Abstract
Glutamine (Gln) and glucose (Glc) represent two important nutrients for proliferating cells, consistent with the observations that oncogenic processes are associated with enhanced glycolysis and glutaminolysis. Gln depletion and Glc depletion have been shown to trigger growth arrest and eventually cell death. Solid tumors often outgrow the blood supply, resulting in ischemia, which is associated with hypoxia and nutrient insufficiency. Whereas oxygen-sensing and adaptive mechanisms to hypoxia have been well-studied, how cells directly sense and respond to Gln and Glc insufficiency remains unclear. Using mRNA profiling techniques, we compared the gene expression profiles of acute Gln-depleted cells, Glc-depleted cells and cells adapted to Gln depletion. Here we report the global changes of the gene expression in those cells cultured under the defined nutrient conditions. Analysis of mRNA profiling data revealed that Gln and Glc depletion triggered dramatic gene expression reprogramming. Either Gln or Glc deletion leads to changes of the expression of cell cycle genes, but these conditions have distinctive effects on transcription regulators and gene expression profiles. Moreover, Gln and Glc depletion triggered distinguishable ER-stress responses. The gene expression patterns support that Gln and Glc have distinctive metabolic roles in supporting cell survival and proliferation, and cells use different mechanisms to sense and respond to Gln and Glc insufficiency. Our mRNA profiling database provides a resource for further investigating the nutrient-sensing mechanisms and potential effects of Glc and Gln abundance on the biological behaviors of cells.
Keywords: ATF4, ATF6, ER stress, XBP1, glucose depletion, glutamine depletion, metabolism
Introduction
Carbon source, nitrogen source and molecular oxygen play critical roles in supporting cell function and proliferation through providing of ATP, reducing power and building blocks.1,2 In particular, glucose (Glc) and glutamine (Gln) have been found to be indispensible for most cells in in vitro culture systems.2,3 Ischemia, a common pathological condition occurring in solid tumors and some cardiovascular disorders, leads to the lack of oxygen, glucose and amino acids and deleterious clinical outcomes. In ischemic tissues or solid tumors, the ability of local cells to sense and properly respond to transient lack of carbon source, nitrogen source and oxygen determines their adaptation, survival and proliferation to the dynamic nutrient environments. Oxygen sensing and adaptive response to hypoxia in normal tissues and solid tumors have been well-characterized.4,5 It remains unclear how cells sense and respond to availability of Glc or Gln.
Particularly, tumor cells require increased supply of carbon, nitrogen sources and molecular oxygen. Reprogramming of energy metabolism has been regarded as an emerging hallmark of tumor progression.6 As consequences of oncogenic signaling and adaptation, tumor cells show aberrant metabolic features, which form the biochemical basis for resistance to chemotherapy and radiation. As a major carbon source, glucose is metabolized via glycolysis to generate ATP instead of oxidative phosphorylation by tumor cells, even in aerobic conditions, a phenomenon termed the Warburg effect.7,8 Warburg effect is the result of oncogenic stimulation and loss of tumor suppressor genes that lead to a series of metabolic reprogramming, such as the expression and/or translocation of glucose transporters to the plasma membrane9,10 and upregulation or activation of glycolytic enzymes.11 Warburg effect provides proliferative tumor cells with anabolic carbons and reducing power for increased biosynthetic activity, such as fatty acid synthesis.12,13 Besides Warburg effect, tumor cells also become addicted to active glutaminolysis for proliferation.14,15 Glutaminolysis defines the conversion of glutamine to glutamate catalyzed by glutaminase.14 Gln contains both carbon and nitrogen and can be used as an energetic, biosynthetic and reductive precursor for highly proliferative tumor cells.16,17
Solid tumors usually have heterogeneous blood supply, leading to localized ischemia and uneven nutrient distribution in different regions of tumors. To survive in such stressful conditions, tumor cells resort to transcriptional and metabolic reprogramming. Hypoxia inducible factor (HIF) is the best-known responding molecule to hypoxia.4 Under hypoxic condition, HIF upregulates the expression of angiogenic, glycolytic and other relevant genes that induce the adaptive response to hypoxia.18 Deprivation of amino acids, such as cysteine or leucine, results in a conserved stress response, termed integrated stress response (IRS).19 During IRS, general control nonderepressible kinase 2 (GCN2) senses the abundance of non-aminoacylated tRNAs.20 The direct binding of uncharged tRNAs with GCN2 activates GCN2, which phosphorylates eukaryotic initiation factor 2α (eIF2α). The phosphorylation of eIF2α results in decreased global translation by inhibiting the formation of ternary complex (Met-tRNAiMet•eIF2-GTP), while increasing the translation of selected mRNAs, such as activating transcription factor 4 (ATF4).21-23 Increased ATF4 translation leads to the expression of genes with amino acid response elements (AAREs), including other transcriptional regulators, solute carrier family transporters, aminoacyl-tRNA synthetase, cell cycle progression, DNA damage repair and so forth.24-27 Gln, a nonessential amino acid, has been shown to be essential for most cells in in vitro culture systems. Gln may play roles as both a carbon source and a nitrogen source. However, it remains unclear how cells response to Gln depletion. Similarly, while Glc has been proven to be indispensible for most cells, there is no systemic analysis of gene expression in tumor cells in response to Glc depletion.
We have showed that the proliferation of Hep3B, a tumor cell line derived from hepatocytes, depends on Glc and Gln for proliferation. Starting from Hep3B, we have established a cell line, MM01, which has adapted to Gln-free media.14 We systemically examined and compared the genome-wide mRNA expression profile of Hep3B cells in Gln-depleted media with that of cells cultured in Glc-depleted media and that of adapted MM01 cells. Using ingenuity pathway analysis (IPA), we did functional analysis of the mRNA profiling data, and found Gln- or Glc depletion triggered distinctive gene expression by regulating different sets of transcription factors. Our results will facilitate future research toward the thorough understanding of the nutrient-sensing mechanisms and the effects of Glc and Gln abundance on the biological behaviors of cells.
Results
Either Gln or Glc depletion alters gene expression patterns
We showed previously that short-term (24–48 h) depletion of either Gln or Glc resulted in growth arrest but not death of Hep3B cells.14 As the first step toward the understanding of cellular responses to the Gln depletion, we compared Gln and Glc depletion by gene expression profiling. We chose Hep3B, a tumor cell line originated from hepatocytes, as a model to study, for it has the ability to adaptively utilize alternative metabolic pathways for anabolism of carbon and nitrogen.14 We isolated total RNA samples from Hep3B cells cultured in complete media (control), Gln-free media or Glc-free media for 24 h. Gene expression profiling was performed in triplicate for each condition with microarrays. The heatmap of gene expression was shown in Figure 1A, and detected genes were summarized in Table S1. Compared with control cells, Gln depletion (for 24 h) resulted in differential expression of 2,197 genes (log2 Ratio > 2.0 or < -2.0); among them 638 genes were upregulated. The upregulated genes with a log2 Ratio > 3.0 are listed in Table 1. To validate the results from gene profiling, we selected genes upregulated at different levels (log2 values range from 1.5–4.7) and examined their mRNA levels using qRT-PCR. Results shown in Figure 1B indicate that the upregulation (direction of regulation) can be confirmed by qRT-PCR.
