capaldi A. Subunit function in eukaryote cytochrome c oxidase: a mutation in the nuclear-coded subunit IV allows assembly but alters the function and stability of yeast cytochrome c oxidase. J Biol Chem. 1991;266:7688–7693. [PubMed] [Google Scholar]
- Luethy MH, David NR, Elthon TE, Miernyk JA, Randall DD. Characterisation of a monoclonal antibody recognising the E1-alpha subunit of plant mitochondrial pyruvate dehydrogenase. J Plant Physiol. 1995a;145:443–449. [Google Scholar]
- Luethy MH, Horak A, Elthon TE. Monoclonal antibodies to the α- and β- subunits of the plant mitochondrial F1-ATPase. Plant Physiol. 1993;101:931–937. doi: 10.1104/pp.101.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luethy MH, Miernyk JA, Randall DD. Expression of the plant mitochondrial pyruvate dehydrogenase in Escherichia coli (abstract no. 664) Plant Physiol. 1995b;108:S-129. [Google Scholar]
- Lund AA, Elthon TE. Maize mitochondrial heat-shock and chaperone protein systems (abstract no. 724) Plant Physiol. 1993;102:S-128. [Google Scholar]
- Mayfield SP, Taylor WC. The appearance of photosynthetic proteins in developing maize leaves. Planta. 1984;161:481–486. doi: 10.1007/BF00407079. [DOI] [PubMed] [Google Scholar]
- McIntosh CA, Oliver DJ. NAD-linked isocitrate dehydrogenase. Isolation, purification and characterisation of the protein from pea mitochondria. Plant Physiol. 1992;100:69–75. doi: 10.1104/pp.100.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AL, Siedow JN, Fricaud AC, Vojnikov V, Walters AJ, Whitehouse DG. Regulation of mitochondrial respiratory activity in photosynthetic systems. In: Tobin AK, editor. Plant Organelles: Compartmentation of Metabolism in Photosynthetic Tissue. Cambridge, UK: Cambridge University Press; 1992. pp. 188–210. [Google Scholar]
- Moore AL, Walters AJ, Lennon AM, Watts FZ (1993) Developmental regulation of the protein import apparatus of plant mitochondria. In A Brennicke, U Kuck, eds, Plant Mitochondria: With Emphasis on RNA Editing and Cytoplasmic Male Sterility. Weinheim, Germany, pp 291–298
- Morgan CL, Turner SM, Rawsthorne S. Coordination of the cell specific distribution of the 4 subunits of glycine decarboxylase and of serine hydroxymethyltransferase in leaves of C3–C4 intermediate species from different genera. Planta. 1993;190:468–473. [Google Scholar]
- Oliver DJ. Formate oxidation and oxygen reduction by leaf mitochondria. Plant Physiol. 1981;68:703–705. doi: 10.1104/pp.68.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DJ. The glycine decarboxylase complex from plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:323–337. [Google Scholar]
- Ougham HJ, Jones TWA, Evans MLL. Leaf development in Lolium temulentum L.: progressive changes in soluble polypeptide complement and isoenzymes. J Exp Bot. 1987;38:1689–1696. [Google Scholar]
- Rios R, Debuyser J, Henry Y, Ambard-Bretteville F, Remy R. 2-dimensional electrophoretic comparison of mitochondrial polypeptides from different wheat (Triticum aestivum L.) tissues. Plant Sci. 1991;76:159–166. [Google Scholar]
- Rogers WJ, Jordan BR, Rawsthorne S, Tobin AK. Changes to the stoichiometry of glycine decarboxylase subunits during wheat (Triticum aestivum L.) and pea (Pisum sativum L.) leaf development. Plant Physiol. 1991;96:952–956. doi: 10.1104/pp.96.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnyder H, Nelson CJ. Diurnal growth of tall fescue leaf blades. I. Spatial distribution of growth, deposition of water, and assimilate import in the elongation zone. Plant Physiol. 1988;86:1070–1076. doi: 10.1104/pp.86.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W, Brennicke A. The plant mitochondrial genome: physical structure, information content, RNA editing, and gene migration to the nucleus. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:61–78. [Google Scholar]
