Abstract
Increased interactions between pro-apoptotic BH3-only proteins and anti-apoptotic Bcl-2 family proteins at mitochondria result in tumor initiation, progression and resistance to traditional chemotherapy. Drugs that mimic the BH3 region are expected to release BH3-only proteins from anti-apoptotic proteins, inducing apoptosis in some cancer cells and sensitizing others to chemotherapy. Recently, we applied fluorescence lifetime imaging microscopy and fluorescence resonance energy transfer to measure protein:protein interactions for the Bcl-2 family of proteins in live MCF-7 cells using fluorescent fusion proteins. While the BH3-proteins bound to Bcl-XL and Bcl-2, the BH3 mimetic ABT-737 inhibited binding of only Bad and tBid, but not Bim. We have extended our studies by investigating ABT-263, a clinical drug based on ABT-737. We show that the inhibitory effects and pattern of the two drugs are comparable for both Bcl-XL and Bcl-2. Furthermore, we show that mutation of a conserved residue in the BH3 region in Bad and tBid disrupted their interactions with Bcl-XL and Bcl-2, while the corresponding BimEL mutant showed no decrease in binding to these anti-apoptotic proteins. Therefore, in MCF-7 cells, Bim has unique binding properties compared with other BH3-only proteins that resist displacement from Bcl-XL and Bcl-2 by BH3 mimetics.
Keywords: BH3-only proteins, Bim, Bcl-XL, Bcl-2, ABT-737, ABT-263, protein-protein interactions, mitochondria, live MCF-7 cells
Introduction
Apoptosis is a common mechanism used by multicellular organisms to remove cells that have damaged regulatory pathways controlling cellular proliferation and homeostasis. The Bcl-2 family of proteins integrates various cellular signals and controls mitochondrial outer membrane permeabilization, an event widely believed to commit cells to apoptosis.1 Anti-apoptotic proteins, including Bcl-XL, Bcl-2, and Mcl-1, inhibit the executer proteins BAX and BAK that, once activated, oligomerize and permeablize mitochondria, and the BH3-only proteins that directly (Bid and Bim) or indirectly (Bad) activate BAX and BAK.2 One of the key common features of cancer cells is the failure of apoptosis that is often caused by the overexpression of anti-apoptotic proteins that neutralize the pro-apoptotic signaling generated by aberrant growth control.3,4 Thus, it is expected that inhibiting anti-apoptotic proteins may provide an effective way to selectively kill cancer cells or sensitize them to chemotherapy.
Based on this concept many anticancer drugs are under development; however, ABT-737 and ABT-263, inhibitors of both Bcl-XL and Bcl-2, are the most successful ones. Designed to mimic the BH3-only protein Bad, ABT-737 has nanomolar affinities for Bcl-XL and Bcl-2 when soluble fragments are used as binding targets and displaces BH3-only proteins from the binding pocket of Bcl-XL.5,6 ABT-737 is specifically toxic to some cancer cell lines without disturbing normal cells,5,7,8 and enhances the cytotoxicity of a variety of chemotherapy reagents to different cancer cell lines.9 To solve the delivery problems of ABT-737, i.e., low oral bioavailability, high nonspecific binding to proteins and low solubility, the orally active derivative ABT-263 was developed. Also called Navitoclax, the binding profile to anti-apoptotic proteins and potent activity against cancer cell lines is similar to ABT-737.10 However, the specificity and selectivity of the drugs to full-length proteins in live cells or mitochondria-like environments were unknown until our recent findings showing that in live MCF-7 cells, ABT-737 can displace Bad and tBid but not Bim from Bcl-XL and Bcl-2.11
Although the activity of ABT-737 in live HEK293T cells was also reported recently by detecting the relocalization of mCherry-fused BH3 proteins, the use of constructs with truncated transmembrane regions limited it from being an effective approach to study the interactions between membrane binding proteins.12 Studies of full-length purified Bcl-2 family proteins with liposomes revealed that significant conformational changes occur when the proteins bind to cellular membranes, as evidenced by dramatic changes in their interacting partners and relative binding affinities.13-15 These changes make it difficult to infer what happens in live cells from measurements made in vitro using soluble protein fragments and peptides.16,17 For this reason, we have developed systems in which physical interactions between Bcl-2 family proteins can be detected using full-length proteins in membranes, membrane-like environments and in live cells.11,15,18-22
