Abstract
Background
Cathepsin L (CTSL1) catalyzes the formation of peptides that influence blood pressure (BP). Naturally occurring genetic variation or targeted ablation of the Ctsl1 locus in mice yield cardiovascular pathology. Here, we searched for genetic variation across the human CTSL1 locus and probed its functional effects, especially in the proximal promoter.
Methods and results
Systematic polymorphism discovery by re-sequencing across CTSL1 in 81 patients uncovered 38 genetic variants, five of which were relatively common (MAF >5%), creating a single linkage disequilibrium block in multiple biogeographic ancestries. One of these five common variants lay in a functional domain of the gene: promoter C-171A (rs3118869), which disrupts a predicted xenobiotic response element (XRE; match C>A). In transfected CTSL1 promoter/luciferase reporter plasmids, C-171A allele influenced transcription (C>A, P = 3.36E-6), and transcription was also augmented by co-exposure to the aryl hydrocarbon receptor (AHR) complex (AHR:ARNT) in the presence of their ligand dioxin (P = 6.81E-8); allele (C vs. A) and AHR:ARNT/dioxin stimulus interacted to control gene expression (interaction P = 0.033). Endogenous Ctsl1, Ahr, and Arnt transcripts were present in chromaffin cells. Promoter functional C-171A genotype also predicted hypertension (P = 1.0E–3), SBP (P = 4.0E–4), and DBP (P = 3.0E–3), in an additive pattern for diploid genotypes (A/A > C/A > C/C) in 868 patients, and the results were extended by validation analysis into an independent population sample of 986 patients.
Conclusion
We conclude that common genetic variation in the proximal CTSL1 promoter, especially at position C-171A, is functional in cells, and alters transcription so as to explain the association of CTSL1 with BP in vivo. At the XRE, endogenous genetic variation plus exogenous aryl hydrocarbon stimulation interact to control CTSL1 gene expression. These results unveil a novel control point whereby heredity and environment can intersect to control a complex trait, and point to new transcriptional strategies for intervention into transmitter biosynthesis and its cardiovascular consequences.
Keywords: autonomic, genetics, hypertension, nervous system
INTRODUCTION
The enzyme cathepsin L (CTSL, CTSL1; EC-3.4.22.15; OMIM 116880) is a cysteine/sulfhydryl protease. Widely expressed, it is found in tissues pertinent to cardiovascular control, including the nervous system, heart, endothelium, and adrenal glands. In previous studies, we found that chromaffin cell CTSL1 is localized to chromaffin granules, in which it cleaves pro-neuropeptide Y to form functional neuropeptide Y [1], and chromogranin A to generate the catecholamine release-inhibitory peptide catestatin [2]. Targeted ablation of the Ctsl1 locus in mice has profound cardiovascular consequences, including dilated cardiomyopathy [3]. Although naturally occurring genetic variation may inactivate the mouse Ctsl1 enzyme [4], little is known about genetic variation at the human CTSL1 locus.
In this study, we searched for naturally occurring common or rare genetic variation across the human CTSL1 locus. As one common variant (C-171A, rs3118869) occurred in a likely functional domain (promoter), we probed its mechanistic consequences, beginning with bioinformatic motif analysis and proceeding to transfected promoter/luciferase reporter plasmids, site-directed muta-genesis, and characterization of trans-acting factors. We developed evidence that variation in the CTSL1 promoter, especially at common variant C-171A, disrupts a particular motif, the xenobiotic response element (XRE), creating a differential gene-by-environment interaction, to alter transcriptional activity and ultimately blood pressure (BP) in humans.
METHODS
Genomics
Systematic polymorphism discovery by re-sequencing
Genomic DNA was prepared from leukocytes in EDTA-anticoagulated blood, using PureGene extraction columns (Gentra; Qiagen) as described previously [5]. Public draft human genome sequences were obtained from the UCSC Genome Bioinformatics website (http://genome.ucsc.edu) and used as a scaffold for primer design. The base position numbers were according to NCBI CTSL1 source clones: RefSeq gene/transcript NM_001912.4, contig/assembly NC_000009 (in GRCh37), and protein NP_001903.1. Promoter positions were numbered with upstream of (−) the CTSL1 exon 1 start (cap) site. The following polymerase chain reaction (PCR) primers were designed by Primer-3 [6] <http://frodo.wi.mit.edu/primer3/> to span approximately 1000 bp of the proximal promoter and between 600 and 1000 bp over each of the eight exons. Promoter region: forward 5′ tggggtaaaggcagaggtaa3′, Reverse 5′-cggttcgtggcttgtttact-3′, Exon 1: forward 5′-gctgaaacagtc cacacagg-3′, reverse 5′-actggctgtagcggtcagag-3′, exon 2: forward 5′-cctaaattgtcatcgcataaactg-3′, reverse 5′-ggaaga gaaaccatggcaag-3′, exon 3–4: forward 5′-cggcatggttagt gaaactt-3′, reverse 5′-agactttgacaacagctcacaca-3′, exon 5: forward 5′-cgcataaactgtttcagctcct-3′, reverse 5′-ggaaga gaaaccatggcaag-3′, exon 6: forward 5′-ggaagtaaaccca gaggtctca-3′, reverse 5′-aaatccgaaaagagccttttga-3′, exon 7: forward 5′-gcttaaatccctggccttct-3′, reverse 5′-cagcctc caaactgtgacct-3′, exon 8: forward 5′-ggaacattgcctgtcctgat-3′, reverse 5′-gcaaccactttgggtgaaat-3′.