- Shah N, Bradbeer JW. The development of the chloroplastic and cytosolic isoenzymes of phosphoglycerate kinase during barley leaf ontogenesis. Planta. 1991;185:401–406. doi: 10.1007/BF00201064. [DOI] [PubMed] [Google Scholar]
- Sibley MH, Anderson LE. Light/dark modulation of enzyme activity in developing barley leaves. Plant Physiol. 1989;91:1620–1624. doi: 10.1104/pp.91.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taanman J, Capaldi RA. Subunit VIa of yeast cytochrome c oxidase is not necessary for assembly of the enzyme complex but modulates the enzyme activity: isolation and characterisation of the nuclear-encoded gene. J Biol Chem. 1993;268:18754–18761. [PubMed] [Google Scholar]
- Tobin AK, Ridley SM, Stewart GR. Changes in the activities of chloroplast and cytosolic isoenzymes of glutamine synthetase during normal leaf growth and plastid development in wheat. Planta. 1985;163:544–548. doi: 10.1007/BF00392711. [DOI] [PubMed] [Google Scholar]
- Tobin AK, Rogers WJ. Metabolic interactions of organelles during leaf development. In: Tobin AK, editor. Plant Organelles: Compartmentation of Metabolism in Photosynthetic Tissue. Cambridge, UK: Cambridge University Press; 1992. pp. 293–323. [Google Scholar]
- Tobin AK, Sumar N, Patel M, Moore AL, Stewart GR. Development of photorespiration during chloroplast biogenesis in wheat leaves. J Exp Bot. 1988;39:833–843. [Google Scholar]
- Tobin AK, Thorpe JR, Hylton CM, Rawsthorne S. Spatial and temporal influences on the cell-specific distribution of glycine decarboxylase in leaves of wheat (Triticum aestivum L.) and pea (Pisum sativum L.) Plant Physiol. 1989;91:1219–1225. doi: 10.1104/pp.91.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R, Mori H, Nishimura M. Purification, cDNA cloning and northern blot analysis of mitochondrial chaperonin 60 from pumpkin cotyledons. Eur J Biochem. 1992;209:453–458. doi: 10.1111/j.1432-1033.1992.tb17309.x. [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. Covalent and non-covalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol. 1993;103:845–854. doi: 10.1104/pp.103.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. Changes in the redox state of the alternative oxidase regulatory sulfhydryl/disulfide system during mitochondrial isolation: implications for inferences of activity in vivo. Plant Sci. 1997;123:19–28. [Google Scholar]
- Viro M, Kloppstech K. Differential expression of the genes for ribulose-1,5-bisphosphate carboxylase and light harvesting chlorophyll a/b protein in developing barley leaves. Planta. 1980;150:41–45. doi: 10.1007/BF00385613. [DOI] [PubMed] [Google Scholar]
- Walker JL, Oliver DJ. Light-induced increases in the glycine decarboxylase multienzyme complex from pea leaf mitochondria. Arch Biochem Biophys. 1986;248:626–638. doi: 10.1016/0003-9861(86)90517-5. [DOI] [PubMed] [Google Scholar]
- Walton NJ. Glyoxylate decarboxylation during glycolate oxidation by pea leaf extracts: significance of glyoxylate and extract concentrations. Planta. 1982;155:218–224. doi: 10.1007/BF00392719. [DOI] [PubMed] [Google Scholar]
- Winning BM, Bourguignon J, Leaver CJ. Plant mitochondrial NAD dependent malic enzyme: cDNA cloning, deduced primary structure of the 59-kDa and 62-kDa subunits, import, gene complexity and expression analysis. J Biol Chem. 1994;269:4780–4786. [PubMed] [Google Scholar]
- Zelitch I. Control of plant productivity by regulation of photorespiration. BioScience. 1992;42:510–516. [Google Scholar]