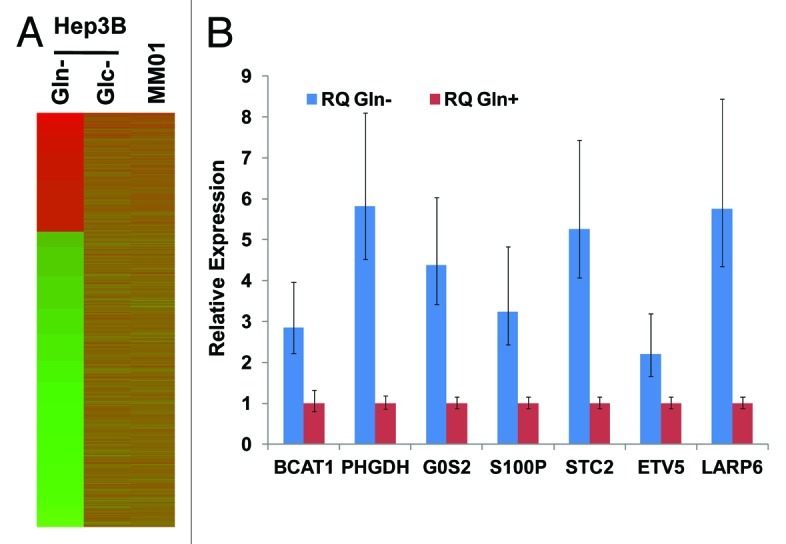
Figure 1. Gene expression in Hep3B cells with different treatments. (A) Heatmap of gene expression pattern in Gln- or Glc-depleted Hep3B and MM01 cells. (B) Validation of the upregulation of representative genes identified by mRNA profiling. TaqMan primers were purchased from Invitrogen. Levels of mRNA were determined by qRT-PCR. RNA samples isolated from Hep3B cells cultured in complete media were used as control, and mRNA levels in the control were arbitrarily defined as 1. Relative levels of mRNA in Gln-depleted cells were shown. For each sample and tested gene, qRT-PCR was performed at triplicate, and standard deviations were indicated. All data presented were statistically significant (p < 0.001).
Table 1. Genes most upregulated by Gln depletion (log2Ratio > 3).
| Rank | Log2 | p value | Genes | Names | Access Numbers |
|---|---|---|---|---|---|
| 1 |
4.712066 |
0.00E+00 |
S100P |
S100 calcium binding protein P |
NM_005980.2 |
| 2 |
4.116244 |
0.00E+00 |
FAM65C |
family with sequence similarity 65, member C |
NM_080829.2 |
| 3 |
4.04208 |
8.55E-44 |
STC2 |
stanniocalcin 2 |
NM_003714.2 |
| 4 |
3.897732 |
0.00E+00 |
TMEM198 |
transmembrane protein 198 |
NM_001005209.1 |
| 5 |
3.88152 |
0.00E+00 |
OSTBETA |
organic solute transporter β |
NM_178859.3 |
| 6 |
3.828137 |
0.00E+00 |
ZNF395 |
zinc finger protein 395 |
NM_018660.2 |
| 7 |
3.825807 |
0.00E+00 |
MEF2B|MEF2B |
MEF2B read through transcript |myocyte enhancer factor 2B |
NR_027308.1,NR_027307.1 |
| 8 |
3.68936 |
0.00E+00 |
ETV5 |
ets variant 5 |
NM_004454.2 |
| 9 |
3.661515 |
0.00E+00 |
WWC1 |
WW and C2 domain containing 1 |
NM_001161662.1,NM_001161661.1 |
| 10 |
3.647815 |
1.34E-40 |
CYP2B6 |
cytochrome P450, family 2, subfamily B, polypeptide 6 |
NM_000767.4|NR_001278.1 |
| 11 |
3.586737 |
0.00E+00 |
PACS2 |
phosphofurin acidic cluster sorting protein 2 |
NM_015197.2,NM_001100913.1 |
| 12 |
3.541464 |
1.57E-39 |
FOXP4 |
forkhead box P4 |
NM_001012426.1,NM_001012427.1 |
| 13 |
3.496547 |
1.40E-45 |
HEY1 |
hairy/enhancer-of-split related with YRPW motif 1 |
NM_001040708.1,NM_012258.3 |
| 14 |
3.457144 |
0.00E+00 |
MAPK8IP2 |
mitogen-activated protein kinase 8 interacting protein 2 |
NM_016431.3,NM_012324.3 |
| 15 |
3.444888 |
0.00E+00 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
NM_006705.3 |
| 16 |
3.378495 |
0.00E+00 |
GPT2 |
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
NM_001142466.1,NM_133443.2 |
| 17 |
3.373006 |
0.00E+00 |
ZSWIM4 |
zinc finger, SWIM-type containing 4 |
NM_023072.2 |
| 18 |
3.343561 |
0.00E+00 |
MBD6 |
methyl-CpG binding domain protein 6 |
NM_052897.3 |
| 19 |
3.337839 |
2.27E-42 |
TSC22D3 |
TSC22 domain family, member 3 |
NM_001015881.1,NM_198057.2 |
| 20 |
3.304534 |
3.86E-34 |
SCN2B |
sodium channel, voltage-gated, type II, β |
NM_004588.4 |
| 21 |
3.268685 |
0.00E+00 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
NM_001135581.1,NM_003038.3 |
| 22 |
3.265561 |
0.00E+00 |
FOXQ1 |
forkhead box Q1 |
NM_033260.3 |
| 23 |
3.260832 |
5.63E-39 |
SPIRE1 |
spire homolog 1 (Drosophila) |
NM_001128627.1,NM_001128626.1 |
| 24 |
3.256531 |
0.00E+00 |
G0S2 |
G0/G1switch 2 |
NM_015714.3 |
| 25 |
3.254313 |
0.00E+00 |
PPP1R3G |
protein phosphatase 1, regulatory (inhibitor) subunit 3G |
NM_001145115.1 |
| 26 |
3.246922 |
0.00E+00 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
NM_002333.3 |
| 27 |
3.223594 |
0.00E+00 |
RASIP1 |
Ras interacting protein 1 |
NM_017805.2 |
| 28 |
3.220771 |
0.00E+00 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
NM_001166356.1,NM_005412.5 |
| 29 |
3.206867 |
0.00E+00 |
FAM134A |
family with sequence similarity 134, member A |
NM_024293.4 |
| 30 |
3.201466 |
0.00E+00 |
PI4KB |
phosphatidylinositol 4-kinase, catalytic, β |
NM_002651.1 |
| 31 |
3.200373 |
0.00E+00 |
ANKRD11 |
ankyrin repeat domain 11 |
NM_013275.4 |
| 32 |
3.159755 |
1.40E-45 |
GPR56 |
G protein-coupled receptor 56 |
NM_001145774.1,NM_001145772.1 |
| 33 |
3.159169 |
7.43E-37 |
IDS |
iduronate 2-sulfatase |
NM_000202.5,NM_001166550.1 |
| 34 |
3.127714 |
0.00E+00 |
TIGD2 |
tigger transposable element derived 2 |
NM_145715.2 |
| 35 |
3.117394 |
1.57E-35 |
KLHL29 |
kelch-like 29 (Drosophila) |
NM_052920.1 |
| 36 |
3.114287 |
0.00E+00 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
NM_020904.2,NM_001161354.1 |
| 37 |
3.088076 |
0.00E+00 |
TBC1D2 |
TBC1 domain family, member 2 |
NM_018421.3 |
| 38 |
3.074656 |
2.21E-29 |
GEFT |
RhoA/RAC/CDC42 exchange factor |
NM_182947.2,NM_001111270.1 |
| 39 |
3.070325 |
0.00E+00 |
POP4 |
processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) |
NR_027368.1,NM_006627.2 |
| 40 |
3.061844 |
3.82E-29 |
BATF2 |
basic leucine zipper transcription factor, ATF-like 2 |
NM_138456.3 |
| 41 |
3.059862 |
0.00E+00 |
UBOX5 |
U-box domain containing 5 |
NM_014948.2,NM_199415.1 |
| 42 |
3.057593 |
1.52E-31 |
TRIOBP |
TRIO and F-actin binding protein |
NM_001039141.2,NM_007032.5 |
| 43 |
3.054969 |
1.10E-32 |
PLA1A |
phospholipase A1 member A |
NM_015900.2 |
| 44 |
3.045242 |
0.00E+00 |
C1orf216 |
chromosome 1 open reading frame 216 |
NM_152374.1 |
| 45 |
3.032083 |
4.42E-33 |
PELI3 |
pellino homolog 3 (Drosophila) |
NM_001098510.1,NM_145065.2 |
| 46 |
3.025387 |
9.42E-31 |
FBLL1 |
fibrillarin-like 1 |
NR_024356.2 |
| 47 |
3.01661 |
0.00E+00 |
FOXC1 |
forkhead box C1 |
NM_001453.2 |
| 48 |
3.015883 |
7.71E-26 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
NM_017778.2 |
| 49 |
3.012984 |
0.00E+00 |
FBXO31 |
F-box protein 31 |
NM_024735.3,NR_024568.1 |
| 50 |
3.005587 |
1.41E-27 |
TCEB2 |
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
NM_207013.1,NM_007108.2 |
| 51 | 3.00214 | 4.82E-41 | KCNMB3 | potassium large conductance calcium-activated channel, subfamily M β member 3 | NM_171830.1,NM_014407.3 |
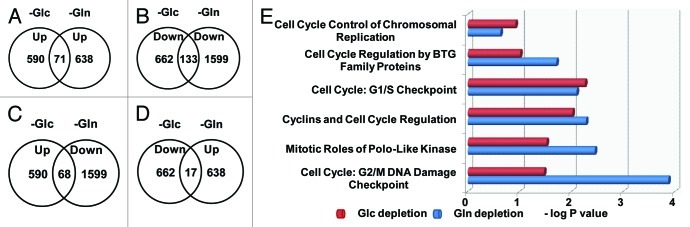