Target sequences were amplified by PCR from 20 ng genomic DNA in a final volume of 25 μl, which also contained 0.1 U of Taq DNA polymerase (Applied Biosystems), 200 nmol/l of each dNTP, 300 nmol/l of each primer, 50 mmol/l KCl, and 2 mmol/l MgCl2. PCR was performed in an MJ PTC-225 thermal cycler, starting with 12 min of denaturation at 95°C, followed by 45 cycles at 95°C for 30 s, 63°C for 1 min (annealing), and 72°C (extension) for 1 min, and then a final extension of 8 min at 72°C. PCR products were treated with exonuclease I and shrimp alkaline phosphatase to remove primers and then dNTPs prior to cycle sequencing with BigDye terminators (Applied Biosystems). Sequence was determined on an ABI-3100 dideoxy capillary sequencer. Polymorphism and heterozygosity were visualized on the ABI tracings using Codon Code Aligner <http://www.codoncode.com/aligner> [23].
Single nucleotide polymorphism genotyping
Diploid genotypes were determined by two extension-based techniques, either the MALDI (matrix assisted laser desorption ionization) mass spectrometry method of Sequenom, or the luminescent base incorporation method of Pyrosequencing (Biotage), in which genotypes were verified by visual inspection, with exclusion of artifactual data from further analysis, or by the TaqMan primer/probe system on an ABI-7900HT device (Applied Biosystems). Reproducibility of diploid genotypes was verified with blinded replicate samples, indicating 98.8% concordance.
Functional studies of CTSL1 promoter genetic variation
CTSL1 promoter haplotype/luciferase reporter design and construction
Human CTSL1 promoter fragments, corresponding to CTSL1-1400/+33 bp in CTSL1 (NCBI CTSL1 source clones: RefSeq gene/transcript NM_001912.4, contig/assembly NC_000009 (in GRCh37), and protein NP_001903.1), were PCR-amplified from genomic DNA (after re-sequencing), and cloned into the promoter-less firefly luciferase reporter plasmid pGL3-Basic (Promega, Madison, Wisconsin, USA), as described [5]. Site-directed mutagenesis (QuikChange, Stratagene) created the required variant at position C-171A. Supercoiled plasmids were purified on columns (Qiagen, Valencia, California, USA) prior to transfection and verified by sequencing. Promoter positions are numbered upstream (−) of the transcriptional start (cap) site.
Luciferase reporter assays of CTSL1 promoter C-171A variants
PC12 rat pheochromocytoma cells were transfected (at 50–60% confluence, 1 day after 1: 4 splitting) with 500 ng of supercoiled promoter/firefly luciferase reporter plasmid and 10 ng of the Renilla luciferase expression plasmid pRL-TK (Promega Inc.) as an internal control per well, by the liposome method (Superfect; Qiagen). The firefly and Renilla luciferase activities in cell lysates were measured 16–24 h after transfection, using the Dual Luciferase reporter assay system (Promega), and the results were expressed as either the ratio of firefly/Renilla luciferase activity, or firefly activity/total protein in the lysate, as described [5]. Each experiment included six replicates. Results were expressed as mean ± SEM. Statistical significance was calculated using Student t-test or ANOVA, and significance was established at the P <0.05 level. Inspection of the NCBI GEO (Gene Expression Omnibus) database <http://www.ncbi.nlm.nih.gov/geo/> indicates that transcripts for CTSL1 are abundantly expressed in the adrenal gland (GEO dataset GDS1464) as well as PC12 chromaffin cells (GDS2555).