To quantify these interactions in live cells, fluorescent fusion proteins (Venus-Bcl-XL/Bcl-2 and mCherry-BH3s) were expressed in MCF-7 cells, and fluorescence resonance energy transfer (FRET) was detected between them by fluorescence lifetime imaging microscopy (FLIM).23-25 Compared with other fluorescence techniques, lifetime measurement is not affected by spectral bleed-through or by changes in the excitation intensity,26 which makes FLIM FRET robust for measurements in live cells undergoing stress or other manipulations. By selecting regions of interest (ROIs) in the images at the areas enriched in mitochondria, we were able to measure the interactions not only in live cells, but also in the cellular site where apoptotic regulation occurs. When we applied this technique to study the interactions of BH3 proteins with Bcl-XL and Bcl-2, we observed that the interactions between Bcl-XL/Bcl-2 and Bim are not as sensitive to mutations of the conserved BH3 region of Bim (a double alanine substitution) as were the BH3-only proteins Bad and tBid. Consistent with this result, ABT-737 selectively inhibited binding of Bcl-XL/Bcl-2 to Bad and tBid but not to the major isoforms of Bim. These findings using FLIM FRET provided unique insights into the protein:protein interactions between Bcl-XL/Bcl-2 and Bim in MCF-7 cells that were not detected by other means.11 Here we have extended our investigations to a mutation of another conserved residue in the BH3 region, the glycine/serine in the LXXXG/SDX motif (the residue is a glycine in Bid and Bim and a serine in Bad). The G/S to E mutants disrupted Bad and tBid but not Bim binding to Bcl-XL/Bcl-2. We also studied the effect of ABT-263 on these interactions. ABT-263 has comparable activity to ABT-737 in disrupting the complexes between Bcl-XL/Bcl-2 and Bad/tBid in MCF-7 cells and displays the same inhibitory pattern; i.e., ABT-263 does not prevent the binding of Bim to Bcl-XL/Bcl-2. Taken together, our results suggest that FLIM FRET interaction studies can be used to guide drug-modification to change the specificities of the drugs in a biologically relevant context.
Results and Discussion
Rationale for using FLIM FRET to measure compartment-specific protein interactions
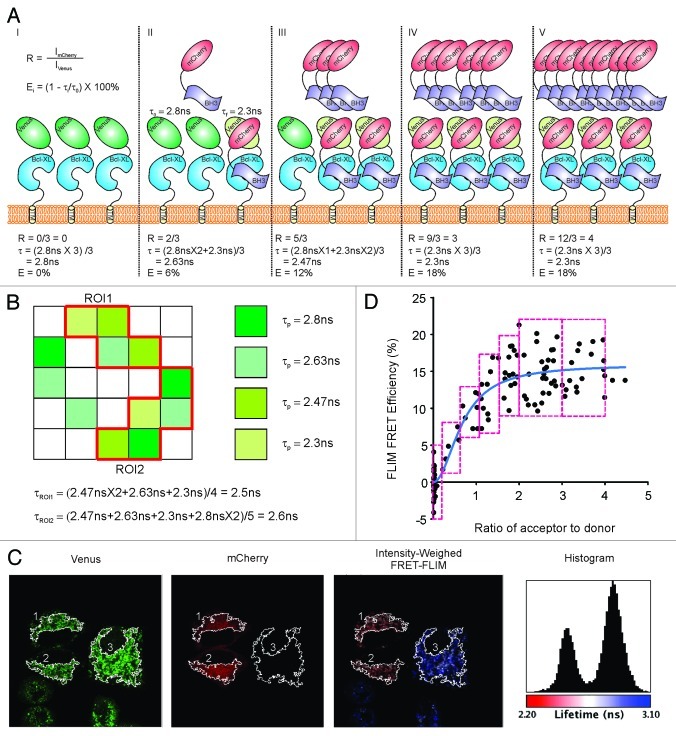
To study the interactions between Bcl-XL and BH3-only proteins in live cells, we established a MCF-7 cell line stably expressing Venus-Bcl-XL, in which we induced variable expression of mCherry-BH3-only proteins (mCherry-BH3s) by transient transfections. The process by which FLIM FRET can be used to measure Bcl-XL:BH3 interactions is illustrated in Figure 1. When imaging these cells, any one bright pixel may correspond to a different number of fluorescent donor or acceptor molecules. Indeed, due to the limited resolution of microscopy, some pixels may contain large numbers of donors and acceptors. In this simplified example, we illustrate five pixels (I–V) in which there are three Venus-Bcl-XL molecules (Fig. 1A). In the first pixel (no expression of mCherry-BH3s), the lifetime of Venusτ0 (“non-FRETing”) is around 2.8 ns (Fig. 1A, I). Subsequent pixels (II–V) illustrate the formation of Bcl-XL:BH3 complexes by which the Venus FRET donor is close to its matching acceptor mCherry (less than 100 Å26) in a fixed distance and alignment. This distance and alignment determine the spectroscopic FRET efficiency and can be different for every pair of proteins. When mCherry-Bad is expressed in the cells by transient transfection, the energy transfer between bound pairs of fluorescent proteins results in a lifetime decrease of Venus to 2.3 ns (τf) (Fig. 1A II–V). Because the expression of mCherry-BH3s will vary in different parts of the cell, the amount available to bind to Venus-Bcl-XL will vary accordingly. Thus, there will be different ratios of Venus-Bcl-XL molecules undergoing FRET in each pixel of the FLIM image. The pixel lifetime (τp) is the mean value of the lifetimes for each fluorescent donor molecule (Venus-Bcl-XL) in that pixel, and the decrease of τp from τ0 is proportional to the fraction of bound or “FRETing” Venus-Bcl-XL in that pixel. As a result, τp for Venus in II-V pixels vary, and the relative values of the FLIM FRET efficiency (calculated by E% = (1-τp/τ0) × 100%) represent the fraction of Venus-Bcl-XL formed complexes with mCherry-BH3s in that pixel (Fig. 1A, II–V).