On the other hand, Glc depletion (for 24 h) causes differential expression of 1,252 genes (log2 Ratio > 1.0 or < -1.0); among them, 590 were upregulated, while 662 genes were downregulated. We noted that comparing to Glc depletion, Gln depletion apparently caused a much greater change of gene expression profiling, underlining the important roles of Gln in normal cell function and metabolism.3 To understand whether Gln depletion and Glc depletion cause similar stresses to cells and lead to the regulation of the same group of genes, we next compared the genes regulated by Gln depletion with those regulated by Glc depletion (Fig. 2). Among the genes upregulated by Glc depletion or Gln depletion, 71 overlapped genes represent a very small portion (Fig. 2A). Similarly, only 133 genes were downregulated by both Glc depletion and Gln depletion (Fig. 2B). Interestingly, 68 genes upregulated by Glc depletion were downregulated by Gln depletion (Fig. 2C), and 17 genes upregulated by Gln depletion were downregulated by Glc depletion (Fig. 2D). Since both Gln depletion and Glc depletion cause similar growth inhibition, we particularly analyzed the expression of several groups of cell cycle regulators. Data shown in Figure 2E and Table S2 indicate that under either Gln depletion or Glc depletion, the expression patterns of cyclins and other cell cycle regulators were significantly changed. However, Gln depletion caused more significant changes in genes controlling the G2/M checkpoint, whereas Glc depletion primarily affected the expression of genes involved in G1/S. To understand if these changes were related to cell cycle progression, we performed cell cycle analysis and found either Gln or Glc depletion increased cell population in G2/M phase (Fig. S1), which is consistent with IPA prediction (Fig. 2E). The distinct gene expression patterns and cell cycle changes indicate that both Gln depletion and Glc depletion represent nutrient depletion stresses, but cells respond in different ways.
Figure 2. Gln depletion and Glc depletion alter gene expression. (A–D) RNA samples from Hep3B cultured in complete media were used as control. For Glc depletion, log2 Ratio > 1 (upregulation) or < -1.0 (downregulation) was used as cut-off. Since Gln depletion caused dramatic change of gene expression levels, log2 Ratio > 2.0 (upregulation) or < -2.0 (downregulation) was set as cut-off. We noted that only a small portion of genes were upregulated under both conditions, or downregulated under both conditions. (E) Expression of cell cycle regulators in Gln- and Glc-depleted cells. Gene expression was analyzed by IPA. P value of the reprogramming of each group of cell cycle regulators were calculated according to established criteria and algorithm, and presented in -log as x-axis. Our data show that under both nutrient conditions, the expression of cyclins and cell cycle regulators was significantly changed. In particular, Gln depletion significantly altered the expression profiling of genes regulating G2/M and G1/S checkpoints, and Glc depletion mainly caused the reprogramming of genes regulating G1/S checkpoint.
Functional responses of transcription regulators to Gln depletion
It has been well known that amino acid starvation triggers the activation of GCN2-eIF2α axis, which generally inhibits the translation of proteins, but specifically stimulates IRES-initiated ATF4 translation, resulting in the upregulation of ATF4 target genes.26 Considering the breadth of altered gene expression, we wondered if other transcription regulators were implicated. Because the functions of most transcription regulators are regulated by a variety of physiological or pathological alterations, mRNA levels of transcription regulators are not the most reliable indicators of their functional status. We analyzed our gene expression data set with the IPA program, which predicts the functional activation or inhibition of transcription regulators based on the expression directions (up- or downregulated) of their known downstream genes. Using established algorithms and criteria, the IPA program calculates regulation z-scores of a given transcription regulator based on the response of all known downstream targeted genes, and uses the z-score as an objective parameter to estimate confidence level. Based on this analysis, 25 transcription regulators were scored smaller than -2.00, thus, were predicted to be inhibited. Among these putatively inhibited transcription regulators are HNF1, HNF4, SREBF2 and FOXO1, indicating that upon Gln depletion, normal functions of hepatocytes are generally repressed to conserve Gln and its metabolic derivatives. Although it was not surprising to observe that many transcription regulators were repressed upon Gln depletion, we were puzzled to find that the function of eight transcription regulators were predicted to be activated (regulation z-score > 2.00) (Table 2). Among these activated regulators are the renowned cell cycle repressor RB, and MDM2, which encodes a protein regulating p53 stability, suggesting that the two pivotal cell cycle regulators, pRb and p53, are involved in Gln depletion-triggered growth arrest. This observation is consistent with the reprogramming of G1/S and G2/M checkpoint regulators (Fig. 2E).
Table 2. Functional responses of transcription regulators to Gln depletion.
| Transcription Regulator |
Predicted State |
Regulation z-score |
p value of overlap |
|---|---|---|---|
| PLAG1 | Activated | 2.685 | 2.67E-02 |
| TCF3 |
Activated |
2.633 |
1.54E-01 |
| TOB1 |
Activated |
2.514 |
5.02E-02 |
| CUX1 |
Activated |
2.217 |
5.17E-01 |
| MDM2 |
Activated |
2.146 |
5.18E-01 |
| HEY2 |
Activated |
2.076 |
1.08E-01 |
| Rb |
Activated |
2.041 |
4.67E-03 |
| MXD1 |
Activated |
2.016 |
8.51E-02 |
| KDM5B |
|
1.803 |
1.90E-06 |
| GFI1 |
|
1.696 |
1.53E-03 |
| NKX2–1 |
|
-1.666 |
2.12E-02 |
| Cyclin E |
|
-1.718 |
4.30E-02 |
| HIF1A |
|
-1.788 |
4.21E-02 |
| PGR |
|
-1.843 |
8.73E-09 |
| TFDP1 |
Inhibited |
-2.028 |
7.70E-03 |
| Tcf 1/3/4 |
Inhibited |
-2.049 |
1.70E-02 |
| MYCBP |
Inhibited |
-2.049 |
7.60E-02 |
| Creb |
Inhibited |
-2.236 |
7.64E-02 |
| BRD7 |
Inhibited |
-2.254 |
3.01E-02 |
| NRF1 |
Inhibited |
-2.261 |
1.07E-02 |
| XBP1 |
Inhibited |
-2.562 |
3.48E-10 |
| HNF4A |
Inhibited |
-2.662 |
6.92E-60 |
| MYC |
Inhibited |
-2.744 |
1.00E-08 |
| ELF4 |
Inhibited |
-2.750 |
7.48E-02 |
| FOXO1 |
Inhibited |
-2.780 |
1.45E-02 |
| EGR1 |
Inhibited |
-3.028 |
6.99E-02 |
| HNF1A |
Inhibited |
-3.116 |
2.53E-04 |
| MYCN |
Inhibited |
-3.440 |
2.65E-04 |
| NFE2L2 | Inhibited | -3.565 | 3.67E-05 |
Transcription regulators with z-scores between 1.65–2.0, or -1.65–-2.0 were listed for reference.