Exogenous/cotransfected factors
Eukaryotic expression plasmids containing cDNAs encoding transcription factors aryl hydrocarbon receptor (AHR) and aryl hydrocarbon nuclear translocator (ARNT) (human) were from OriGene Technologies, Inc. (Rockville, Maryland, USA), or Open Biosystems (Huntsville, Alabama, USA), respectively. cDNAs were either obtained in pCMV expression plasmids, or subcloned into pcDNA-3.1 (Invitrogen, Carlsbad, California, USA). 2,3,7,8-Tetrachlorodi-benzo-p-dioxin (TCDD, ‘dioxin’; 10 μg/ml in toluene) was purchased from Sigma–Aldrich (SIGMA #48599; CAS #1746–01–6; SUPELCO Analytical, Bellefonte, Pennsylvania, USA).
Trans-activation: cotransfection of transcription factors with CTSL1 promoter haplotype/reporters
50 ng of each transcription factor plasmid, or 50 or 100 ng pcDNA-3.1 empty vector (control), were co-transfected into PC12 cells, along with 500 ng of CTSL1 promoter/luciferase reporter variants. After 24 h, cells were lysed and luciferase activities were assayed as described above and normalized by total protein. Reaction of the CTSL1 promoter to trans-activation was expressed as fold-change of the mean value of luciferase activity between the transcription factor-transfected group and the mock-transfected (empty vector, pcDNA-3.1) group. Dioxin (2,3,7,8-tetrachlorodibenzo-p-dioxin, or 2,3,7,8-TCDD, also referred to as TCDD) was used at 5 nmol/l final concentration.
Endogenous gene expression
Total RNA was extracted from PC12 cells, and real-time PCR was done with fluorescent reporter-tagged oligonucleotide primers on an ABI-7700 TaqMan platform (Life Technologies, Carlsbad, California, USA), with normalization of data to beta-actin expression. Ct (threshold cycle) is determined for both the specific target mRNA/cDNA as well as beta-actin, and the difference in Ct (from target mRNA versus beta-actin mRNA) is normalized to the average for that state (e.g., control versus experimental), and expressed on a % change (difference) scale. The negative control probes were targeted to human elastin, with a sequence that does not occur in the rat isoform.
Human patients
Polymorphism discovery and biogeographic ancestry
Initially, a series of n = 81 individuals (2n = 162 chromosomes) was studied by resequencing of CTSL1 for systematic polymorphism discovery in four groups collected at University of California at San deigo (UCSD), selected for this purpose solely on the basis of biogeographic ancestry: n = 25 individuals of European ancestry (white), n = 24 patients of sub-Saharan African ancestry (black, African American), n = 16 individuals of east Asian ancestry (China, Japan, Korea, south-east Asia, Phillipines), and n = 16 patients of Hispanic (Mexican American) ancestry. Ethnicity was established by self-identification by the participants as well as their parents and grandparents. None of the patients had a history of renal failure. Definitions of patient characteristics are according to previous reports from our laboratory [5]. Patients were volunteers from southern California, and each patient gave informed, written consent; the protocol was approved by the UCSD Human Research Protection Program.
Blood pressure: trait extremes
We first studied 868 white (European biogeographic ancestry, by self identification) patients, selected from more than 53 000 individuals within a large adult primary care (Kaiser Permanente) population in San Diego, as previously described [7]. In this primary care population, ~81% attended the clinic, and ~46% consented to participation in the study, with collection of blood for preparation of genomic DNA. From consented participants, the patients in this study were selected, based upon measurement of DBP, to represent the highest and lowest ~5th DBP percentiles in that population. A second sampling of n = 986 different individuals from BP extremes of the same source population allowed for replication. The statistical power of association between bi-allelic DNA markers and human quantitative trait loci can be substantially augmented by the sampling individuals from opposite (upper and lower) ends of the trait distribution [8], and analyses of the quantitative trait in extreme patients (as opposed to dichotomization of the trait) further enhances power [7,9]. This population sample afforded us more than 90% power to detect genotype association with a trait when the genotype contributes as little as 2.5% to the total variation in males; the power is even higher in the women. To accomplish this, lower BP individuals were selected from the bottom 5th percentiles of DBP, whereas the higher BP group was selected from the top 5th percentiles of DBP. Both SBP and DBP differed significantly between the BP extreme groups (P <0.001). ~57% of patients in the higher BP samples were taking one or more antihypertensive medications (including diuretics and angiotensin-converting enzyme (ACE) inhibitors), whereas none in the lower BP group were on such treatment. In subsequent analyses, BP results were also adjusted for the effects of antihypertensive treatment, by adding a fixed value (10/5 mmHg) to each treated BP, as described [10]; although such adjustments are necessarily imperfect, their value is reinforced by restoration of familial (sibling/sibling) BP correlations [10]. Further BP/single nucleotide polymorphism (SNP) association studies were validated in an independent sample of 986 different individuals from the same primary healthcare provider, using the same recruitment criteria as the cohort described above.