Figure 1. Diagram interpretation for FLIM FRET data analysis. (A) Scenarios for individual pixels (I–V) that may have different lifetimes of Venus-Bcl-XL due to different FLIM FRET efficiency (%) between donor (Venus) and acceptor (mCherry) resulting from different percentages of Bcl-XL bound to BH3-only proteins. Bcl-XL is in blue, BH3-only protein is in lavender, mCherry is in red, “non-FRETing” Venus whose lifetime τ0 is 2.8 ns is in dark green, and the “FRETing” Venus with a lifetime τf at 2.3 ns is in light green. R stands for the ratio of the intensities of the acceptor to donor, and E is the FLIM FRET efficiency. (B) Mean lifetime of ROIs. Pixels with different lifetimes for Venus-Bcl-XL in the 5 × 5 image are shown in different colors as indicated, and the pixels in white are those without expression of Venus-Bcl-XL. The equations to calculate the mean lifetime for each ROI are listed. (C) Representative images of FLIM FRET measurement. From left to right are the intensity images from the Venus channel and the mCherry channel, the intensity-weighed FLIM FRET image in which fluorescence lifetimes are presented in a continuous pseudocolor scale ranging from 2.2 to 3.1 ns, and the lifetime histogram of Venus-Bcl-XL fluorescence lifetimes using the same pseudocolor scale. (D) Binding curve fitting for FLIM FRET Efficiency (%) over the ratio of acceptor to donor. The red boxes show the bin size and standard error calculation, and the blue line shows the fitted binding curve.
Similar to the pixel lifetime (τp), the lifetime for a selected region of an image (τroi) is averaged among the pixels in that ROI (Fig. 1B), thus the extent of energy transfer between the FRET pair in that ROI reduces the lifetime of the fluorescence donor in a manner proportional to the fraction of Venus-Bcl-XL that is in complexes with an interacting mCherry-BH3. Therefore the value of the FLIM FRET efficiency of that ROI is an indicator of the percentage of Venus-Bcl-XL bound to mCherry-BH3s in that region of a cell or an image (Fig. 1B). Since the Bcl-2 family of proteins function primarily at the outer mitochondrial membrane,14 ROIs enriched in mitochondria were selected for further analysis from FLIM images (Fig. 1C). For ROIs #1 and #2, mCherry-BH3 acts as FRET acceptor, and the lifetimes of Venus-Bcl-XL decrease to 2.5 ns (as shown in red) in the intensity-weighed FLIM image and the FLIM histogram. In contrast, ROI #3 shows non-detectable expression of mCherry-BH3, thus the lifetime of Venus-Bcl-XL in that ROI corresponds to the 2.8 ns peak in the histogram and is displayed in blue in the intensity-weighed FLIM image. After collecting intensities and lifetime values for > 500 ROIs, the FLIM FRET efficiency (E%) and the ratio of the intensities of the acceptor to donor (R) are calculated as E% = (1-τroi/τ0) × 100% and R = ImCherry/IVenus respectively. These data are shown in Figure 1D plotted as E (proportional to bound Venus-Bcl-XL) vs R (the relative expression of mCherry-BH3). These data are then distributed in different bins (red boxes) and fit with an improved hyperbolic function (see details in Materials and Methods section, blue curve in Fig. 1D). This curve provides an estimate of the maximal FLIM FRET efficiency (Emax), a feature unique to each pair of interacting partners that is determined by the distance and alignment between the fused FRET donor and acceptor and the binding affinities (Kd) between Bcl-XL and BH3-only proteins in the analyzed cellular compartment. The fitted Kd is in the same unit scale as the ratio R, which is an artificial number depending on the laser settings. Thus, Kd may vary when different instruments or settings are employed.
