Abstract
Treatment of wounded military personnel at military medical centers is often complicated by colonization and infection of wounds with pathogenic bacteria. These include nosocomially transmitted, often multidrug-resistant pathogens such as Acinetobacter baumannii-calcoaceticus complex, Pseudomonas aeruginosa and extended spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. We analyzed the efficacy of multivalent adhesion molecule (MAM) 7-based anti-adhesion treatment of host cells against aforementioned pathogens in a tissue culture infection model. Herein, we observed that a correlation between two important hallmarks of virulence, attachment and cytotoxicity, could serve as a useful predictor for the success of MAM7-based inhibition against bacterial infections. Initially, we characterized 20 patient isolates (five from each pathogen mentioned above) in terms of genotypic diversity, antimicrobial susceptibility and important hallmarks of pathogenicity (biofilm formation, attachment to and cytotoxicity toward cultured host cells). All isolates displayed a high degree of genotypic diversity, which was also reflected by large strain-to-strain variability in terms of biofilm formation, attachment and cytotoxicity within each group of pathogen. Using non-pathogenic bacteria expressing MAM7 or latex beads coated with recombinant MAM7 for anti-adhesion treatment, we showed a decrease in cytotoxicity, indicating that MAM7 has potential as a prophylactic agent to attenuate infection by multidrug-resistant bacterial pathogens.
Keywords: Acinetobacter, Klebsiella, MAM7, Pseudomonas, adhesin, anti-adhesion treatment, bacterial attachment, multidrug-resistance, wound infections
Introduction
Wound treatment and healing is often complicated by colonization and infection of wounds with bacterial pathogens. Colonization screening at time of admission performed on military medical personnel wounded in combat during missions in Iraq and Afghanistan has shown that the majority of wound infections are caused by a limited number of bacterial species.1 Besides Gram-positive methicillin-resistant Staphylococcus aureus (MRSA), there are a number of Gram-negative bacteria routinely isolated from wounds, including Acinetobacter baumannii-calcoaceticus complex (ABC), Pseudomonas aeruginosa and extended spectrum β-lactamase (ESBL)-producing Enterobacteriacea such as Escherichia coli and Klebsiella pneumoniae. Together, these are the causal agents of a large proportion of wound-associated infections, which are responsible for prolonged wound healing, thereby leading to extended periods of care.2-5 Screening performed over a period of 2003–2009 revealed that an increasing number of isolates could be classified as multidrug-resistant (MDR), additionally complicating the care and treatment of infected patients.1,6,7 Attempts have been made to prevent infection by administering antimicrobials in the field, but this has only lead to limited success.8 Thus, there is a clear need for alternative approaches to prevent infections or treat existing infections.
We have recently identified a novel group of adhesins, termed multivalent adhesion molecule (MAM) 7, which is widely found in Gram-negative pathogens. These adhesins are constitutively expressed and involved in initial attachment of pathogens to host cells, by interacting with both the host cell receptor phosphatidic acid and the co-receptor fibronectin.9,10 We have previously explored the use of non-pathogenic bacteria expressing MAM7 on their surface (BL21-MAM7) or recombinant, purified MAM7 immobilized on polymer beads (bead-MAM7), which mimics the bacterial surface display of the adhesin, as a way of inhibiting bacterial attachment.9,11 Pre-treatment of host cells with MAM7-based inhibitors has proved to be effective in ameliorating the effects of bacterial infection with a range of important clinical pathogens, including Vibrio parahaemolyticus, Vibrio cholerae, Yersinia pseudotuberculosis and enteropathogenic E. coli (EPEC). In the present study, we set out to explore the potential of MAM7-based inhibitors against infection with patient-derived bacterial isolates associated with hard-to-treat wound infections, since MAM7 homologs have been identified in sequenced strains from all five bacterial species used. We further compared the susceptibilities of the bacterial isolates to conventional antimicrobials with their susceptibility to MAM7-based inhibition in vitro. Lastly, we characterized several features described to contribute to pathogenicity, such as biofilm formation, host cell attachment and cytotoxicity toward host cells, caused by each of the isolates. Based on our findings, we propose that isolates that show a correlation between their ability to attach to host cells and to cause cytotoxicity are generally more susceptible to MAM7-based anti-adhesion treatment. However, this hypothesis is based on a limited set of isolates tested and our studies will have to be extended to include a larger variety of bacterial strains to be tested further.
Results
Isolation and genotyping of bacterial strains associated with patient wounds
Bacterial isolates were collected from infected patients treated at the San Antonio Military Medical Center at Fort Sam Houston, TX, following their evacuation from Afghanistan and Iraq, during a time period spanning the years 2006 to 2010.12,13 We characterized a total of 20 different isolates obtained either by superficial (three cases) or deep wound culture (14 cases), blood culture (two cases) or urine culture (one case), each of which belonged to one of the four most predominant species of Gram-negative bacteria leading to wound infections, ABC, P. aeruginosa and ESBL-producing K. pneumoniae and E. coli (each represented by five isolates, respectively). Pulsed field gel electrophoresis (PFGE) was used for genotypic characterization of all isolates and revealed a high degree of genotypic diversity within each of the four groups, with each of the five isolates within each group representing a distinct genotype (Table 1).
Table 1. Details of isolation, genotyping and phenotypic characterization of bacterial isolates.