Neuropeptide Y secretion
Since NPY may be cleaved from its precursor pro-NPY by CTSL1 [1], we also measured NPY in plasma of 417 individuals of white (European or Hispanic) ancestry, from 242 nuclear families ascertained at UCSD were subjected to genome-wide genotyping with the Illumina-610-Quad array, encompassing ~590K SNP genotypes, after which an additional ~2 million Hap-Map-2 SNPs were imputed with MACH v. 1.0.16 (http://www.sph.umich.edu/csg/abecasis/MaCH/). SNP-on-phenotype effects with tested in MERLIN v1.1.2 (http://www.sph.umich.edu/csg/abecasis/merlin/) in order to explicitly account for family structure. Plasma NPY was measured by RIA based on synthetic human [125I]-NPY1–36-amide, as previously described [11] with reagents from Eurodiagnostica (Malmo, Sweden).
Statistics and informatics
Clinical trait genetic analyses
Statistical analyses were carried out with SPSS-11.5 (Chicago, Illinois, USA). SNP marker-on-trait analyses for continuous traits (e.g., SBP in mmHg) were performed by two-way ANOVA (independent variable: diploid genotype; dependent variable: BP trait), adjusting for age and sex. Dichotomous variables (BP status) were analyzed by proportions and chi-square test. For BP analyses, all subjects were of the same biogeographic ancestry (European), by self-identification. To validate genetic association results from the initial cohort into a second cohort, meta-analysis was carried out with the command META, testing fixed effect (i.e., genotype) models in STATA-12 (College Station, Texas, USA), after separate study regression analysis in SPSS, incorporating separate study genotype and phenotype (BP; age and sex as covariates) data to derive significance as well as pooled genotype effect size (beta, or slope per allele) and its SE.
Promoter and cellular analyses
Parametric, general linear model (typically, two-way ANOVA) analyses were used to test the associations of promoter variants and luciferase reporter activity for each construct. Interaction of promoter variant C-171A with stimuli (e.g., AHR:ARNT complex) was carried out in the univariate tests of the general linear model, wherein there was a single dependent variable (e.g., luciferase activity), and the interactive (multiplicative) effects of the two independent variables (e.g., SNP and trans-activator) could be modeled. Each factor (independent variable, or the interaction between two independent variables) was assessed for significance using F-tests, with significance defined as P <0.05. Effects of co-transfected transcription factors were also analyzed by both two-tailed t-test and univariate tests of the general linear model.
Linkage disequilibrium
Patterns of LD were analyzed and visualized by the software Haploview [12]. LD blocks were derived by the 4 gamete criterion and visualized as standard D′/logarithm (base 10) of odds (D′/LOD score) plot in Haploview, from unphased diploid genotypes of subjects from four diverse biogeographic ancestry groups systematically sequenced across the CTSL1 locus: white (European ancestry, 2n = 50 chromosomes), black (sub-Saharan African ancestry, 2n = 48 chromosomes), Hispanic (Mexican American, 2n = 26 chromosomes), and east Asian (2n = 36 chromosomes). Common variants (minor allele frequency >5%) were used to establish linkage disequilibrium.
Bioinformatics: computational prediction of transcription factor binding motifs overlying CTSL1 promoter common variants
Multiple sequence alignments were performed by Clustal-W [13] <http://www.ebi.ac.uk/Tools/clustalw2/>. Potential transcription factor binding motifs at C-171A were predicted from the TRANSFAC position weight matrix database, beginning with the web-based methods at Chip-Mapper <http://bio.chip.org/mapper> and JASPAR [14] <http://jaspar.genereg.net/>. We also studied potential motifs with Mac-Vector 9.0.2. <http://www.macvector.com/>, separately evaluating each allele of bi-allelic variants.