Therefore, FLIM FRET is useful not only to confirm the physical interactions between proteins, but can also be used to accurately measure these interactions. Therefore, this system can be used as a platform in live cells to compare quantitatively the efficiency of drugs or mutations that disrupt protein:protein interactions. Both of these effects can be measured as an increase of the fluorescent donor lifetime and a decrease in the FLIM FRET efficiency.
ABT-263 is a comparable alternative for ABT-737
In our previous publication,11 we tested the inhibitory effects of ABT-737 using FLIM FRET in MCF-7 cells overexpressing either Venus-Bcl-XL or Venus-Bcl-2. As noted in the previous section, one cannot directly compare the binding curves for BH3-only proteins binding to Bcl-XL with Bcl-2 because of differences in the expression levels, in the folding fidelities of the two Venus-fused proteins and potential differences in distance and alignment of the FRET pair. However, for a Venus-fused Bcl-XL and one mCherry-BH3 protein, it is possible to compare the extent to which different small molecules inhibit binding. Therefore, to compare the activities of ABT-263 with ABT-737 in MCF-7 cells, we performed FLIM FRET measurements in MCF-7 cells expressing Venus-Bcl-XL or Venus-Bcl-2 transfected with different mCherry-BH3 constructs in the presence of these drugs. Due to laser maintenance and realignment, the settings for laser power differ slightly from those we used previously. These settings affect the exact value of the artificial ratio of acceptor to donor (R) and, thus, the measured dissociation coefficient (Kd). For this reason we repeated the binding curves for the DMSO control and for 5 μM ABT-737 under the same conditions as the measurements for ABT-263 for the current comparison.
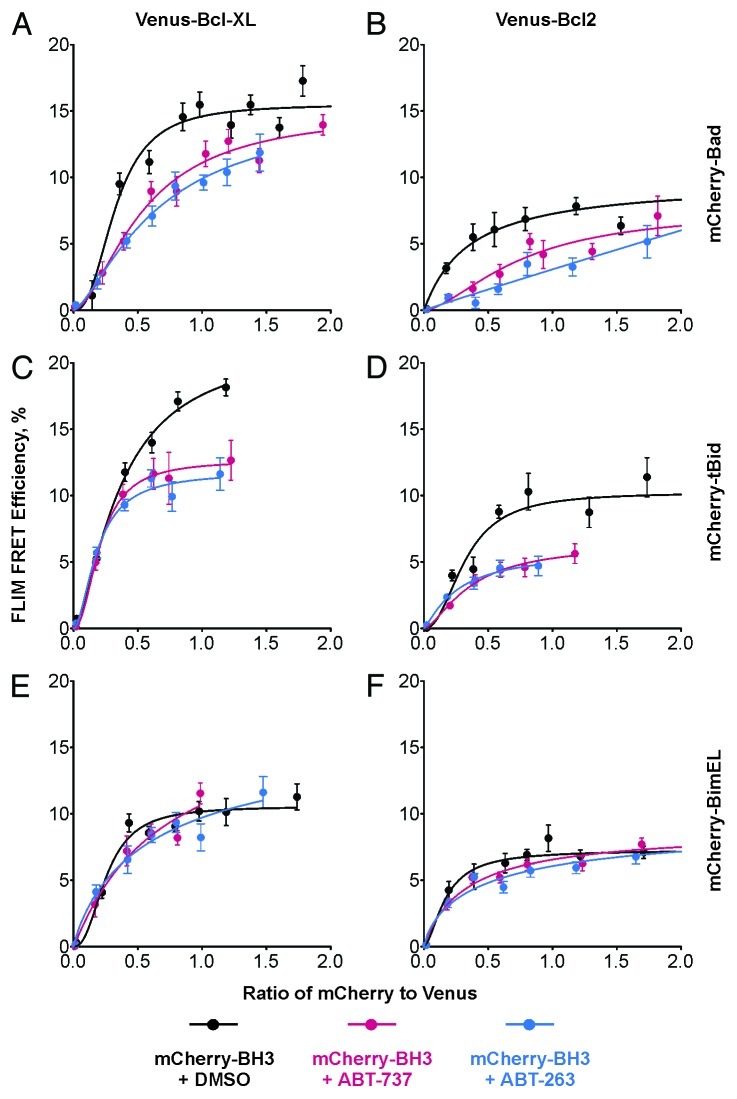
As shown in Figure 2, the binding of the mCherry-BH3s to either Venus-Bcl-XL or Venus-Bcl-2 (black curves) generated typical binding curves except the one between Venus-Bcl-XL and mCherry-tBid. We could not determine if this curve came to saturation, as the population did not contain enough cells expressing high levels of mCherry-tBid (Fig. 2B), possibly because these levels were toxic to cells. Treatment of the cells with ABT-737 (red curves) or ABT-263 (blue curves) reduced the binding between Venus-anti-apoptotic proteins and mCherry-Bad or mCherry-tBid