In addition, PLAG1, TCF3, TOB1, Cux1, HEY2 and MXD1 were scored as activated transcription regulators, and their involvement in nutrient depletion has not been reported before. Particularly, PLAG1 (pleomorphic adenoma gene 1) encodes a zinc finger protein, and its expression is developmentally regulated. Chromosomal translocation of PLAG1 has been observed frequently in pleomorphic adenomas of the salivary glands.28 IPA analysis predicts the activation of PLAG1 by Gln depletion (Table 3). TCF3, also known as E2A, E47, ITF1, VDIR and bHLHb21, has been identified as a component of multiple transcription complexes.29 IPA analysis suggests TCF3 activation upon Gln depletion (Table S3). Cux1 (CUTL1, CDP, CDP/Cux) is an Rb-interacting protein and has been found to regulate morphogenesis, differentiation and cell cycle progression, particularly at S-phase. It also has been considered as a tumor suppressor gene in uterine leiomyomas and breast cancers.30 TOB1 (transducer of ERBB2) encodes a member of the tob/btg1 family of anti-proliferative proteins. It has been reported that when exogenously expressed in cultured cells, TOB1 suppresses cell growth.31 HEY2 (hairy/enhancer-of-split related with YRPW motif protein 2) is also known as cardiovascular helix-loop-helix factor 1 (CHF1).32 HEY2 is a member of the hairy and enhancer of split-related (HESR) family of basic helix-loop-helix (bHLH)-type transcription regulator. The encoded protein forms either homo- or hetero-dimers and interacts with histone deacetylase complexes, such as Sirtuin 1 and nuclear receptor corepressor 1, to repress the transcription of a variety of genes.33 MXD1 encodes MAD protein, which competes with Myc for Max to form heterodimers. Since MAD also recruits HDAC2 and Sin3A, MAD represses Myc-dependent transcription.34,35 Consistent with this finding, the functions of Myc and N-Myc were found to be inhibited (Table 1). The precise biological significance of the activation of these transcription regulators upon Gln depletion remains to be determined.
Table 3. Regulation directions of PLAG1 target genes upon Gln-depletion.
| Genes | Log2 Ratio | Findings | Prediction* |
|---|---|---|---|
| VEGFA |
2.146 |
Upregulates (1) |
Activated |
| SMARCD3 |
1.868 |
Upregulates (1) |
Activated |
| S100A2 |
1.297 |
Upregulates (1) |
Activated |
| ROM1 |
1.070 |
Upregulates (1) |
Activated |
| PTP4A3 |
1.515 |
Upregulates (1) |
Activated |
| PPIC |
-1.529 |
Downregulates (1) |
Activated |
| MLXIP |
-1.283 |
Downregulates (1) |
Activated |
| MAP2 |
2.668 |
Upregulates (1) |
Activated |
| KCNJ4 |
2.302 |
Upregulates (1) |
Activated |
| GJB1 |
-2.954 |
Downregulates (1) |
Activated |
| FLNC |
2.186 |
Upregulates (2) |
Activated |
| ETV4 |
2.472 |
Upregulates (1) |
Activated |
| CNKSR1 |
2.006 |
Upregulates (1) |
Activated |
| CLTB |
1.810 |
Upregulates (1) |
Activated |
| C11orf9 |
1.435 |
Upregulates (1) |
Activated |
| ATF5 |
1.194 |
Upregulates (2) |
Activated |
| ACVR2B |
2.082 |
Upregulates (1) |
Activated |
| TGM2 |
-1.069 |
Upregulates (1) |
Inhibited |
| PLEC |
-2.381 |
Upregulates (1) |
Inhibited |
| PIGF |
-1.218 |
Upregulates (1) |
Inhibited |
| COL9A3 | -5.620 | Upregulates (2) | Inhibited |
PLAG1 function predicted based on directions of target gene expression.
Functional responses of transcription regulators to Glc depletion
Both Glc and Gln are indispensible for most cells, and both share a common metabolic role as carbon sources for proliferative biosynthesis. However, the alteration of gene expression triggered by Glc depletion is clearly distinct from that of Gln depletion (Fig. 1A and 2). In addition, Gln is also an important nitrogen source for the synthesis of nitrogenous biomolecues.14 To predict the functional states of transcription regulators and identify potential transcription regulators involved in response to Glc depletion, we used IPA to analyze the microarray data. As shown in Table 4, 16 transcription regulators have z-scores greater than 2.0 (thus being predicted to be activated at functional levels). The activated CDKN2A, TP73, RB1, YY1 and TP63 are proliferative suppressors, consistent with the observed cell growth arrest associated with Glc depletion. In addition, both E2F1 and E2F2 are found among the regulators repressed by Glc depletion. Other functionally inhibited transcription regulators play roles in normal metabolic regulation in hepatocytes. These findings suggest that the cellular stresses caused by Glc depletion are different from that triggered by Gln depletion.
Table 4. Functional responses of transcription regulators to Glc(-).
| Transcription Regulator |
Predicted State |
Regulation z-score |
P value of overlap |
|---|---|---|---|
|
XBP1 |
Activated |
4.115 |
3.16E-09 |
|
ATF4 |
Activated |
4.014 |
2.61E-16 |
|
KDM5B |
Activated |
3.811 |
1.53E-07 |
|
CDKN2A |
Activated |
3.131 |
9.50E-13 |
|
TP73 |
Activated |
2.685 |
3.35E-04 |
|
RB1 |
Activated |
2.600 |
1.56E-13 |
|
YY1 |
Activated |
2.437 |
1.71E-08 |
|
SMARCA4 |
Activated |
2.417 |
4.36E-06 |
|
RBPJ |
Activated |
2.355 |
7.42E-02 |
|
ATF6 |
Activated |
2.295 |
6.37E-05 |
|
HDAC1 |
Activated |
2.279 |
1.59E-04 |
|
DDIT3 |
Activated |
2.257 |
4.12E-04 |
|
TP63 |
Activated |
2.233 |
4.28E-06 |
|
TCF3 |
Activated |
2.224 |
1.12E-04 |
|
NFE2L2 |
Activated |
2.028 |
3.77E-07 |
|
HDAC2 |
|
1.984 |
4.20E-02 |
|
STAT4 |
|
1.942 |
5.96E-03 |
|
NRIP1 |
|
1.831 |
1.69E-03 |
|
BRCA1 |
|
1.805 |
1.82E-04 |
|
BTG2 |
|
1.711 |
7.41E-04 |
|
IRF4 |
|
1.658 |
1.40E-02 |
|
NR4A2 |
|
-1.654 |
1.61E-02 |
|
MLXIPL |
|
-1.797 |
8.96E-03 |
|
GLI1 |
|
-1.825 |
1.26E-04 |
|
MED1 |
|
-1.875 |
3.28E-02 |
|
E2F4 |
|
-1.921 |
2.94E-17 |
|
NFkB (complex) |
|
-1.952 |
8.59E-09 |
|
GLI2 |
Inhibited |
-2.023 |
8.84E-02 |
|
RUNX2 |
Inhibited |
-2.072 |
2.61E-03 |
|
E2F1 |
Inhibited |
-2.130 |
4.42E-15 |
|
HNF1A |
Inhibited |
-2.175 |
4.45E-10 |
|
Hdac |
Inhibited |
-2.287 |
3.70E-04 |
|
E2F2 |
Inhibited |
-2.424 |
1.52E-08 |
|
FOXM1 |
Inhibited |
-2.655 |
5.99E-07 |
| TBX2 | Inhibited | -3.943 | 4.47E-09 |
Notably, ATF4 and ATF6, two regulators known to be involved in ER-stress responses, were found among these activated regulators, consistent with reports from other independent studies.36,37 Although XBP1 was predicted to be functionally inhibited by Gln depletion, it was significantly activated by Glc depletion (z-score: 4.12) (Table S4).