RESULTS
Systematic polymorphism discovery across CTSL1
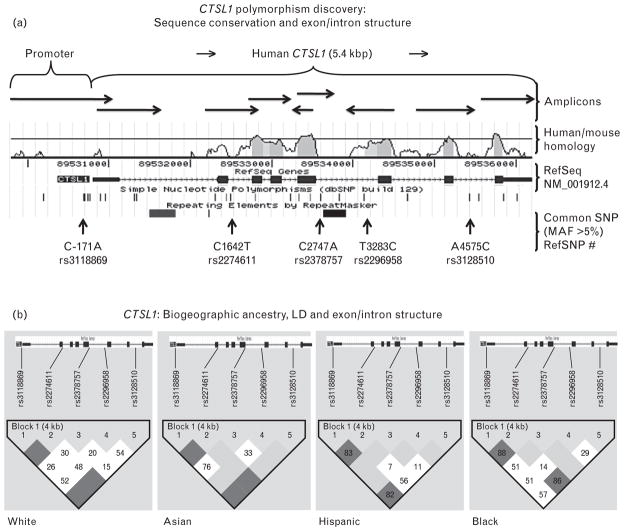
Located on chromosome 9q21-q22, CTSL1 spans eight exons (seven coding) and seven introns. We re-sequenced each exon, adjacent intron/exon borders, ~1000 bp of the proximal promoter, and any other regions that displayed substantial homology across mammalian species (Fig. 1a) in 2n = 162 chromosomes derived from four biogeographic ancestry groups (supplemental Table 1, http://link-s.lww.com/HJH/A188). We identified 38 variants (37 SNPs, 1 Ins/Del) in individuals of diverse biogeographic ancestries (supplemental Table 2, http://links.lww.com/HJH/A188). Of these SNPs, 33 are rare (MAF <5%, supplemental Table 2, http://links.lww.com/HJH/A188), including four in the open reading frame within coding exon 5 (all synonymous): Gln134Gln, Cys135Cys, Gly136Gly, and Gly149Gly. Of the five common (MAF >5%) SNPs, one is located in the proximal promoter (C-171A), and the rest are intronic (Fig. 1a); information on common variant frequencies in different biogeographic ancestry groups is provided in supplemental Table 3, http://links.lww.com/HJH/A188. Our results are consistent with CTSL1 polymorphism data for common SNPs in public databases such as dbSNP <http://www.ncbi.nlm.nih.gov/SNP>, though we provide new information on less common variants at the locus, as well as more precise estimates of allele frequencies in subjects of multiple biogeographic ancestries.
FIGURE 1.
Human CTSL1 genomics. (a) Systematic polymorphism discovery at the human protease cathepsin L (CTSL1) locus: Exon/intron structure, amplicons and common (MAF >5%) single nucleotide polymorphisms (SNPs) identified in 81 individuals (2n = 162 chromosomes). Horizontal arrows indicate amplicon pointing in the direction of sequencing. The blue blocks of the reference gene indicate exons; lines between the blocks indicate introns. Shown above the reference gene, is the sequence conservation between human and mouse Ctsl1. RefSNP numbers refer to common SNPs discovered at UCSD and the vertical arrows show the position of the SNPs with reference to the gene. (b) Human protease cathepsin L (CTSL1): patterns of linkage disequilibrium (LD) across the locus in four biogeographic ancestry groups (Asian, n = 16, white, n = 25, black, n = 24, and Hispanic, n = 16). Linkage disequilibrium values are shown as D′. Along the top of the graphs are markers (common minor alleles) in order along the CTSL1 gene. Linkage disequilibrium color scheme is represented as standard D′/LOD; the darker scale level represents higher D′ values between pairs of markers. CTSL1 exon/intron structure is shown above each group. Plots were created in Haploview, using the 4-gamete criterion.
Biogeographic ancestry and linkage disequilibrium
CTSL1 common allele frequencies did not differ across the 4 biogeographic ancestry groups (supplemental Table 3, http://links.lww.com/HJH/A188). To visualize patterns of marker-on-marker association, pair-wise LD correlations among the five common (MAF >5%) SNPs were quantified by the 4-gamete rule (4th gamete observed at frequency >0.124) across the CTSL1 locus. In each biogeographic ancestry group, a single block of LD across the ~5.4 kbp locus was maintained (Fig. 1b).