similarly (Fig. 2A, B, D and E). ABT-263 displays comparable activity to ABT-737 in inhibiting these interactions in MCF-7 cells, suggesting that they have similar binding affinities for the anti-apoptotic proteins, or that the reported slightly lower affinity of ABT-263 compared with ABT-737 may be compensated for in live cells by improved specificity.10 The increased values of the Kd for Venus-Bcl-XL:mCherry-Bad or Venus-Bcl-2:mCherry-Bad binding in the presence of the drugs indicates that the drugs function as competitive inhibitors of Bcl-XL and Bcl-2, as expected for BH3 mimetics (Fig. 2A and D). The inhibitory effects of ABT-737 and ABT-263 on binding of mCherry-tBid with Venus-Bcl-XL or Venus-Bcl-2 resulted in a decreased Emax and/or a higher Kd (Fig. 2B and E). The drop in Emax is possibly due to missing data at high ratios of mCherry-tBid to Venus fused anti-apoptotic proteins, which affects curve fitting and the calculated Kd value. In contrast to the inhibition of mCherry-Bad or mCherry-tBid binding to Bcl-XL and Bcl-2, both ABT-737 and ABT-263 did not disrupt the binding of mCherry-BimEL to Venus-Bcl-XL or Venus-Bcl-2 (Fig. 2C and F). Very minor changes in Kds were observed in ROIs containing mitochondria, indicating that BimEL binds anti-apoptotic proteins at mitochondria either with much higher affinity than in solution, or by a different mechanism in MCF-7 cells.
Figure 2. Both ABT-737 and ABT-263 inhibit the interactions between Bcl-XL/Bcl-2 and Bad or tBid but not BimEL. The binding of (A and D) mCherry-Bad, (B and E) mCherry-tBid and (C and F) mCherry-BimEL to (A-C) Venus-Bcl-XL or (D-F) Venus-Bcl-2 in the presence of vehicle control (DMSO) are shown in black, and the inhibition by ABT-737 and ABT-263 are shown in red and blue respectively. The error bar indicates standard error.
Binding of mCherry-BimEL to Venus-Bcl-XL exhibits a higher tolerance to mutations in the BH3 region than Bad or tBid
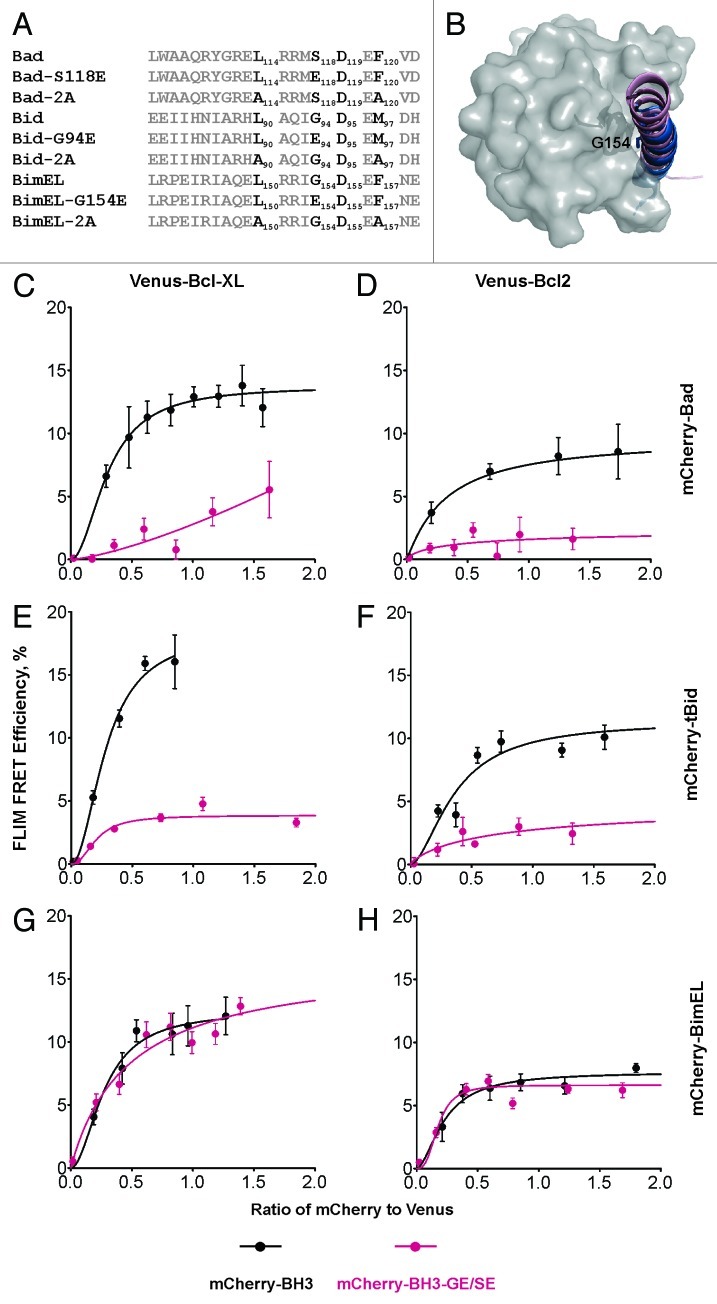
To further examine the differences in binding to Venus-Bcl-XL or Venus-Bcl-2 by mCherry-BimEL, mCherry-Bad and mCherry-tBid, we introduced point mutations in the BH3 region of these proteins. As reported previously, the 2A mutations (Fig. 3A) abolished binding of mCherry-Bad and mCherry-tBid to either Venus-Bcl-XL or Venus-Bcl-2, but only had minor effects on the interaction of the three major isoforms of Bim with