Altered gene expression in cells adapted to long-term Gln depletion
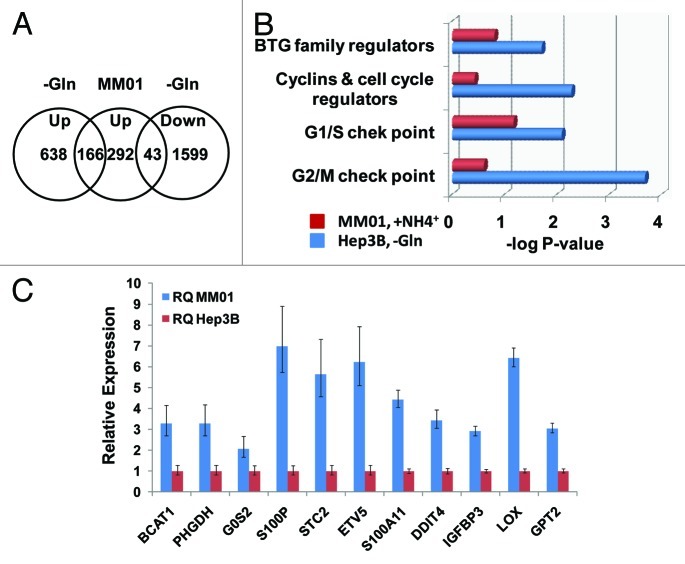
By culturing Hep3B cells in Gln-free media supplemented with ammonia, we established a cell line MM01 with a slow growing phenotype, which represents a result of long-term adaptation to Gln insufficiency.14 Comparing the gene expression of MM01 cells with acute Gln-depeleted Hep3B cells, we expected to reveal insight into the adaptive mechanisms. Microarray analysis reveal only 292 genes were upregulated, while 398 genes were downregulated in MM01 cells. Among the upregulated genes, 166 were also upregulated, while 43 were downregulated under acute Gln-depleted conditions (Fig. 3A). To gain insight into the growth inhibition upon acute Gln depletion, we compared the gene expression of cell cycle regulators. Based on IPA analysis, acute Gln depletion caused significant reprogramming of three groups of cell cycle-related genes: G1/S checkpoint, G2/M checkpoint and cyclins and cell cycle regulators (Fig. 3B). While in the adapted MM01 cells, these groups of genes were not significantly altered. The above results may explain why MM01 cells proliferate continually but slowly. Upregulation of selected genes was further confirmed by qRT-PCR, shown in Figure 3C. Data analysis by IPA predicted the responses of several transcription regulators, which are summarized in Table 5. In addition to ATF4, the other transcription regulators predicted to be upregulated include SMAD2, SMAD3, ESR1, PPARA, DDIT3, TP73 and HIC1. Comparing with cells failing to proliferate during acute Gln depletion (Table 2), the activation of ATF4, SMAD2, SMAD3, ESR1 and PPARα in adapted cells is particularly intriguing. IPA analysis suggests the activation of PPARα in MM01 cells (Table S5).
Figure 3. Gene expression and cell cycle regulation in MM01 cells. In contrast to acute Gln-depleted cells, MM01 cells have adapted to Gln-free media. (A) Upregulated genes in MM01 and their expression states in acute Gln-depleted cells. In MM01 cells, 292 genes were identified as upregulated (log2 Ratio > 1.0). Among them, 166 were found upregulated, while 43 downregulated in acute Gln-depleted cells. (B) Functional analysis of cell cycle regulators in acute Gln-depleted Hep3B and MM01 cells. In acute Gln-depleted cells, all four groups of cell cycle regulatory genes were significantly reprogrammed (-log p values were given). On the other hand, the same groups of genes were not significantly affected. Note that the expression levels of some regulators of G1/S checkpoint in MM01 were affected, but collectively, change of this group of genes remain statistically insignificant (p = 0.084). (C) Validation of the upregulation of selected genes in MM01 cells. Representative genes identified to be upregulated by microarray analysis were picked up for further validation. Levels of mRNA were determined by qRT-PCR. RNA samples isolated from Hep3B cells cultured in complete media were used as control, and mRNA levels in the control sample were arbitrarily defined as 1.
Table 5. Functional responses of transcription regulators in MM01 cells.
| Transcription Regulator |
Predicted State |
Regulation z-score |
p value of overlap |
|
|---|---|---|---|---|
|
SMAD2 |
Activated |
2.845 |
4.30E-03 |
|
|
ATF4 |
Activated |
2.755 |
4.97E-11 |
|
|
ESR1 |
Activated |
2.382 |
2.99E-07 |
|
|
PPARA |
Activated |
2.302 |
3.33E-13 |
|
|
DDIT3 |
Activated |
2.268 |
1.97E-02 |
|
|
SMAD3 |
Activated |
2.161 |
6.03E-04 |
|
|
TP73 |
Activated |
2.152 |
1.45E-02 |
|
|
HIC1 |
Activated |
2.124 |
2.69E-02 |
|
|
PLAG1 |
|
1.913 |
1.97E-02 |
|
|
PRDM1 |
|
1.757 |
3.33E-02 |
|
|
NCOR1 |
|
1.664 |
6.86E-04 |
|
|
PGR |
|
-1.752 |
4.60E-06 |
|
|
FOS |
|
-1.863 |
9.95E-03 |
|
|
MYOD1 |
|
-1.961 |
2.71E-02 |
|
|
TFAP2A |
Inhibited |
-2.024 |
1.77E-02 |
|
|
NFKB1 |
Inhibited |
-2.025 |
2.19E-03 |
|
|
GATA4 |
Inhibited |
-2.081 |
2.43E-03 |
|
|
CEBPA |
Inhibited |
-2.170 |
1.03E-07 |
|
|
SMAD7 |
Inhibited |
-2.239 |
1.91E-02 |
|
|
HNF1A |
Inhibited |
-2.814 |
4.43E-18 |
|
| HNF4A | Inhibited | -3.135 | 5.54E-17 | |
The status of ER-stress signaling pathways in different nutrient conditions
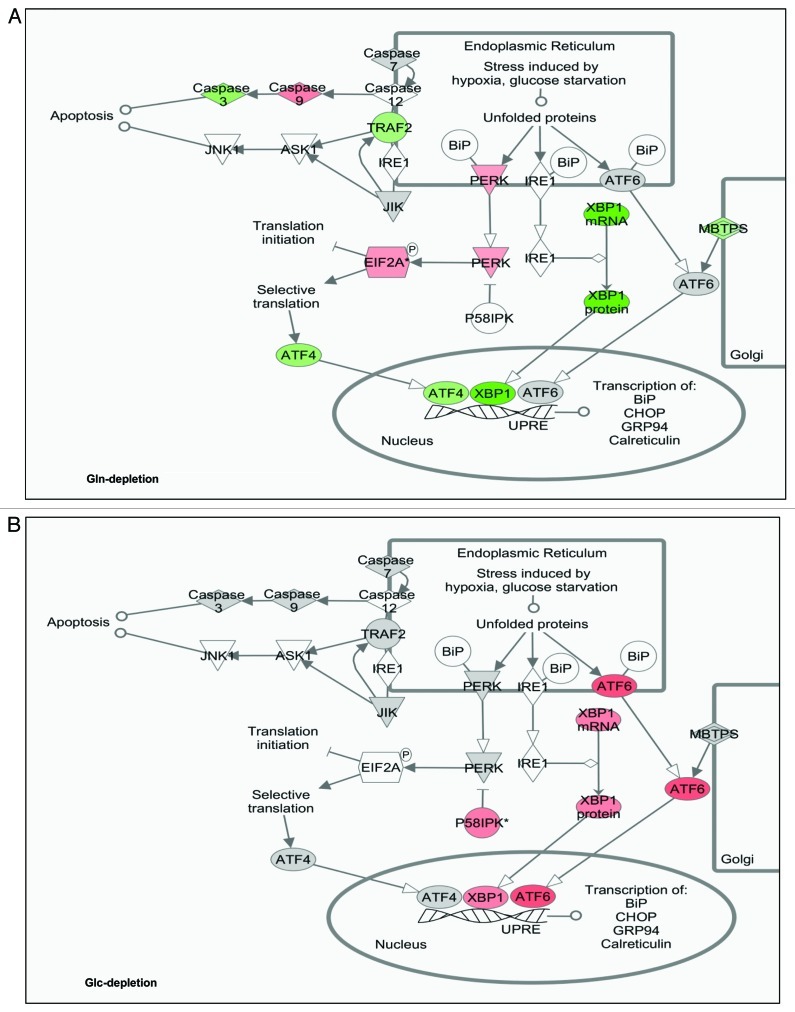
It has been well-established that nutrient depletion triggers ER stress responses, which can be mediated by PERK-eIF2α or GCN2-eIF2α.26,38 Thus, we examined potential ER-stress signaling pathways in different nutrient-insufficient conditions. We first studied the effects of Gln depletion on the functional regulation of ER stress-related factors. Overlapping of the RNA profiling data to the ER stress pathways revealed that Gln depletion triggered eIF2α activation (Fig. 4A), which is consistent with our previously reported phosphorylation of eIF2α upon Gln depletion.14 However, functionally, ATF4 was predicted to be repressed in our experimental setting (Fig. 4A); this result might be ascribed to the fact that ATF4 mRNA level was dramatically decreased under this nutrient condition (log2Ratio = -6.13). While no evidence to indicate that IRE1 or ATF6 function was affected, XBP1 was predicted to be repressed at mRNA, protein and functional levels. On the other hand, under Glc depletion conditions, all ATF4, XBP1 and ATF6 pathways were activated (Fig. 4B), showing a pattern dramatically different from that of Gln depletion. The states of ER stress signaling pathways under all three nutrient conditions are summarized in Table 6.