Genetic variation in the proximal human CTSL1 promoter: motifs
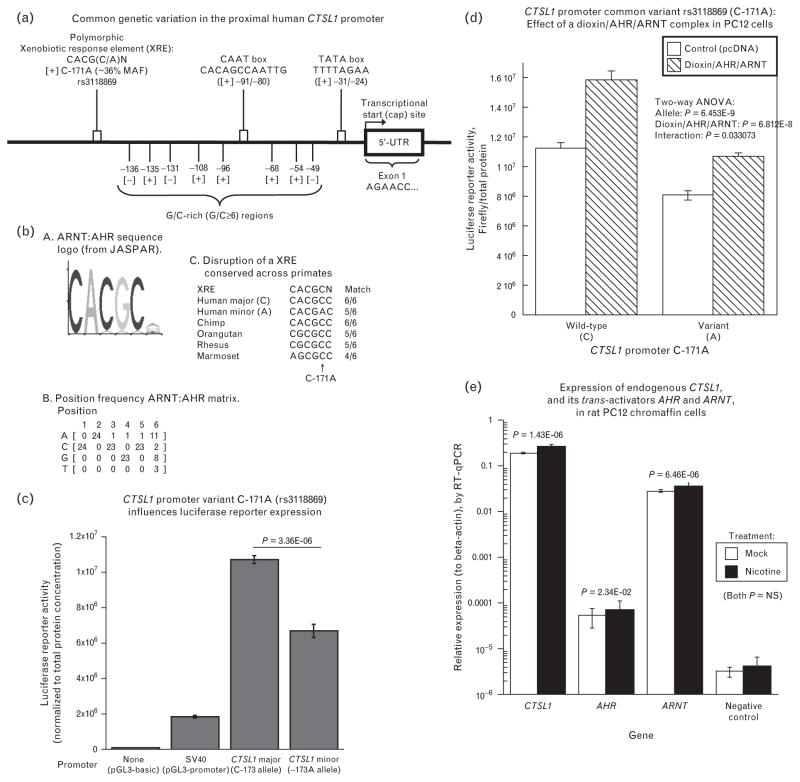
Motifs identified included a TATA box (TTTTAGAA, −31/−24), and CAAT box (CACAGCCAATTG, −91/−80). Eight G/C-rich (consecutive G/C ≥6 bp) regions were noted in the proximal promoter, from positions −49 to −136 bp (Fig. 2a). We identified nine polymorphisms in the promoter (supplemental Table 2, http://links.lww.com/HJH/A188), one of which was common (at MAF >5%): C-171A (rs3118869, MAF ~25%). Of note, the very proximal ‘core’ promoter (−91/−1 bp) was devoid of common variation. At promoter variant C-171A, we identified a motif likely to be disrupted by the sequence change: a xenobiotic response element (or XRE).
FIGURE 2.
Human CTSL1 promoter functional genetic variation. (a) Schema of the proximal human CTSL1 promoter region showing major regulatory elements. Shown from right to left on the schematic are a Cap site, TATA box, CAAT box, and xenobiotic response element (XRE) spanning common promoter variant C-171A. In brackets are the positions of the regulatory elements on the gene. The sequence of the xenobiotic response element is shown spanning the C-171A polymorphism. (C/A): Polymorphic base []: Strand (+ or −) MAF: Minor allele frequency. Based on Biochem. J. 2002; 361: 173–184. (b) Human CTSL1 common promoter polymorphism C-171A (rs3118869): Disruption of a potential xenobiotic response element (XRE). a. Aryl hydrocarbon nuclear translocator (ARNT): aryl hydrocarbon receptor (AHR) sequence logo (from JASPAR). Graph indicates relative likelihood of a specific allele at a given position. AHR: Aryl Hydrocarbon Receptor; ARNT: ARyl hydrocarbon receptor Nuclear Translocator. The polymorphic C/A site is at position 5. b. Position frequency ARNT: AHR matrix, indicating the probability of finding a particular allele at a specific location in the consensus sequence of a xenobiotic response element. c. Disruption of a XRE conserved across primates. In bold is the polymorphic base (C/A). N indicates any base (IUPAC code). (c) Human CTSL1 common promoter polymorphism C-171A (rs3118869): Basal; activity of promoter variants. PC12 cells were transfected with either ~1.4 kbp human CTSL1 promoter/luciferase reporter plasmids (C-171 vs. C-171A alleles), or a negative control plasmid (pGL3-Basic) without a eukaryotic promoter, or a positive control (pGL3-promoter, with luciferase expression driven by the SV40 early promoter), and then incubated for 24 h at 37°C with 6% CO2. Luciferase reporter activity, normalized to total protein, is expressed as the average of 5–6 replications per condition. (d) Interactive effect of CTSL1 promoter variant and the aryl hydrocarbon receptor (dioxin/AHR/ARNT) complex on human CTSL1 transcriptional activity in PC12 chromaffin cells. PC12 cells were cotransfected with ~1.4 kbp human CTSL1 promoter/luciferase reporter, pcDNA (empty pCMV vector control), or with AHR and ARNT (each driven by a pCMV promoter), and incubated with or without dioxin (5 nM final concentration), for 24 h at 37°C with 6% CO2. Luciferase reporter activity, normalized to total protein, is expressed as the average of six replicaties each for wild-type and mutant. (e) Quantification of endogenous transcripts by reverse transcriptase-PCR in rat PC12 chromaffin cells, for Ctsl1 expression and its trans-acting factors Ahr and Arnt. Transcripts were quantified in cells exposed for 24 h to either 100 μmol/l nicotine (activator of the physiological nicotinic cholinergic secretory pathway), or mock stimulation. Results were normalized to expression of beta-actin. In these rat transcript studies, the negative control primers were directed to human elastin. Experiments were performed with five replicate cell plates.