| Species |
Isolate No. |
Year of isolation |
Source |
PFGE type |
Biofilm formation (%) |
Hemolysis |
|---|---|---|---|---|---|---|
| ABC | ||||||
| |
1 |
2006 |
Blood |
ABC PFT 4 |
31 |
N |
| |
2 |
2006 |
Blood |
ABC PFT 3 |
109 |
N |
| |
3 |
2007 |
Wound culture superficial |
ABC PFT 1 |
222 |
N |
| |
4 |
2007 |
Wound culture deep |
ABC PFT 2 |
38 |
N |
| 5 | 2008 | Wound culture deep | ABC PFT 5 | 55 | N |
| P. aeruginosa | ||||||
|---|---|---|---|---|---|---|
| |
1 |
2008 |
Wound culture deep |
PA PFT 7 |
62 |
N |
| |
2 |
2007 |
Wound culture deep |
PA PFT 2 |
192 |
N |
| |
3 |
2009 |
Wound culture deep |
PA PFT 1 |
183 |
N |
| |
4 |
2007 |
Wound culture deep |
PA PFT 4 |
149 |
N |
| 5 | 2008 | Wound culture superficial | PA PFT 5 | 182 | N |
| K. pneumoniae | ||||||
|---|---|---|---|---|---|---|
| |
1 |
2007 |
Wound culture deep |
KP PFT 1 |
420 |
N |
| |
2 |
2008 |
Wound culture deep |
KP PFT 2 |
132 |
N |
| |
3 |
2009 |
Wound culture deep |
KP PFT 7 |
129 |
N |
| |
4 |
2007 |
Wound culture deep |
KP PFT 3 |
58 |
N |
| 5 | 2008 | Wound culture deep | KP PFT 9 | 246 | N |
| E. coli | ||||||
|---|---|---|---|---|---|---|
| |
1 |
2007 |
Wound culture deep |
EC PFT 3 |
24 |
N |
| |
2 |
2007 |
Wound culture superficial |
EC PFT 4 |
20 |
N |
| |
3 |
2009 |
Wound culture deep |
EC PFT 1 |
20 |
N |
| |
4 |
2010 |
Wound culture deep |
EC PFT 7 |
25 |
N |
| 5 | 2008 | Urine culture | EC PFT 2 | 180 | N |
Clinical isolates from patient wounds vary in their ability to form biofilms, attach to host cells and cause cytotoxicity
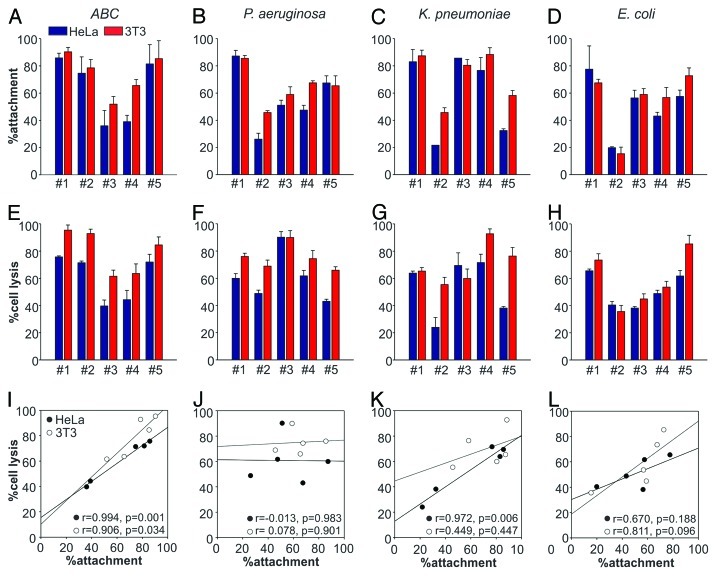
Next, we studied the bacterial isolates for their ability to form biofilms and attach to cultured HeLa epithelial cells and 3T3 fibroblasts using serial dilution plating assays. K. pneumoniae and P. aeruginosa isolates showed the greatest tendency to form biofilms (197 ± 142% and 154 ± 54%, respectively), followed by ABC (105 ± 77%) and E. coli (54 ± 71%) (Table 1). In contrast, ABC isolates had the greatest ability to attach to host cells, followed by K. pneumoniae, P. aeruginosa and E. coli (Fig. 1A–D). However, we observed a large variability in attachment properties within each group, with E. coli isolates showing the most variability (attachment ranged from 15% for isolate #2 to 73% for isolate #5). In addition, we tested all 20 isolates for their ability to cause cytotoxicity in host cells within a 4 h infection experiment using lactate dehydrogenase (LDH) release assays. ABC isolates caused the highest overall cytotoxicity, followed by P. aeruginosa, K. pneumoniae and E. coli (Fig. 1E–H). The variability was largest among the K. pneumonia isolates, with cell lysis ranging from 24% for isolate #2 to 69% for isolate #3. Moreover, we noted two interesting features when comparing attachment and cytotoxicity profiles of the tested isolates. First, in most cases the strains showed a slightly higher attachment and cytotoxic effect on 3T3 fibroblasts. Second, in the case of all groups of pathogens except P. aeruginosa, we observed positive correlation between host cell attachment and pathogen-induced cytotoxicity (Fig. 1I–L).
Figure 1. Host cell attachment and lysis by bacterial isolates. Attachment (top) and cell lysis (LDH release), (middle) caused by five different isolates each of ABC (A and E), P. aeruginosa (B and F), K. pneumoniae (C and G) and E. coli (D and H) tested on HeLa (blue) and 3T3 (red) cells. Values given are means ± standard error (n = 3) from a representative experiment performed in triplicate. Correlation between attachment and cytotoxicity on HeLa (●) and 3T3 (○) cells across different isolates (I–L). Pearson correlation coefficients (r values) and significance levels for correlation coefficients (p values) were used to analyze the correlation between strain attachment and cytotoxicity.
Isolates display a high degree of resistance against a wide range of commonly used antimicrobials
One of the greatest problems faced in treatment of Gram-negative wound infections is the increasing number of MDR bacterial isolates found in patients. We performed antimicrobial susceptibility testing (AST) by determining the minimal inhibiting concentration (MIC) of a panel of antimicrobials on all 20 bacterial isolates using the BD PhoenixTM Automated Microbiology System (Becton Dickinson; Table 2). Antimicrobial panels included several drugs from each of the four most commonly used classes of antimicrobials, aminoglycosides, β-lactams, carbapenems and fluoroquinolones. The panels also included antimicrobials from other classes, such as trimethoprim/sulfamethoxazole, but resistance to these was not considered in the classification of isolates as MDR.7 All tested ABC and P. aeruginosa isolates were found to be MDR. According to the CDC definition, all isolates of ESBL-producing K. pneumoniae and E. coli are also considered MDR. The K. pneumoniae and E. coli we tested indeed displayed a high degree of resistance, being non-susceptible to most aminoglycosides, β-lactams and fluoroquinolones tested (Table 2). All K. pneumoniae and E. coli isolates were susceptible to the tested carbapenems (imipenem, meropenem and ertapenem).
Table 2. Strain susceptibility against commonly screened antibiotics.