Figure 4. States of ER-stress signaling pathways under Gln- and Glc-depleted cells. Changes of ER-stress signaling pathways were generated by IPA. Three transcription factors, ATF4, ATF6 and XBP1, have been reported to play critical roles in ER-stress response. Activation of PERK leads to phosphorylation of eIF2α, hence translationally activating ATF4. ER stress also activates IRE1, which facilitates the splicing of XBP1 RNA. Grey-filled factors indicate no significant change; red filled activated; green filled inhibited and non-filled factors not covered in the data set analyzed. (A) Functional states of ER-stress signaling pathways in Gln-depleted cells. (B) Functional states of ER-stress signaling pathways in Glc-depleted cells.
Table 6. States of ER-stress signaling pathways at various nutrient conditions.
| Nutrients | *eIF2α | ATF4 | ATF6 | XBP1 | Cell Fates | |
|---|---|---|---|---|---|---|
|
Gln(-), 24 h |
|
Activation |
Inhibition |
Insignificant |
Inhibition |
Arrest/death |
|
MM01** |
+NH4+ |
Activation |
Activation |
Insignificant |
Insignificant |
Adapted |
| Glc (-), 24 h | Activation | Activation | Activation | Activation | Arrest/death |
Based on eIF2α phosphorylation detected in western blots, all other predication was based on IPA analysis.
Adapted to Gln-free media, long-term survival and proliferates at slow rate.
Discussion
Molecular mechanisms mediate oxygen sensing, and subsequent adaptive responses have been well-characterized;18 it becomes eminent to address how cells sense and respond to the dynamic change of availability of carbon source and nitrogen source. We hypothesized that cells sense carbon source and nitrogen source availability via a paradigm similar to that of oxygen sensing. Besides the systemic and hormonal regulation, cells also have an innate ability to sense the low availability of carbon and nitrogen sources and to regulate the expression of genes to support the cell survival and proliferation. In this study, we take advantage of the cell culture system, in which all nutrient conditions can be clearly defined, as our first step to test our hypothesis. We exploited the mRNA profiling and IPA program to study the systemic gene expression reprogramming pattern and the complicated regulatory networks and hierarchies. Based on our studies, we found several shared characteristics of gene expression in different nutrient conditions: (1) transcription regulators controlling normal cell functions were generally repressed; (2) transcription regulators controlling ER stresses were affected; and (3) cell cycle regulators were reprogrammed, and, particularly in Gln-free or Glc-free media, cell growth was inhibited.14 More importantly, although both Gln depletion and Glc depletion cause growth inhibition, the gene expression that occurred under each condition has distinctive features. This difference was reflected not only by the upregulated genes under each nutrient condition, but also by the predicted functional response of transcription factors. Moreover, we also tested the effects of Gln or Glc depletion on cell cycle progression. Because Hep3B cells easily form clumps during trypsinization, we used HeLa, another cell line which has been demonstrated to grow in Gln-dependent manner, and we found both Gln and Glc depletion causes the arrest of non-synchronized HeLa cells in G2/M phase. In a recent paper, Sergio et al. cultured synchronized HeLa cells in Glc-free or Gln-free media for 18 h, and their data demonstrated that both Glc and Gln were indispensible nutrients for cell cycle progression through G1 phase.39 Therefore, we conclude that the low availability of either Gln or Glc restricts cell cycle progression.
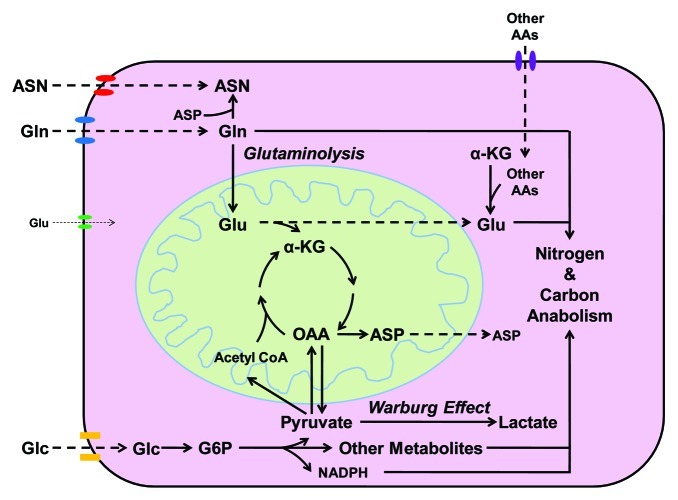
Amino acid starvation response has been studied, primarily based on results obtained from essential amino acid starvation. However, lack of non-essential amino acids has not been intensively studied in the past, partly because of the concept that non-essential amino acids can be synthesized by cells. Considering the metabolic roles of Gln shown in Figure 5 and the inefficient uptake of glutamate (Glu) by most types of cells, Gln depletion will also result in decreased levels of Asn and Glu, and decreased Glu, in turn, reduces the uptake of cystine, leading to lack of intracellular cysteine. Indeed, 24 h Gln depletion also caused eIF2α phosphorylation (Fig. S2),14 indicating Gln depletion may trigger general inhibition of translation via eIF2α activation. However, it is unclear if other pathways also participate in tumor cells’ sensing and responding to the lack of a specific non-essential amino acid and activate its biosynthesis. In addition to acute Gln insufficiency, we also utilized MM01 cells surviving in Gln-free media supplemented with ammonia as an adapted cell line to long-term Gln insufficiency.14 MM01 cells grow slowly compared with Hep3B cells cultured in regular complete media; if ammonia is removed, MM01 cells will show more slowly proliferative phenotype and eventually die out (data not shown). In this paper, we only compared the genes upregulated in acute Gln-insufficient Hep3B cells and in adapted MM01 cells; further studies on the genes upregulated in both conditions may identify critical regulators involved in cellular response to Gln insufficiency. In this study, we found ATF4 and DDIT3 were activated in MM01 cells, suggesting successful establishment of ER-stress response. Moreover, SMAD2, SMAD3 and ESR1 (ERα) were predicted to be functionally activated, suggesting the involvement of TGFβ and estrogen signaling pathways for Hep3B adaptation to Gln insufficiency.40 The activation of TP73 and HIC,41,42 which encode two tumor suppressors, might attenuate the cell proliferation rate to match the decreased Gln availability, thus contributing to the slow-growing phenotype.