CTSL1 C-171A promoter polymorphism: xenobiotic response element sequence conservation/alignment
C-171A is located in a region highly conserved across sequenced primates (supplemental Table 4, http://links.lww.com/HJH/A188), with the C/C allele ancestral in the human lineage, as judged by the chimp sequence (Fig. 2b and c). In this conserved local region (Fig. 3c), there is a partial consensus match for a XRE site (CACGCN; position −171 in bold) with a better match for the C allele (6/6 bp match) than the A allele (5/6 bp match) (Fig. 2b).
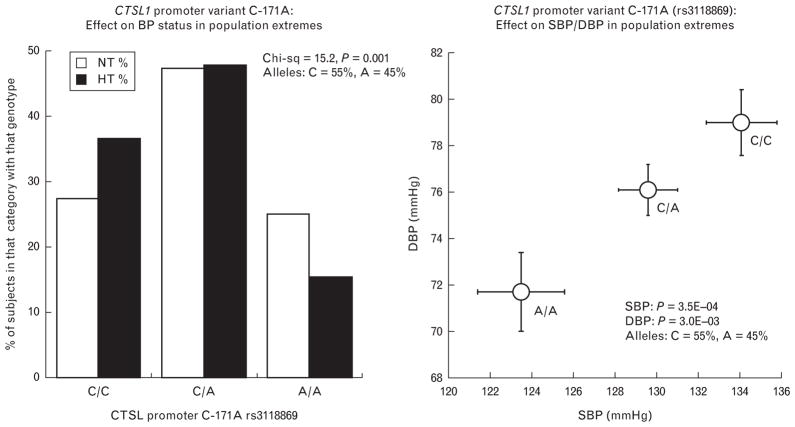
FIGURE 3.
Human CTSL1 promoter genetic variation and blood pressure (BP). (a) Effect of CTSL1 promoter variant C-171A on BP status (dichotomous trait) in population trait extremes (KWE1 cohort) from Southern California. On the X-axis are the major/major (C/C), major/minor (C/A) and the minor/minor (A/A) allele diploid genotypes. The Y-axis represents the percentage of patients in each allele type category, with the hypertensive (HT) category shaded in black, and normotensive (NT) in white. Analysis was by chi-square test. (b) Association of CTSL1 promoter C-171A variant with BP quantitative traits (mmHg) in population trait extremes (KWE1 cohort) from Southern California. On the X-axis is SBP, whereas on the Y-axis is DBP. Analyses were by univariate ANOVA, with age and sex as covariates.
Transfected CTSL1 promoter: basal expression and effect of exogenous (cotransfected) aryl hydrocarbon receptor:aryl hydrocarbon nuclear translocator complex
During CTSL1 promoter/luciferase reporter transfections into chromaffin cells (Fig. 2c), the C allele displayed greater expression than the A allele (C>A, P = 3.36E–6).
Co-transfection of subunits of the human AHR:ARNT heterodimer, plus exposure to the ligand dioxin, revealed increased activation by the trans-acting complex (P = 6.812E–8), wherein the C allele also exhibited an even greater increment after AHR:ARNT/dioxin stimulation (interaction P = 0.033 on ANOVA).
Endogenous gene expression
By quantitative PCR, rat PC12 chromaffin cells expressed mRNAs encoding not only the target gene Ctsl1, but also the two transcription factors recognizing the XRE motif: Arnt and Ahr (Fig. 2e). The abundance of these transcripts was not affected by activation of the physiological secretory pathway of chromaffin cells with the nicotinic cholinergic agonist nicotine).