| Bacterial species |
Isolate# |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABC | Amik. | AmoxC. | Amp. | Az. | Cefo. | Ceft. | Cefu. | Ceph. | Cipro. | Gati. | Gent. | Im. | Lev. | Mero. | Nf. | Pip. | Tet. | Tob. | TmSm. | |||
| |
1 |
> 32 R |
> 16/8 R |
> 16 R |
> 16 R |
> 32 R |
> 16 R |
> 16 R |
> 16 R |
> 2 R |
> 4 R |
> 8 R |
2 S |
> 4 R |
2 S |
> 64 R |
> 64 R |
> 8 R |
8 I |
> 2/38 R |
|
|
| |
2 |
> 32 R |
> 16/8 R |
> 16 R |
> 16 R |
> 32 R |
> 16 R |
> 16 R |
> 16 R |
> 2 R |
> 4 R |
> 8 R |
> 8 R |
> 4 R |
> 8 R |
> 64 R |
> 64 R |
4 S |
> 8 R |
> 2/38 R |
|
|
| |
3 |
32 I |
> 16/8 R |
> 16 R |
> 16 R |
> 32 R |
8 S |
> 16 R |
> 16 R |
> 2 R |
4 I |
> 8 R |
> 8 R |
4 I |
> 8 R |
> 64 R |
> 64 R |
> 8 R |
> 8 R |
> 2/38 R |
|
|
| |
4 |
> 32 R |
> 16/8 R |
> 16 R |
> 16 R |
> 32 R |
> 16 R |
> 16 R |
> 16 R |
> 2 R |
> 4 R |
> 8 R |
2 S |
> 4 R |
≤ 1 S |
> 64 R |
> 64 R |
> 8 R |
> 8 R |
> 2/38 R |
|
|
| 5 | > 32 R | > 16/8 R | > 16 R | > 16 R | > 32 R | > 16 R | > 16 R | > 16 R | > 2 R | > 4 R | > 8 R | > 8 R | > 4 R | > 8 R | > 64 R | > 64 R | 8 I | 4 S | > 2/38 R |
| P. aeruginosa | Amik. | Amp. | AmpS. | Az. | Cefa. | Cefe. | Cx. | Cd. | Co. | Cefu. | Cipro. | Gent. | Im. | Lev. | Mero. | Nf. | PipT. | Tet. | Tob. | TmSm | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
1 |
≤ 8 S |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
16 I |
> 16 R |
> 16 R |
> 32 R |
> 16 R |
> 2 R |
4 S |
> 8 R |
> 4 R |
4 S |
> 64 R |
> 64/4 R |
> 8 R |
≤ 2 S |
> 2/38 R |
|
| |
2 |
16 S |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
8 S |
> 32 R |
> 16 R |
> 2 R |
4 S |
> 8 R |
> 4 R |
> 8 R |
> 64 R |
> 64/4 R |
> 8 R |
> 8 R |
> 2/38 R |
|
| |
3 |
16 S |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 32 R |
> 16 R |
> 2 R |
8 I |
> 8 R |
> 4 R |
> 8 R |
> 64 R |
> 64/4 R |
> 8 R |
≤ 2 S |
> 2/38 R |
|
| |
4 |
32 I |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 32 R |
> 16 R |
> 2 R |
> 8 R |
> 8 R |
> 4 R |
> 8 R |
> 64 R |
32/4 S |
> 8 R |
> 8 R |
> 2/38 R |
|
| 5 | 32 I | > 16 R | > 16/8 R | > 16 R | > 16 R | > 16 R | > 16 R | > 16 R | > 32 R | > 16 R | > 2 R | > 8 R | 2 S | > 4 R | 4 S | > 64 R | > 64/4 R | > 8 R | > 8 R | > 2/38 R |
| K. pneumoniae | Amik. | AmoxC. | Amp. | Az. | Cefa. | Cefe. | Cefo. | Cx. | Cd. | Co. | Cipro. | Ert. | Gent. | Im. | Lev. | Mero. | Nf. | PipT. | Tet. | Tob. | TmSm | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
1 |
≤ 8 S |
16/8 R |
> 16 R |
> 16 R |
NA |
> 16 R |
> 32 R |
NA |
> 16 R |
NA |
> 2 R |
NA |
> 8 R |
≤ 1 S |
> 4 R |
≤ 1 S |
> 64 R |
NA |
> 8 R |
8 I |
> 2/38 R |
| |
2 |
≤ 8 S |
16/8 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 32 R |
≤ 4 S |
> 2 R |
> 32 R |
> 2 R |
≤ 0.5 S |
> 8 R |
≤ 1 S |
≤ 1 S |
≤ 1 S |
> 64 R |
32/4 I |
> 8 R |
> 8 R |
≤ 0.5/9.5 S |
| |
3 |
≤ 8 S |
8/4 R |
> 16 R |
> 16 R |
> 16 R |
2 R |
≤ 4 S |
> 16 R |
> 2 R |
16 R |
1 S |
≤ 0.5 S |
8 I |
≤ 1 S |
≤ 1 S |
≤ 1 S |
> 64 R |
16/4 S |
> 8 R |
8 I |
≤ 0.5/9.5 S |
| |
4 |
≤ 8 S |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 32 R |
8 S |
> 2 R |
> 32 R |
≤ 0.5 S |
≤ 0.5 S |
> 8 R |
≤ 1 S |
≤ 1 S |
≤ 1 S |
> 64 R |
> 64/4 R |
> 8 R |
8 I |
≤ 0.5/9.5 S |
| 5 | > 32 R | > 16/8 R | > 16 R | > 16 R | NA | 2 R | > 32 R | NA | > 16 R | NA | ≤ 0.5 S | NA | > 8 R | ≤ 1 S | ≤ 1 S | ≤ 1 S | > 64 R | NA | > 8 R | > 8 R | > 2/38 R |
| E. coli | Amik. | Amp. | AmpS. | Az. | Cefa. | Cefe. | Cx. | Cd. | Co. | Cefu. | Cipro. | Gent. | Im. | Lev. | Mero. | Nf. | PipT. | Tet. | Tob. | TmSm | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
1 |
≤ 8 S |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
8 S |
> 16 R |
> 32 R |
> 16 R |
> 2 R |
> 8 R |
≤ 1 S |
> 4 R |
≤ 1 S |
32 S |
32/4 I |
> 8 R |
> 8 R |
> 2/38 R |
|
| |
2 |
≤ 8 S |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
≤ 4 S |
> 16 R |
> 32 R |
> 16 R |
> 2 R |
> 8 R |
≤ 1 S |
> 4 R |
≤ 1 S |
≤ 16 S |
≤ 2/4 S |
≤ 2 S |
> 8 R |
> 2/38 R |
|
| |
3 |
≤ 8 S |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
8 S |
> 16 R |
> 32 R |
> 16 R |
> 2 R |
> 8 R |
≤ 1 S |
> 4 R |
≤ 1 S |
≤ 16 S |
32/4 I |
> 8 R |
> 8 R |
> 2/38 R |
|
| |
4 |
≤ 8 S |
> 16 R |
> 16/8 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 16 R |
> 32 R |
> 16 R |
> 2 R |
≤ 2 S |
≤ 1 S |
> 4 R |
≤ 1 S |
≤ 16 S |
16/4 I |
> 8 R |
≤ 2 S |
> 2/38 R |
|
| 5 | ≤ 8 S | > 16 R | > 16/8 R | 8 R | > 16 R | 8 R | 8 S | 4 R | > 32 R | > 16 R | > 2 R | ≤ 2 S | ≤ 1 S | > 4 R | ≤ 1 S | ≤ 16 S | 32/4 I | > 8 R | > 8 R | > 2/38 R |