Venus-Bcl-XL or Venus-Bcl-2.11 Here, we extended this study to the conserved G/S in the LXXXG/SD motif of the BH3 regions. According to the 3D structures obtained for soluble versions of the proteins, this glycine localizes along the binding interface of the Bcl-XL:BH3 complexes (2BZW and 2YXJ, Figure 3B) and is critical for the interactions between tBid and Bcl-XL.27 Consistent with these results our data demonstrated that both the G94E mutant of mCherry-tBid and the S118E mutant of mCherry-Bad did not bind to either Venus-Bcl-XL or Venus-Bcl-2 (Fig. 3C, D, F and G, red curves). In comparison, the corresponding G154E mutant of mCherry-BimEL behaved like the wild-type protein (Fig. 3E and 3H, red curve), reinforcing the notion that Bim is much more tolerant of mutations in the BH3 region. This result is consistent with the results obtained using the BH3 mimetics ABT-737 and ABT-263. At present, it is not clear if this unexpected binding characteristic of Bim is an inherent property of the membrane-bound form of the protein, or if it is due to a novel binding surface not previously characterized, or the result of an unidentified posttranslational modification or additional binding partner(s) that stabilize the complex in MCF-7 cells. Thus, further exploration of the interaction between anti-apoptotic proteins and Bim in live cells will be required to determine the molecular mechanism(s) underlying the unusual binding properties of Bcl-XL:Bim and Bcl-2:Bim complexes in mitochondria of MCF-7 cells.
Figure 3. The effects of mutations in BH3 regions of BH3-only proteins on their interactions with Bcl-XL and Bcl-2. (A) Sequence alignments for the BH3 regions from the indicated proteins. The LXXXG/SD motif and the conserved residues that were mutated are in black and labeled as subscripts. (B) G154 and S118 localize along the interacting interfaces in the complexes. G154 is shown and labeled in the aligned structures of BH3 peptides in the complexes Bcl-XL:Bim-BH3 (1PQ1, pink) and Bcl-XL:Bad-BH3 (2BZW, blue). Bcl-XL is shown in gray surface. (C–H) The binding of (C and F) mCherry-Bad, (D and G) mCherry-tBid and (E and H) mCherry-BimEL to (C-E) Venus-Bcl-XL or (F-H) Venus-Bcl-2 are shown in black, and the curves for mutants (mCherry-Bad-S118E, mCherry-tBid-G94E, and mCherry-BimEL-G154E) binding to Venus-Bcl-XL or Venus-Bcl-2 are in red. The error bar indicates standard error.