Figure 5. Distinct metabolic roles of Glc and Gln in cell proliferation. Glc utilization provides cells with ATP, NADPH and carbon metabolites, which fulfill the major needs of energy, reducing power and carbon skeletons for anabolic activities. Gln is a substrate for the synthesis of Asn and nucleotides. It also serves as a precursor of Glu, which cannot be taken by cells efficiently. Glu serves as a central hub of nitrogen storage and redistribution, amino groups from other amino acids can be transferred to α-KG to form Glu. On the other hand, Glu serves as a major donor of amino group in the biosynthesis of other non-essential amino acids. In addition, Glu also serves as substrate for the synthesis of proteins, nucleotides, glutathione, polyamine and other nitrogenous molecules. Maintaining a relatively high intracellular concentration of Glu is also important for cells to uptake other nutrients; for example, the release of Glu down the concentration gradient provides the energy for cells to uptake cystine. Finally, the carbon skeleton of Gln or Glu may be eventually utilized by cells as a carbon source. Gln and Glc utilization work together to facilitate the biosynthesis of nitrogenous biomolecules.
In the well-studied pancreatic β cells, the supply of Glc directly affects the ATP levels, which mediates the insulin production and release. However, it has been unknown how other cells directly respond to the lack of Glc (or G6P). In our study, we observed a strong functional activation of ATF6, which may be partly responsible for the upregulation and functional activation of XBP1 pathway (Fig. 4B). This ER-stress response pattern is different from that caused by Gln depletion, suggesting that Glc depletion triggered a cellular stress different from that triggered by Gln depletion. It has been reported that ATF6 is activated by unfolded protein response,43 suggesting Glc supply is important to prevent cells from unfolded protein stress.
In conclusion, we have established a genome-wide gene expression profiling database of Hep3B cells under defined nutrient conditions. Analysis of the gene expression patterns under those conditions revealed that Gln and Glc represent two important nutrients for cell survival and proliferation, but each has distinctive metabolic roles. Lack of either Glc or Gln will lead to the inhibition of cell cycle progression and eventual cell death through related, but not identical, regulatory hierarchies. Imbalanced availability of Glc and Gln results in cellular stresses and adaptive responses that may also participate in oncogenic processes by selecting cancer cells more adaptive to poor nutrient conditions. It is possible to identify the molecules that facilitate tumor cell survival in nutrient-deprived conditions. Inhibiting or knocking-down of these molecules may provide putative targets for tumor treatment.
Materials and Methods
Cell culture media and reagents
Hep3B cells (ATCC HB-8064) were cultured in Eagle's minimal essential medium supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere, and the cells were passed twice a week. Gln-independent MM01 cells were established by culturing Hep3B cells in Gln-free DMEM (15-0130-CV) with 10% dialyzed FBS (Atlantic Biologicals, dialyzed 10 kD) and 0.8 mM ammonia. MM01 cells were kept at 37°C in a humidified 5% CO2 atmosphere and the culture medium was changed every 2 d, and cells were passed when reaching 90% confluence. For long-term glutamine-deprivation study, MM01 cells were split and passed in a ratio of 1:3, the next day, cells were cultured in fresh medium for 24 h before RNA isolation. For acute glutamine-deprivation study, Hep3B cells were passed in a ratio of 1:3 and cultured in Gln-free DMEM with 10% dialyzed FBS and 4 mM Gln for adaptation; the next day, cells were cultured in fresh medium (Gln-free DMEM with 10% dialyzed FBS) for 24 h before RNA isolation. For acute glucose-deprivation study, Hep3B cells were passed in a ratio of 1:3 and cultured in glucose-free DMEM with 10% dialyzed FBS for adaptation; the next day, cells were cultured in fresh medium (glucose-free DMEM with 10% dialyzed FBS) for 24 h before RNA isolation. Hep3B cells cultured in Gln-free DMEM with 10% dialyzed FBS and 4 mM Gln were used as control cells.
RNA isolation
MM01 and Hep3B cells were treated as above for 24 h and then washed with ice-cold PBS twice. Total RNA was extracted from the cells using RNeasy Mini Kit (Qiagen). RNA concentration was determined using Nanodrop spectrofotometer (Nanodrop Technologies) at 260/280 nm.
Microarray analysis
Gene expression profiling was performed by OneArrays (Phalanx Biotech Group). Human OneArrays contain 32,275 oligonucleotides, 29,187 human genome probes and 1,088 experimental control probes formed as 60-mer sense-strand DNA elements. The samples from Hep3B and MM01 cells were hybridized with array chips, and signal intensity was measured and processed by GenePix Pro version 4.0 (Axon). The GenePix data was analyzed using Array Studio (Omicsoft Corp). Significant expression differences were defined as a p value < 0.05 and displayed as log2 ratio values > 1.00 or < -1.00.
Quantitative real-time PCR (qRT-PCR) confirmation of selected genes
Total RNA was first reverse-transcribed to cDNA using SuperScript II Reverse Transcriptase (Invitrogen). PCR for β-actin was used to confirm cDNA quality. Then cDNA was diluted with ratio of 1/50 and used for quantitative analysis using validated gene-specific TaqMan probes (Applied Biosystem). β-actin was used as a housekeeping gene for normalization of RNA loading. Relative quantification (RQ) studies were made from collected data (threshold cycle numbers, referred to as Ct) using StepOne™ Software v2.1 (Applied Biosystems).
Ingenuity pathway analysis
The microarray data set was analyzed using Ingenuity pathway analysis (IPA, www.ingenuity.com Ingenuity Systems, Inc.). IPA can be used to generate gene function or metabolism network according to gene ontology, relevance to canonical pathways and molecular networks based on previously published findings on mammalian biology. The following are the basic processes: (1) Filter the microarray data for each group; genes with detected p value less than 0.05 were included for further comparative analysis; for differentially expressed genes, log2 Ratio values with p < 0.05 were included as significant data. (2) Upload data to IPA for each observation, log2 Ratio values > 1.00 or < -1.00 were used as cut-off during upload, if not otherwise specified. (3) Biostatistic analysis of the association between our microarray data and the known canonical pathways through (a) calculating the percentage of genes from our microarray data in regard of the total number of genes in one specific canonical pathway or (b) determining the probability (overlap p value) of the association between our data and the canonical pathway by the Fischer’s exact test; overlaping p value measures significant overlap between the genes in an uploaded data set and the genes regulated by a transcription factor (it sets p values less than 0.01 as biostatistical significance). (4) Calculate network scores (Z score). These scores reflect the casual association of our data with the canonical pathways; the higher score means a lower probability of casual association. Our data set was uploaded into the IPA program (Content version: 11904312, Release Date: 2011-12-15). Hypothetical networks from our experiments were then built using the core analysis algorithm, one of the several algorithms integrated within Ingenuity systems. The data and networks were then generated and exported from IPA and reformatted for presentation.