Given is the interpretation of minimal inhibiting concentrations in μg/mL in terms of S, susceptible, I, intermediate or R, resistant, according to CLSI breakpoint standards.38 Abbreviations: Amik, Amikacin; AmoxC., Amoxillin-Clavulanate; Amp., Ampicillin; AmpS., Ampicillin-Sulbactam; Az., Aztreonam; Cd., Ceftazidime; Cefa., Cefazolin; Cefe., Cefepime; Cefo., Cefotaxime; Ceft., Ceftazidime; Cefu., Cefuroxime; Ceph., Cephalotin, Co., Ceftriaxone; Cx., Cefoxitin; Cipro., Ciprofloxacin; Ert., Ertapenem; Gati., Gatifloxacin; Gent., Gentamicin; Im., Imipenem; Lev., Levofloxacin; Mero., Meropenem; Nf., Nitrofurantoin; Pip., Piperacillin; PipT., Piperacillin-Tazobactam; Tet., Tetracycline; Tob., Tobramycin; TmSm., Trimethoprim-Sulfamethoxazole; NA, data not available
Prophylactic anti-adhesion treatment decreases pathogen-mediated cytotoxicity for a large number of clinical isolates
We have previously demonstrated that a widely conserved adhesin found in Gram-negative bacteria, termed multivalent adhesion molecule (MAM) 7 can be used to block bacterial attachment sites on host cells, thus diminishing infection by gastrointestinal pathogens such as Vibrio parahaemolyticus, Vibrio cholerae, Yersinia pseudotuberculosis and EPEC in tissue culture models of infection.9 Herein, we investigated if such treatment could be successfully extended to include relevant wound-associated pathogens and tested if MAM7-based anti-adhesion treatment would affect the cytotoxicity mediated by the clinical isolates on HeLa or 3T3 cells using LDH release assays. We used MAM7 either expressed on the surface of non-pathogenic E. coli (BL21-MAM7) or recombinant, purified MAM7 protein immobilized on 1 μm latex beads to mimic surface display on bacteria (bead-MAM7). Cultured cells were pre-incubated with BL21-MAM7 at a multiplicity of infection (MOI) of 100 or bead-MAM7 at a dosage of 7.5 mg protein/106 beads/well for 30 min. Excess inhibitor was subsequently removed by washing and cells were infected with each of the 20 isolates at an MOI of 10. Cytotoxicity toward HeLa and 3T3 cells, either without pre-incubation or following anti-adhesion treatment with BL21-MAM7 or MAM7-beads, was assessed following 4 h of infection (Figs. 2 and 3, respectively). Overall, we observed a large decrease in cytotoxicity when cells were pre-incubated with MAM7-based inhibitors, with the extent of inhibition mediated by either BL21-MAM7 or MAM7-beads being very similar. We have previously shown that pre-incubation with either GST-beads or BL21MAM7ΔN1–44 does not ameliorate the effects of subsequent bacterial infections.9,11 The highest protective effect toward HeLa cells was observed with treatment against ABC (mean inhibition of 76 ± 8% with BL21-MAM7 and 68 ± 6% with MAM7-beads) and K. pneumoniae isolates (mean inhibition of 85 ± 4% and 71 ± 18% with BL21 and beads, respectively) (Fig. 2A and C). In contrast, the inhibitory effect against P. aeruginosa (48 ± 30% and 43 ± 24% for bacteria and beads, respectively) and E. coli (54 ± 19%/51 ± 20%) was less pronounced for most strains, but the variability of treatment responses between different isolates within these groups was generally high (Fig. 2B and D). For example in the case of P. aeruginosa, cytotoxicity inhibition ranged from > 91% for isolate #1 to 12% for isolate #3. The inhibitory profiles were very similar between HeLa cells and 3T3 cells (Fig. 3).
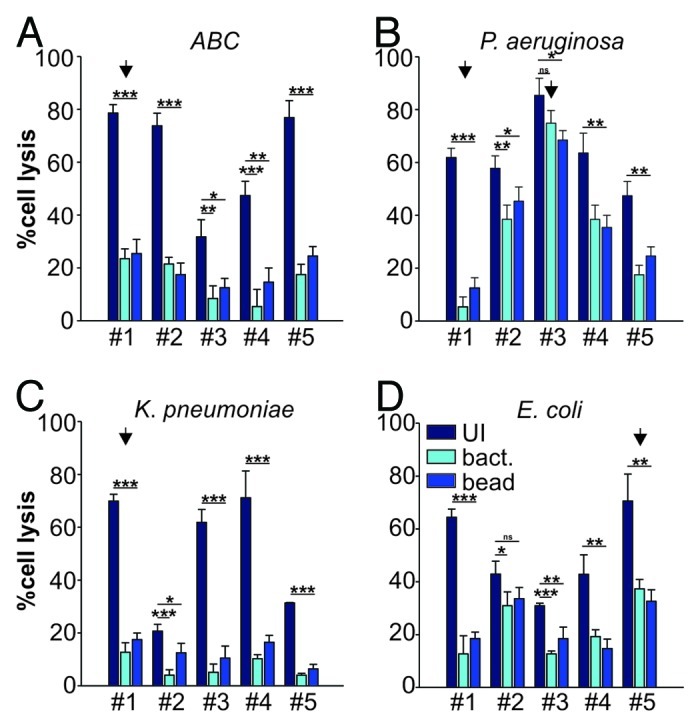
Figure 2. Anti-adhesion treatment of HeLa cells infected with bacterial isolates. Cytotoxocity of ABC (A), P. aeruginosa (B), K. pneumoniae (C) and E. coli (D) isolates was measured following infection of HeLa cells left untreated (dark blue), BL21-MAM7 treated (cyan) or treated with bead-immobilized MAM7 (mid-blue). Values given are means ± standard error (n = 3) from a representative experiment performed in triplicate. Data points marked by an arrow were chosen for visualization using confocal microscopy (Fig. 4). Inhibition data was analyzed for statistical significance using a two-tailed t-test. Levels of significance were indicated as extremely significant (***, p < 0.001), very significant (**, p between 0.001–0.01), significant (*, p between 0.01–0.05) or not significant (ns, p > 0.05).
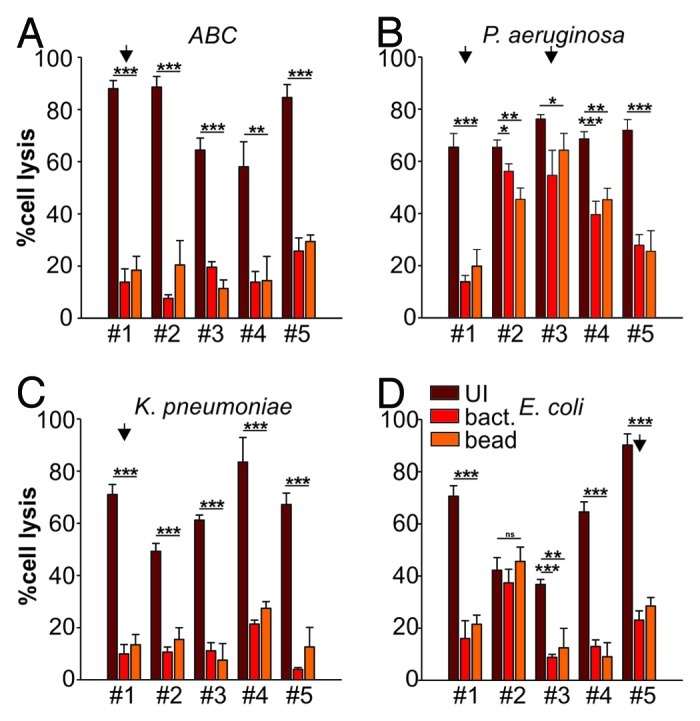
Figure 3. Anti-adhesion treatment of 3T3 cells infected with bacterial isolates. Cytotoxocity of ABC (A), P. aeruginosa (B), K. pneumoniae (C) and E. coli (D) isolates was measured following infection of 3T3 cells left untreated (dark red), BL21-MAM7 treated (red) or treated with bead-immobilized MAM7 (orange). Values given are means ± standard error (n = 3) from a representative experiment performed in triplicate. Data points marked by an arrow were chosen for visualization using confocal microscopy (Fig. 5). Inhibition data was analyzed for statistical significance using a two-tailed t-test. Levels of significance were indicated as extremely significant (***, p < 0.001), very significant (**, p between 0.001–0.01), significant (*, p between 0.01–0.05) or not significant (ns, p > 0.05).