Conclusions
FLIM FRET is a powerful approach to explore protein:protein interactions at specific cellular locations in live cells. By precisely measuring the effects of small-molecule inhibitors and point-mutations, FLIM FRET was able to elucidate the unique features of BimEL binding to either Bcl-XL or Bcl-2. This suggests that the technique can guide drug modification studies to improve inhibition of the binding of BimEL to anti-apoptotic proteins. Moreover the results presented here together with those we published previously that characterize the interaction between Hexokinase II and PEA-15 demonstrate the versatility of this approach and suggest that many other subcellular interactions are amenable to measurement by FLIM FRET.28
Materials and Methods
Constructs and cell lines
The constructs used in this study were generated as described;11 the mutants were obtained using oligonucleotides and DNA amplification using Phusion DNA polymerase (New England Biolab). The MCF-7 human breast cancer cell line stably expressing Venus-Bcl-2 was cultured in α-Minimum Essential Medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone) and 500 μg/ml neomycin. For microscopy, 1 × 105 cells were seeded onto 25 mm diameter coverslips (0.17 mm thickness) and cultured as above for 18–24 h. Transient transfections of the plasmids encoding mCherry-BH3 proteins11 were performed using Fugene HD according to the manufacturer’s protocol (GE). Cells treated with ABT-737 or ABT-263 were incubated at 37°C in a fresh media containing drugs at the indicated concentration 18–24 h before imaging.
Steady-state fluorescence confocal imaging and fluorescence lifetime measurements
The coverslips with cells were washed in PBS and placed in a chamber with 0.5 ml of PBS or PBS-containing drug at the specified concentrations. Steady-state fluorescence images were acquired using a Leica TCS SP5 (Leica Microsystems CMS GmbH) equipped with a 63 × 1.3NA PlanApo glycerol immersion objective. Images of Venus and mCherry were acquired using 514 nm and 561 nm excitation, respectively, and emission was collected between 525–555 nm and 580–670 nm, respectively. The average fluorescence intensity of individual cells or regions was quantified using ImageJ.29
Fluorescence lifetimes were quantified using the Leica TCS SP5 confocal microscope (Leica Microsystems CMS GmbH) equipped with an integrated SPC-830 TCSPC system (Becker and Hickl GmbH) and Chameleon Ultra pulsed laser (Coherent Inc.). The Ti:Sapphire laser was tuned to 960 nm to provide optimal multiphoton-excitation for Venus,24 and the emission was acquired between 510–555 nm. TCSPC-images (256 pixels × 256 pixels × 256 time bins) were recorded for 40 sec using Becker and Hickl SPCM acquisition software with the photon count rate set to avoid photon pileup and the laser power set to minimize photobleaching during acquisition. Using SPCImage software (Becker and Hickl GmbH), the average lifetime per pixel of the TCSPC-data was analyzed with the FLIM pixels binned to ensure a peak photon count > 100 and a total photon count > 1,000. The lifetime and photon count for each pixel were further exported for ImageJ analysis. Lifetime histograms and intensity-weighted fluorescence lifetime images were generated using ImageJ.
To generate binding curves, the ROIs containing mitochondria and ER were selected, and the corresponding intensities in both Venus and mCherry channels and the average lifetime were measured using a customized ImageJ script. Cells were manually discarded from the analysis if the Venus-Bcl-2 signal was too low (and therefore noisy), or close to saturated or the mCherry signal was close to saturated. The ratio of mCherry to Venus was calculated using the intensities of each channel. FLIM FRET efficiency (E%) was calculated for each ROI as: E% = (1-τi/τ0) × 100%, where τi is the mean lifetime for that ROI, and τ0 is the average lifetime of all ROIs not expressing detectable mCherry. FLIM FRET efficiency was the distributed into bins of the same size according to mCherry:Venus ratio (bin size 0.2 for ratios lower than 1 and 0.5 for ratios higher than 1) and plotted (± se) against mCherry:Venus ratio. The binding curves were fitted using GraphPad Prism version 5.0d for Macintosh (GraphPad Software, www.graphpad.com) with the function: E% = Emax × (ImCherry ÷ IVenus)h/[Kdh+(ImCherry ÷ IVenus)h]. Emax is the maximum FLIM FRET efficiency corresponding to saturation of donor binding sites by an acceptor; ImCherry and IVenus are intensities of mCherry and Venus, respectively; Kd is the relative equilibrium dissociation constant; h is the Hill slope.