Western blot analysis
Whole-cell lysate was collected using urea lysis buffer containing 6.6 M urea, 10 mM TRIS-HCl, 1% SDS, 5 mM DTT, 1% Triton X-100, 10% Glycerol and 1x protein inhibitor mix. Same amount of protein was loaded into a Mini-protein TGX precast gel (Bio-rad) and run following the manufacturer’s protocol. The separated proteins were transferred onto Immunoblot PVDF membrane (Bio-rad), blocked with 5% nonfat dry milk powder in TBST buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.2% Tween 20) and probed with anti-Phospho-eIF2α (Cell Signaling), total eIF2α (Cell Signaling) and α-tubulin (Sigma-Aldrich) antibody, and bands were visualized using horseradish peroxidase (HRP)-conjugated secondary antibody combined with chemiluminescence system (Thermo scientific).
Cell cycle analysis
Hep3B and HeLa cells were cultured in complete medium or medium without either Gln or Glc for 18 h. Then, cells were trypsinized and stained using propidium iodide. Each sample was analyzed by flow cytometry with a FACScan flow cytometer (Becton Dickinson Biosciences) with a 488-nm laser. A minimum of 10,000 events were collected to maximize statistical validity of the analysis.
Statistical analysis
SPSS version 20 was used for all calculations. Values were shown as mean ± standard deviation. Student’s t-test was used to analyze data. The significance level was set at 0.05 for each analysis.
Supplementary Material
Acknowledgments
This work is supported in part by grant R01-CA129494 (to NS) from NCI, National Institutes of Health (NIH) and start-up funds from Drexel University.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21944
References
- 1.Dang CV. Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 2010;9:3884–6. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 2.Dang CV, Hamaker M, Sun P, Le A, Gao P. Therapeutic targeting of cancer cell metabolism. J Mol Med (Berl) 2011;89:205–12. doi: 10.1007/s00109-011-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin C, Qie S, Sang N. Carbon source metabolism and its regulation in cancer cells. Crit Rev Eukaryot Gene Expr. 2012;22:17–35. doi: 10.1615/CritRevEukarGeneExpr.v22.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang D, Kong X, Sang N. Effects of histone deacetylase inhibitors on HIF-1. Cell Cycle. 2006;5:2430–5. doi: 10.4161/cc.5.21.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sang N, Fang J, Srinivas V, Leshchinsky I, Caro J. Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1 alpha is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol Cell Biol. 2002;22:2984–92. doi: 10.1128/MCB.22.9.2984-2992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Bayley JP, Devilee P. The Warburg effect in 2012. Curr Opin Oncol. 2012;24:62–7. doi: 10.1097/CCO.0b013e32834deb9e. [DOI] [PubMed] [Google Scholar]
- 8.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–4. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 9.Huang YC, Chang WL, Huang SF, Lin CY, Lin HC, Chang TC. Pachymic acid stimulates glucose uptake through enhanced GLUT4 expression and translocation. Eur J Pharmacol. 2010;648:39–49. doi: 10.1016/j.ejphar.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Welsh GI, Hers I, Berwick DC, Dell G, Wherlock M, Birkin R, et al. Role of protein kinase B in insulin-regulated glucose uptake. Biochem Soc Trans. 2005;33:346–9. doi: 10.1042/BST0330346. [DOI] [PubMed] [Google Scholar]
- 11.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 12.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A, Fang MD, Song WQ, Chen CB, Qi LW, Wang CG. Gene expression profiles of two intraspecific Larix lines and their reciprocal hybrids. Mol Biol Rep. 2012;39:3773–84. doi: 10.1007/s11033-011-1154-y. [DOI] [PubMed] [Google Scholar]
- 14.Meng M, Chen S, Lao T, Liang D, Sang N. Nitrogen anabolism underlies the importance of glutaminolysis in proliferating cells. Cell Cycle. 2010;9:3921–32. doi: 10.4161/cc.9.19.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang CV. Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:369–74. doi: 10.1101/sqb.2011.76.011296. [DOI] [PubMed] [Google Scholar]
- 18.Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin Cancer Res. 2010;16:5928–35. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–33. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 20.Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, et al. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science. 2005;307:1776–8. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- 21.Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics. 2009;38:328–41. doi: 10.1152/physiolgenomics.90396.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–96. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–43. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan J, Lopez MC, Baker HV, Kilberg MS. Expression profiling after activation of the amino acid deprivation response in HepG2 human hepatoma cells. Physiol Genomics. 2010 doi: 10.1152/physiolgenomics.00217.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikalidis AK, Lee JI, Stipanuk MH. Gene expression and integrated stress response in HepG2/C3A cells cultured in amino acid deficient medium. Amino Acids. 2011;41:159–71. doi: 10.1007/s00726-010-0571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JI, Dominy JE, Jr., Sikalidis AK, Hirschberger LL, Wang W, Stipanuk MH. HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol Genomics. 2008;33:218–29. doi: 10.1152/physiolgenomics.00263.2007. [DOI] [PubMed] [Google Scholar]
- 28.Matsuyama A, Hisaoka M, Nagao Y, Hashimoto H. Aberrant PLAG1 expression in pleomorphic adenomas of the salivary gland: a molecular genetic and immunohistochemical study. Virchows Arch. 2011;458:583–92. doi: 10.1007/s00428-011-1063-4. [DOI] [PubMed] [Google Scholar]
- 29.Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 2011;25:1654–61. doi: 10.1101/gad.16800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon NS, Rong Zeng W, Premdas P, Santaguida M, Bérubé G, Nepveu A. Expression of N-terminally truncated isoforms of CDP/CUX is increased in human uterine leiomyomas. Int J Cancer. 2002;100:429–32. doi: 10.1002/ijc.10510. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Liu P, Cui X, Sui Y, Ji G, Guan R, et al. Identification of novel subregions of LOH in gastric cancer and analysis of the HIC1 and TOB1 tumor suppressor genes in these subregions. Mol Cells. 2011;32:47–55. doi: 10.1007/s10059-011-2316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer A, Gessler M. Hey genes in cardiovascular development. Trends Cardiovasc Med. 2003;13:221–6. doi: 10.1016/S1050-1738(03)00082-3. [DOI] [PubMed] [Google Scholar]
- 33.Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–7. doi: 10.1016/S0006-291X(02)03020-6. [DOI] [PubMed] [Google Scholar]
- 34.Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, et al. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol. 2004;11:738–46. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- 35.Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–56. doi: 10.1016/S0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 36.Kode A, Mosialou I, Silva BC, Joshi S, Ferron M, Rached MT, et al. FoxO1 protein cooperates with ATF4 protein in osteoblasts to control glucose homeostasis. J Biol Chem. 2012;287:8757–68. doi: 10.1074/jbc.M111.282897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meex SJ, van Greevenbroek MM, Ayoubi TA, Vlietinck R, van Vliet-Ostaptchouk JV, Hofker MH, et al. Activating transcription factor 6 polymorphisms and haplotypes are associated with impaired glucose homeostasis and type 2 diabetes in Dutch Caucasians. J Clin Endocrinol Metab. 2007;92:2720–5. doi: 10.1210/jc.2006-2280. [DOI] [PubMed] [Google Scholar]
- 38.Ye J, Koumenis C. ATF4, an ER stress and hypoxia-inducible transcription factor and its potential role in hypoxia tolerance and tumorigenesis. Curr Mol Med. 2009;9:411–6. doi: 10.2174/156652409788167096. [DOI] [PubMed] [Google Scholar]
- 39.Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, et al. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci USA. 2011;108:21069–74. doi: 10.1073/pnas.1117500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massagué J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–91. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 41.Lefkimmiatis K, Caratozzolo MF, Merlo P, D’Erchia AM, Navarro B, Levrero M, et al. p73 and p63 sustain cellular growth by transcriptional activation of cell cycle progression genes. Cancer Res. 2009;69:8563–71. doi: 10.1158/0008-5472.CAN-09-0259. [DOI] [PubMed] [Google Scholar]
- 42.Cigognini D, Corneo G, Fermo E, Zanella A, Tripputi P. HIC gene, a candidate suppressor gene within a minimal region of loss at 7q31.1 in myeloid neoplasms. Leuk Res. 2007;31:477–82. doi: 10.1016/j.leukres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.