Visualization of MAM7 inhibitory potential on bacterial infections using confocal microscopy
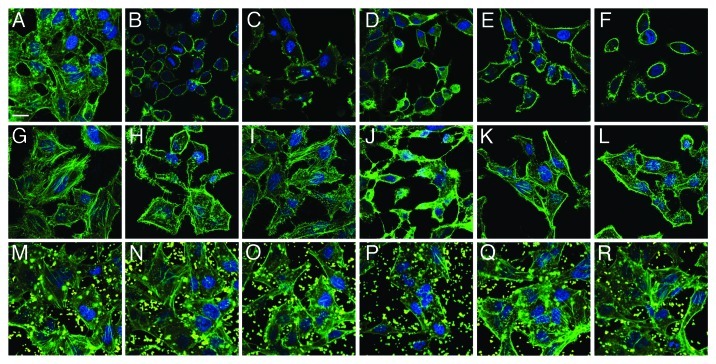
To visualize the detrimental effects of individual pathogens as well as the protective potential of MAM7 inhibitors, we chose representative strains from each of the four groups (marked with arrow in Figs. 2 and 3) and analyzed infections of both HeLa and 3T3 cells as well as inhibition experiments using confocal microscopy (Figs. 4 and 5). This allowed us to further test and correlate results found in LDH release assays with cellular phenotypes. For this purpose, cells were infected with either ABC #1, P. aeruginosa #1, K. pneumoniae #1 or E. coli #5 (all of which showed good responses to anti-adhesion treatment with MAM7). In addition, we analyzed infections with P. aeruginosa #3, which showed the lowest response to inhibition. Comparative microscopic analyses of HeLa cells infected with either P. aeruginosa isolate #1 or #3 demonstrated a large difference between cellular phenotypes following infection with the two isolates, which is in agreement with our findings from LDH release experiments as well as PFGE analyses. While infection with P. aeruginosa #1 caused only limited cell rounding and lysis, even after several hours of infection, and was predominated by an actin phenotype characterized by induction of filopodia and microspikes, isolate #3 causes rapid cell rounding and cell lysis with seemingly no intermediate phenotype (Fig. 4C and D). ABC #1 and E. coli #5 both caused rapid cell rounding (Fig. 4B and F), while K. pneumoniae #1 caused slower and limited rounding and lead to formation of actin protrusions which were distinct from those observed with P. aeruginosa isolate #1 (Fig. 4E). With 3T3 cells, it was harder to discern distinct phenotypes of infection, since upon infection with most bacterial isolates the 3T3 cells underwent rapid deterioration characterized by formation of actin stress fibers and microspikes, followed by cell lysis (Fig. 5). However, with both HeLa and 3T3 cells, pre-incubation with BL21-MAM7 and MAM7-beads markedly slowed down the progress of infection, with only limited cell rounding and lysis visible after infection with either ABC #1, P. aeruginosa #1, K. pneumoniae #1 or E. coli #5. In all these cases, the remaining cellular phenotypes were limited to changes in actin phenotype, such as formation of stress fibers, filopodia or microspikes (Figs. 4 and 5H–R). In contrast, cellular phenotypes following infection with P. aeruginosa isolate #3 did not change upon pre-treatment with either BL21-MAM7 or MAM7-beads on either cell type (Figs. 4 and 5J and P), which is also in agreement with our results from LDH assays.
Figure 4. Visualization of anti-adhesion treatment of HeLa cells for representative examples from each group of bacterial isolate. HeLa cells were either left untreated (top row), pre-incubated with BL21-MAM7 (middle row) or pre-treated with bead-immobilized MAM7 (bottom row). Cells were then left uninfected (controls) (A, G and M) or infected with ABC isolate #1 (B, H and N), P. aeruginosa isolate #1 (C, I and O), P. aeruginosa isolate #3 (D, J and P), K. pneumoniae isolate #1 (E, K and Q) or E. coli isolate #5 (F, L and R) for 4 h. Cells were stained for actin (phalloidin-Alexa488, green) and DNA (Hoechst stain). Fluorescent latex beads are shown in yellow. Scalebar, 20 μm.
Figure 5. Visualization of anti-adhesion treatment of 3T3 cells for representative examples from each group of bacterial isolate. 3T3 cells were either left untreated (top row), pre-incubated with BL21-MAM7 (middle row) or pre-treated with bead-immobilized MAM7 (bottom row). Cells were then left uninfected (controls) (A, G and M) or infected with ABC isolate #1 (B, H and N), P. aeruginosa isolate #1 (C, I and O), P. aeruginosa isolate #3 (D, J and P), K. pneumoniae isolate #1 (E, K and Q) or E. coli isolate #5 (F, L and R) for 4 h. Cells were stained for actin (phalloidin-Alexa488, green) and DNA (Hoechst stain). Fluorescent latex beads are shown in yellow. Scalebar, 20 μm.
Discussion
Bacterial colonization and infection of wounds is a common cause of complication of treatment in military personnel receiving care at military medical facilities following evacuation from combat sites. Wound-associated infections with Gram-negative bacteria are predominantly caused by ABC, P. aeruginosa and ESBL-producing E. coli and K. pneumonia.1 The increasing number of MDR pathogens isolated from patients poses a serious concern and underpins the necessity for alternative measures in infection prophylaxis and therapy. Anti-adhesion treatment has been considered and tested as an alternative to small molecule antimicrobials. In most cases, this is based on administering molecular mimics of host cell receptor structures, such as sugars or sugar mimics.14-17 Following our previous studies exploring the use of MAM7-based inhibitors in anti-adhesion prophylaxis of bacterial infections with gastrointestinal pathogens, we set out to explore the potential of these inhibitory molecules against infection with wound-associated Gram-negative pathogens.
We utilized five representative patient isolates from each of the five above-mentioned Gram-negative bacterial species. We chose a range of phenotypically diverse isolates to ensure our sampling would be representative and our findings would be generally applicable for outbreak strains. PFGE typing showed that all isolates were genotypically distinct. Although this is in part due to a conscious choice of isolates displaying a high degree of phenotypic diversity upon initial characterization (biofilm formation, colony morphology on culture media), which is also reflected in their genotypic diversity, it also reflects the fact that outbreaks are not clonal.
A major problem in the treatment of wound-associated infections is the increasing number of MDR organisms encountered. Admission-associated screening over the period of 2003–2009 has revealed that a wide range of patient isolates are resistant against most antimicrobials commonly used in the clinic and their ability to rapidly acquire additional resistance, such as described in the case of ABC isolates, which developed colistin resistance during the period of testing.1,18 This trend was also confirmed in the present study—all tested isolates have to be considered MDR.
We further analyzed a range of parameters described to be important contributing factors for virulence, using in vitro and tissue culture assays. These included biofilm formation, attachment to host cells and cytotoxicity in tissue culture models of infection.19-22 The majority of strains (64%) were found to form biofilms in vitro and all strains displayed an ability to attach to host cells and elucidate host cell killing. However, the degree of attachment and cytotoxicity varied widely across all tested isolates as well as between isolates within the same species, which is in agreement with the genotypic variance found during PFGE profiling of the isolates.