Acknowledgments
This work was supported by grant FRN12517 from the Canadian Institute of Health Research (CIHR) to D.W.A. and B.L. and by a Tier I Canada Research Chair in Membrane Biogenesis to D.W.A. Q.L. is recipient of a postdoctoral fellowship from the Canadian Breast Cancer Foundation, Ontario Division.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21462
References
- 1.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 3.Slavov N, Dawson KA. Correlation signature of the macroscopic states of the gene regulatory network in cancer. Proc Natl Acad Sci U S A. 2009;106:4079–84. doi: 10.1073/pnas.0810803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–32. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 6.Bruncko M, Oost TK, Belli BA, Ding H, Joseph MK, Kunzer A, et al. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J Med Chem. 2007;50:641–62. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan D, Velankar M, Brahmandam M, Hideshima T, Podar K, Richardson P, et al. A novel Bcl-2/Bcl-X(L)/Bcl-w inhibitor ABT-737 as therapy in multiple myeloma. Oncogene. 2007;26:2374–80. doi: 10.1038/sj.onc.1210028. [DOI] [PubMed] [Google Scholar]
- 8.Hann CL, Daniel VC, Sugar EA, Dobromilskaya I, Murphy SC, Cope L, et al. Therapeutic efficacy of ABT-737, a selective inhibitor of BCL-2, in small cell lung cancer. Cancer Res. 2008;68:2321–8. doi: 10.1158/0008-5472.CAN-07-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang MH, Kang YH, Szymanska B, Wilczynska-Kalak U, Sheard MA, Harned TM, et al. Activity of vincristine, L-ASP, and dexamethasone against acute lymphoblastic leukemia is enhanced by the BH3-mimetic ABT-737 in vitro and in vivo. Blood. 2007;110:2057–66. doi: 10.1182/blood-2007-03-080325. [DOI] [PubMed] [Google Scholar]
- 10.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–8. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 11.Aranovich A, Liu Q, Collins T, Geng F, Dixit S, Leber B, et al. Differences in the mechanisms of proapoptotic BH3 proteins binding to Bcl-XL and Bcl-2 quantified in live MCF-7 cells. Mol Cell. 2012;45:754–63. doi: 10.1016/j.molcel.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Wong C, Anderson DJ, Lee EF, Fairlie WD, Ludlam MJ. Direct visualization of Bcl-2 family protein interactions using live cell fluorescent protein redistribution assays. Cell Death Dis. 2012;3:e288. doi: 10.1038/cddis.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leber B, Lin J, Andrews DW. Still embedded together binding to membranes regulates Bcl-2 protein interactions. Oncogene. 2010;29:5221–30. doi: 10.1038/onc.2010.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–84. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 17.Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008;27(Suppl 1):S128–36. doi: 10.1038/onc.2009.50. [DOI] [PubMed] [Google Scholar]
- 18.Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, et al. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 2005;24:2096–103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Annis MG, Zamzami N, Zhu W, Penn LZ, Kroemer G, Leber B, et al. Endoplasmic reticulum localized Bcl-2 prevents apoptosis when redistribution of cytochrome c is a late event. Oncogene. 2001;20:1939–52. doi: 10.1038/sj.onc.1204288. [DOI] [PubMed] [Google Scholar]
- 20.Billen LP, Kokoski CL, Lovell JF, Leber B, Andrews DW. Bcl-XL inhibits membrane permeabilization by competing with Bax. PLoS Biol. 2008;6:e147. doi: 10.1371/journal.pbio.0060147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dlugosz PJ, Billen LP, Annis MG, Zhu W, Zhang Z, Lin J, et al. Bcl-2 changes conformation to inhibit Bax oligomerization. EMBO J. 2006;25:2287–96. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim PK, Annis MG, Dlugosz PJ, Leber B, Andrews DW. During apoptosis bcl-2 changes membrane topology at both the endoplasmic reticulum and mitochondria. Mol Cell. 2004;14:523–9. doi: 10.1016/S1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Mills JD, Periasamy A. Protein localization in living cells and tissues using FRET and FLIM. Differentiation. 2003;71:528–41. doi: 10.1111/j.1432-0436.2003.07109007.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Periasamy A. Characterization of two-photon excitation fluorescence lifetime imaging microscopy for protein localization. Microsc Res Tech. 2004;63:72–80. doi: 10.1002/jemt.10430. [DOI] [PubMed] [Google Scholar]
- 25.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–6. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–69. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 28.Mergenthaler P, Kahl A, Kamitz A, van Laak V, Stohlmann K, Thomsen S, et al. Mitochondrial hexokinase II (HKII) and phosphoprotein enriched in astrocytes (PEA15) form a molecular switch governing cellular fate depending on the metabolic state. Proc Natl Acad Sci U S A. 2012;109:1518–23. doi: 10.1073/pnas.1108225109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasband WS. ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA. http://imagejnihgov/ij/ 1997-2011.