It is evident that for many pathogens, close contact has to be maintained with host cells in order to establish a successful infection. Many virulence factors are either soluble, secreted toxins, which directly bind and translocate across the host cell plasma membrane or form pores in the plasma membrane. Both these processes are concentration dependent and therefore require close contact with host cells to avoid loss by diffusion.23-25 Other important virulence factors are injected directly into the host cell’s cytoplasm, either by type III, type IV or type VI secretion machinery, which also requires the bacterium to attach to host cells.26-28 For this reason, we analyzed the correlation between host cell attachment and cytotoxicity across each group of pathogen. We generally found a positive correlation between attachment and cytotoxicity for ABC, K. pneumoniae and E. coli isolates. Interestingly, P. aeruginosa isolates showed no significant correlation between these two factors. One explanation for this would be a higher potency of virulence mechanisms in some P. aeruginosa strains compared with other pathogens or other strains within the same species (and thus only limited need for attachment to elucidate a high degree of cytotoxicity). In this light, it would be interesting to explore if some of the isolates are hypervirulent compared with others. Another explanation for the atypical behavior of some P. aeruginosa isolates would be that they use additional virulence mechanisms which do not strictly depend on direct contact between bacteria and host cells, such as bacterial outer membrane vesicles (OMVs) or toxins which are active even at low concentrations (and thus depend less on close proximity between bacteria and host cells).29 OMVs are shed by a wide variety of bacterial species and can be enriched in certain bacterial proteins.30-32 In some cases, this may be employed as a mechanism of translocating virulence factors across the host membrane, my means of vesicle/plasma membrane fusion and endocytosis of bacterial components.33,34 We have tested all isolates for the presence of hemolysins using blood agar plating assays (Table 1), but failed to detect any hemolytic activity in any of the isolates. However, this does not fully exclude the presence of toxins with other functions and this option will be more thoroughly explored in future studies.
Previously, we have shown that pre-treatment of host cells with MAM7, either presented on the surface of non-pathogenic bacteria or immobilized on polymer beads, markedly decreased the effects of infection with gastro-intestinal pathogens, such as V. cholerae, V. parahaemolyticus, Y. pseudotuberculosis and EPEC.9 We further showed that the mechanism of MAM7-based attachment inhibition is likely to be competition for a limited number of host cell receptors (sites rich in both phosphatidic acid and fibronectin), rather than steric hindrance, since the number of attached BL21-MAM7 or MAM7-beads plateaus above an MOI of 20 and the MAM7 dose required to achieve inhibition can be kept relatively low.9,10 Thus, we hypothesized that wound-associated Gram-negative pathogens utilizing a MAM7 homolog for host cell attachment, such as strains belonging to the species analyzed in this study, might also be responsive to the same mode of treatment and explored the potential of MAM7-based inhibitors in anti-adhesion treatment of such infections. We observed a significant decrease of cytotoxic effects on cultured cells following pre-treatment with MAM7 across all five tested species. Two isolates of P. aeruginosa (P. aeruginosa isolates #2 and 3) and one isolate of ESBL-producing E. coli (isolate #2), however, showed very limited responses to MAM7-treatment. Most prominently, the highly cytotoxic isolate P. aeruginosa #3 showed almost no response to MAM7 inhibition. Overall, we noticed that isolates with weak or no correlation between attachment and cytotoxicity were less responsive to treatment. Given that MAM7 inhibits the attachment of bacteria to host cells, it is conclusive that strains that display a high degree of cytotoxicity even at comparably low levels of attachment, such as P. aeruginosa #3, would be less susceptible to anti-adhesion treatment.
It has been noted in the literature that a strain’s ability to form biofilms can be a crucial parameter in determining its resistance toward small-molecule antimicrobials. This is due to the special architecture of biofilms, which are composed of a conglomerate of bacteria embedded in a matrix of secreted bacterial molecules, such as polysaccharides, proteins and DNA and form a diffusion barrier limiting delivery of antimicrobials to the pathogen.39 We tested whether the isolates’ ability to form a biofilm had any influence on their susceptibility to MAM7-based treatment. No significant correlation was found between the amount of biofilm formed and the efficiency of MAM7-based inhibition of cytotoxicity (Table 3). However, it would be interesting to re-evaluate these findings if the use of MAM7-based inhibition was extended from prophylactic treatment as presented here (where the timeframe of the experiment is likely shorter than that required to form biofilm), to a potential therapeutic use in the future (where biofilm structure would likely be pre-formed prior to treatment).
Table 3. Analysis of correlation between biofilm formation and MAM7-based inhibition.
| |
|
BL21-MAM7 |
Bead-MAM7 |
||
|---|---|---|---|---|---|
| Species | Correlation coefficient (r) | Significance (p) | Correlation coefficient (r) | Significance (p) | |
|
ABC |
HeLa |
-0.329 |
0.589 |
-0.486 |
0.407 |
| |
3T3 |
-0.274 |
0.655 |
0.530 |
0.358 |
|
P. aeruginosa |
HeLa |
-0.785 |
0.116 |
-0.903 |
0.357 |
| |
3T3 |
-0.777 |
0.122 |
-0.615 |
0.270 |
|
K. pneumoniae |
HeLa |
-0.335 |
0.582 |
0.173 |
0.780 |
| |
3T3 |
0.656 |
0.229 |
0.460 |
0.436 |
|
E. coli |
HeLa |
-0.180 |
0.772 |
0.120 |
0.847 |
| 3T3 | 0.221 | 0.721 | 0.205 | 0.742 | |
We conclude that anti-adhesion prophylaxis with MAM7-based inhibitors shows promise in the fight against a number of important wound-associated pathogens that might be hard to treat with conventional small molecule antimicrobials. Thus, experiments presented here, investigating the efficacy of MAM7-based inhibition using tissue culture models of infection, should be extended to include relevant animal models of infection in the future, even though this is beyond the scope of the present study. Furthermore, we will aim future studies at improving MAM7-based inhibitors in terms of affinity and avidity, with the hope to extend their application from prophylaxis to therapy. Most importantly, we demonstrate that the correlation between two important hallmarks of virulence that can easily be tested for in tissue culture, attachment and cytotoxicity could serve as a useful predictor for the success of MAM7-based inhibition against bacterial infections, and potentially other molecules used in prophylactic anti-adhesion treatment.
Materials and Methods
Bacterial strains: isolation and growth conditions
Bacterial isolates were acquired from patients treated at the San Antonio Military Medical Center within the period from 2006 to 2010. Cultures were obtained from sites as indicated in Table 1 and passed twice on blood agar plates at 35°C prior to biofilm, PFGE and susceptibility experiments. All isolates were grown in LB or LB agar or in DMEM at 37°C for attachment and infection experiments, unless otherwise indicated.
Bacterial genotyping
Clonal relationships between bacterial strains of each genus were assessed by PFGE according to the FDA method “Procedure for PFGE of Gram-negative rods” (Version 1, 10/30/2007) and as previously described using the CHEF-DRIII system (Bio-Rad Laboratories).35 The endonuclease ApaI was used for ABC PFGE, XbaI for K. pneumoniae and E. coli PFGE, and SpeI was used for P. aeruginosa PFGE. Gel images were analyzed using BioNumerics software (Applied Maths). PFGE patterns were interpreted and grouped into pulsed-field types (PFTs) using established criteria.36
Biofilm assays
Biofilm formation was examined by measuring the ability of bacteria to adhere and accumulate biomass on the bottom of sterile 96-well flat-bottom polystyrene plates. Briefly, fresh bacterial suspensions were prepared in either TSB or LB from overnight cultures and adjusted to an OD600 of 0.25–0.3 (approximately 3 × 108 CFU/mL). One hundred microliters aliquots of bacterial suspension were inoculated into individual wells of a 96-well flat-bottomed polystyrene plate and incubated at 37°C for 48 h. Following incubation, plates were gently washed with 1X phosphate buffered saline (PBS; pH 7.4) and stained with 100 µL of 0.1% Crystal Violet (CV) for 30 min. Excess crystal violet was removed by washing, and biofilm biomass was quantified by measuring the corresponding optical density at 570 nm of the supernatant following the solubilization of CV in 95% ethanol. For each strain tested, biofilm experiments were performed in triplicate. Percentage of biofilm formation was calculated as (OD570 sample/OD570control)*100. The control strain used was Staphylococcus epidermidis ATCC 12228.
Hemolysis assays
To access production of hemolysins, strains were grown in LB at 37°C overnight, streaked onto blood agar plates containing 5% sheep erythrocytes and again grown overnight at 37°C. Hemolysis was accessed visually (appearance of zone of clearance around colonies).
Attachment assays
Bacterial attachment to host cells was essentially tested for as previously described.9 Briefly, mammalian cells were cultured in DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (HeLa cells) or 10% bovine calf serum (3T3 fibroblasts), 5 mM sodium pyruvate and 50 μg/mL penicillin and steptomycin at 37°C with 5% CO2. Cells were washed with PBS (phosphate-buffered saline) prior to the addition of bacteria in DMEM without antibiotics. Bacteria were added to give a multiplicity of infection (MOI) of 10. To determine the bacterial titer after incubation, bacteria were added to empty wells. Plates were centrifuged (1,000 × g, 22°C, 5 min) prior to incubation at 37°C for 1 h. Cells were washed three times with PBS and lysed by adding 0.5% Triton X-100 in PBS. Input samples and Triton lysates were mixed by pipetting up and down several times, serially diluted, plated on LB agar and enumerated by colony counting. Percentage attachment was determined as (number of attached cells/bacterial titer after incubation)*100.
Lactate dehydrogenase (LDH) release assays
To measure cytotoxicity, tissue culture cells were washed with PBS prior to the addition of bacteria in DMEM without antibiotics at an MOI of 10. Infections were started by centrifugation of plates (1,000 × g, 22°C, 5 min) prior to incubation at 37°C. Two hundred microliters of supernatant was removed in triplicate from each well four hours after infection, centrifuged (1,000 × g, 22°C, 5 min), and 100 μl of the supernatant transferred to a fresh 96-well plate for assays. To quantitate cell lysis, the amount of lactate dehydrogenase (LDH) released into the culture medium was measured colorimetrically using the LDH cytotoxicity detection kit (Takara) according to the manufacturer’s protocol. Percentage cell lysis was calculated as [(Ainfected cells-Auntreated cells)/(ATriton-lysed cells-Auntreated cells)]*100.
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was performed using the BD PhoenixTM Automated Microbiology System (Becton Dickinson) according to the manufacture’s guidelines. Results are accessed through the EpiCenter Database (Becton Dickinson) connected to the Phoenix system.
MAM7-inhibition experiments
For inhibition experiments, tissue culture cells were pre-incubated with either E. coli BL21 expressing V. parahaemolyticus MAM7 (MOI of 100) or recombinant, bead-immobilized MAM7 for 30 min as described previously.9,11 Generation of BL21-MAM7, cloning of expression constructs for GST and GST-MAM7 fusion protein and protein purifications have been described elsewhere.9,10 Purified proteins were immobilized on 1 μm fluorescent orange latex beads (Sigma) as described by El Shazly et al.37 For inhibition experiments, a total of 7.5 μg protein/106 beads/well in PBS was used. After removing excess bacteria or beads by three PBS washes, cultured cells were infected with clinical isolates for four hours and cytotoxicity measured as described above for LDH release assays.
Fluorescence microscopy
Cells were seeded onto coverslips at 150,000 cells/mL and subjected to infection experiments the next day. Following infection experiments, cells were washed with PBS and fixed with 3.2% paraformaldehyde in PBS for 15 min. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and treated with Hoechst (Sigma) and Alexa Fluor 488-phalloidin (Molecular Probes) for 10 min to stain for DNA and F-actin, respectively. Coverslips were mounted onto 10% (w/v) glycerol and 0.7% (w/v) propyl gallate in PBS, sealed with nail polish and viewed using a Zeiss LSM510 META Laser Scanning Confocal Microscope. Images were processed using ImageJ and Photoshop software.
Acknowledgments
We would like to thank the Orth lab for critical reading and comments on the manuscript. K.O. and A.M.K. are supported by grants from NIH-Allergy and Infectious Disease (R01-AI056404 and R01-AI087808) and the Welch Foundation (I-1561). K.O. is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease and a W.W. Caruth Jr. Biomedical Scholar. This work was also supported by the Armed Forces Health Surveillance Center including the Global Emerging Infectious System. The views expressed herein are those of the authors and do not reflect the official policy or position of the Department of the Army, Department of Defense or the US Government. K.M. and C.M. are employees of the US government. This work was prepared as part of their official duties and, as such, there is no copyright to be transferred.
Glossary
Abbreviations:
- MAM
multivalent adhesion molecule
- MRSA
methicillin-resistant Staphylococcus aureus
- ABC
Acinetobacter baumannii-calcoaceticus complex
- ESBL
extended spectrum β-lactamase
- EPEC
enteropathogenic Escherichia coli
- MDR
multidrug-resistant
- MOI
multiplicity of infection
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest to disclose.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/20816
References
- 1.Hospenthal DR, Green AD, Crouch HK, English JF, Pool J, Yun HC, et al. Prevention of Combat-Related Infections Guidelines Panel Infection prevention and control in deployed military medical treatment facilities. J Trauma. 2011;71(Suppl 2):S290–8. doi: 10.1097/TA.0b013e318227add8. [DOI] [PubMed] [Google Scholar]
- 2.Murray CK, Roop SA, Hospenthal DR, Dooley DP, Wenner K, Hammock J, et al. Bacteriology of war wounds at the time of injury. Mil Med. 2006;171:826–9. doi: 10.7205/milmed.171.9.826. [DOI] [PubMed] [Google Scholar]
- 3.Yun HC, Murray CK, Roop SA, Hospenthal DR, Gourdine E, Dooley DP. Bacteria recovered from patients admitted to a deployed U.S. military hospital in Baghdad, Iraq. Mil Med. 2006;171:821–5. doi: 10.7205/milmed.171.9.821. [DOI] [PubMed] [Google Scholar]
- 4.O’Shea MK. Acinetobacter in modern warfare. Int J Antimicrob Agents. 2012;39:363–75. doi: 10.1016/j.ijantimicag.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Masini BD, Waterman SM, Wenke JC, Owens BD, Hsu JR, Ficke JR. Resource utilization and disability outcome assessment of combat casualties from Operation Iraqi Freedom and Operation Enduring Freedom. J Orthop Trauma. 2009;23:261–6. doi: 10.1097/BOT.0b013e31819dfa04. [DOI] [PubMed] [Google Scholar]
- 6.Calhoun JH, Murray CK, Manring MM. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin Orthop Relat Res. 2008;466:1356–62. doi: 10.1007/s11999-008-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antimicrobial Resistance. Issues and Options. In: Harrison PF, Lederberg J, eds. Forum on Emerging Infections. Washington, DC: National Academy Press, 1998:8-74. [PubMed] [Google Scholar]
- 8.Murray CK, Hospenthal DR, Kotwal RS, Butler FK. Efficacy of point-of-injury combat antimicrobials. J Trauma. 2011;71(Suppl 2):S307–13. doi: 10.1097/TA.0b013e318227af79. [DOI] [PubMed] [Google Scholar]
- 9.Krachler AM, Ham H, Orth K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc Natl Acad Sci U S A. 2011;108:11614–9. doi: 10.1073/pnas.1102360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krachler AM, Orth K. Functional characterization of the interaction between bacterial adhesin multivalent adhesion molecule 7 (MAM7) protein and its host cell ligands. J Biol Chem. 2011;286:38939–47. doi: 10.1074/jbc.M111.291377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krachler AM, Ham H, Orth K. Turnabout is fair play: use of the bacterial Multivalent Adhesion Molecule 7 as an antimicrobial agent. Virulence. 2012;3:68–71. doi: 10.4161/viru.3.1.18172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray CK, Yun HC, Griffith ME, Thompson B, Crouch HK, Monson LS, et al. Recovery of multidrug-resistant bacteria from combat personnel evacuated from Iraq and Afghanistan at a single military treatment facility. Mil Med. 2009;174:598–604. doi: 10.7205/milmed-d-03-8008. [DOI] [PubMed] [Google Scholar]
- 13.Tribble DR, Conger NG, Fraser S, Gleeson TD, Wilkins K, Antonille T, et al. Infection-associated clinical outcomes in hospitalized medical evacuees after traumatic injury: trauma infectious disease outcome study. J Trauma. 2011;71(Suppl):S33–42. doi: 10.1097/TA.0b013e318221162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firon N, Ashkenazi S, Mirelman D, Ofek I, Sharon N. Aromatic alpha-glycosides of mannose are powerful inhibitors of the adherence of type 1 fimbriated Escherichia coli to yeast and intestinal epithelial cells. Infect Immun. 1987;55:472–6. doi: 10.1128/iai.55.2.472-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joosten JA, Loimaranta V, Appeldoorn CC, Haataja S, El Maate FA, Liskamp RM, et al. Inhibition of Streptococcus suis adhesion by dendritic galabiose compounds at low nanomolar concentration. J Med Chem. 2004;47:6499–508. doi: 10.1021/jm049476+. [DOI] [PubMed] [Google Scholar]
- 16.Salminen A, Loimaranta V, Joosten JA, Khan AS, Hacker J, Pieters RJ, et al. Inhibition of P-fimbriated Escherichia coli adhesion by multivalent galabiose derivatives studied by a live-bacteria application of surface plasmon resonance. J Antimicrob Chemother. 2007;60:495–501. doi: 10.1093/jac/dkm251. [DOI] [PubMed] [Google Scholar]
- 17.Thomas RJ. Receptor mimicry as novel therapeutic treatment for biothreat agents. Bioeng Bugs. 2010;1:17–30. doi: 10.4161/bbug.1.1.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawley JS, Murray CK, Griffith ME, McElmeel ML, Fulcher LC, Hospenthal DR, et al. Susceptibility of acinetobacter strains isolated from deployed U.S. military personnel. Antimicrob Agents Chemother. 2007;51:376–8. doi: 10.1128/AAC.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dallo SF, Weitao T. Insights into acinetobacter war-wound infections, biofilms, and control. Adv Skin Wound Care. 2010;23:169–74. doi: 10.1097/01.ASW.0000363527.08501.a3. [DOI] [PubMed] [Google Scholar]
- 20.Schierle CF, De la Garza M, Mustoe TA, Galiano RD. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen. 2009;17:354–9. doi: 10.1111/j.1524-475X.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- 21.Davis SC, Ricotti C, Cazzaniga A, Welsh E, Eaglstein WH, Mertz PM. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 2008;16:23–9. doi: 10.1111/j.1524-475X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 22.Vance RE, Rietsch A, Mekalanos JJ. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect Immun. 2005;73:1706–13. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda S, Kodama T, Okada N, Okayama K, Honda T, Iida T. Association of Vibrio parahaemolyticus thermostable direct hemolysin with lipid rafts is essential for cytotoxicity but not hemolytic activity. Infect Immun. 2010;78:603–10. doi: 10.1128/IAI.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YR, Lee SE, Kook H, Yeom JA, Na HS, Kim SY, et al. Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells. Cell Microbiol. 2008;10:848–62. doi: 10.1111/j.1462-5822.2007.01088.x. [DOI] [PubMed] [Google Scholar]
- 25.Zrimi J, Ng Ling A, Giri-Rachman Arifin E, Feverati G, Lesieur C. Cholera toxin B subunits assemble into pentamers--proposition of a fly-casting mechanism. PLoS One. 2010;5:e15347. doi: 10.1371/journal.pone.0015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cambronne ED, Roy CR. Recognition and delivery of effector proteins into eukaryotic cells by bacterial secretion systems. Traffic. 2006;7:929–39. doi: 10.1111/j.1600-0854.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- 27.Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–83. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 28.Winnen B, Schlumberger MC, Sturm A, Schüpbach K, Siebenmann S, Jenny P, et al. Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS One. 2008;3:e2178. doi: 10.1371/journal.pone.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2011;286:1269–76. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, et al. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11:3424–9. doi: 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- 32.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–84. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, et al. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One. 2011;6:e17027. doi: 10.1371/journal.pone.0017027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun. 2010;78:5054–61. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akers KS, Mende K, Yun HC, Hospenthal DR, Beckius ML, Yu X, et al. Tetracycline susceptibility testing and resistance genes in isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex from a U.S. military hospital. Antimicrob Agents Chemother. 2009;53:2693–5. doi: 10.1128/AAC.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Shazly S, Ahmad S, Mustafa AS, Al-Attiyah R, Krajci D. Internalization by HeLa cells of latex beads coated with mammalian cell entry (Mce) proteins encoded by the mce3 operon of Mycobacterium tuberculosis. J Med Microbiol. 2007;56:1145–51. doi: 10.1099/jmm.0.47095-0. [DOI] [PubMed] [Google Scholar]
- 38.Cockerill FR, Wikler MA, Alder J, Dudley MN, Eliopoulos GM, Ferraro MJ, et al. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Infromational Supplement. Clinical and Laboratory Standards Institute, 2012. [Google Scholar]
- 39.Percival SL, Hill KE, Malic S, Thomas DW, Williams DW. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair Regen. 2011;19:1–9. doi: 10.1111/j.1524-475X.2010.00651.x. [DOI] [PubMed] [Google Scholar]