Human CTSL1 genetic variation and blood pressure
High BP may result from both genetic and environmental factors. To determine whether genetic variation at the CTSL1 gene is associated with hypertension, we genotyped the high-frequency SNP discovered in a likely functional domain of the gene: the proximal promoter at C-171A (rs3118869). In a sample of 868 Caucasian individuals with the most extreme BP values in a large primary care population, we found a significant association of C-171A with the dichotomous BP status trait (P = 1.0E–3, Fig. 3a): the diploid genotype distribution diagram indicates that C/C homozygotes are more likely to be hypertensive, whereas A/A homozygotes are more likely to be normotensive. We then found significant associations of promoter C-171A with the quantitative BP traits: both systolic (P = 3.5E–4) and DBP (P = 3.0E–3, Fig. 3b). These BP associations also appeared to be additive, with separation of the effects of all three genotype classes (A/A <A/C <C/C) on both BP traits. Sample characteristics are given in supplemental Table 5, http://links.lww.com/HJH/A188. Extension of the study into a second, independent sample of 986 additional individuals from the population was performed to validate the initial association results (supplemental Table 6, http://links.lww.com/HJH/A188): the effects of the gene were directionally coordinate (by beta, or regression slope) in each population sample, and the results remained significant for both SBP/DBP (P = 0.001/P = 0.012). Effect size was available as an output of this analysis, in the form of the slope (or beta) for effect of the minor (C) allele for CTSL1 promoter variant C-171A (rs3118869) on BP in additive regression models (supplemental Table 6, http://link-s.lww.com/HJH/A188): The additive, per-allele effect on SBP was −2.60 ± 0.78 mmHg (that is, a decline in SBP of ~2.6 mmHg for every copy of the minor (A) allele), with an effect size on DBP of −1.48 ± 0.59 mmHg (that is, a decline in DBP of ~1.5 mmHg for every copy of the minor (A) allele). In these patients, diploid genotype frequencies for promoter variant C-171A (rs3118869) did not deviate from Hardy–Weinberg equilibrium for the entire population sample (P = 0.374), as well as sample-1 (P = 0.847) or sample-2 (P = 0.375).
Finally, as CTSL1 may cleave pro-neuropeptide Y (pro-NPY) to yield NPY [1], we studied the effect of CTSL1 genetic variation on a more proximate trait: secretion of NPY. CTSL1 was typed at tagging variant C1642T (rs2274611) in Intron-2 (Fig. 1a). CTSL1 genotype had a significant (P = 0.043) effect on plasma NPY: major allele homozygotes (C/C) displayed lower NPY (71.2 ± 2.6 pM, n = 125) than either heterozygotes (C/T, 80.2 ± 2.3, n = 203) or minor allele homozygotes (T/T, 80.2 ± 3.0, n = 89).
DISCUSSION
Overview
Human protease CTSL1 represents a control point for generation of biologically active peptides that exert circulatory control, including neuropeptide Y [1] and catestatin [2]. In this study, we explored whether and how common genetic variation at CTSL1 influences expression of the gene and ultimately cardiovascular traits. We present evidence from several approaches (genomic, bioinformatic, transfection) in which we found that promoter variant C-171A conferred functional changes upon the CTSL1 expression, and that particular transcription factors were implicated. In addition we present evidence of previously unexpected gene-by-environment interaction in the regulation of CTSL1.
CTSL1 promoter variant C-171A: blood pressure and xenobiotic response element
Hypertension, the most potent risk factor for cardiovascular disease, [15] is a complex trait determined by interplay between gene and environment. Already more than ~25% of adults in industrialized countries exhibit hypertension, and numbers in the developing world are on the rise.
Here, we describe a novel contributor to BP regulation: genetic variation at the CTSL1 locus. Since one common CTSL1 variant (C-171A) lay in a likely functional domain, the proximal promoter (Fig. 1a, 2a), we typed this variant in hypertension, and noted an effect on BP (Fig. 3a, b), with the C allele associated with higher BP. During tests of the transfected CTSL1 promoter, we noted increased activity of the C allele on reporter expression (Fig. 2c); the results are compatible with increased expression of CTSL1 augmenting formation of pressor peptide, such as NPY. Indeed, we found that genetic variation at CTSL1 influenced NPY secretion.
C-171A is located in a XRE region (Fig. 2a, b), with the C (major/ancestral) allele enhancing the binding motif for the AHR:ARNT heterodimeric transcription factor complex (Fig. 2b) [16]. In PC12 cells exposed to dioxin in the presence of co-transfected AHR:ARNT, the CTSL1 promoter was activated (Fig. 2d), with an enhanced response by the C allele.
Of note for the physiological significance of these results, query of the NCBI GEO (Gene Expression Omnibus) database <http://www.ncbi.nlm.nih.gov/geo/> indicates that transcripts for both AHR (aryl hydrocarbon receptor) and ARNT (aryl hydrocarbon receptor nuclear translocator Arnt1), which are basic helix-loop-helix (bHLH)/PER-ARNT-SIM (PAS) family transcription factors, are expressed endogenously in PC12 chromaffin cells, by inspection of the following GEO transcript datasets: GDS2255 [17], GDS1234 [18], GDS1236 [19], or GDS1038 [20]. Indeed, we experimentally detected expression of transcripts for not only Ctsl1 but also both transcription factors (Ahr and Arnt) in PC12 cells (Fig. 2e).
Results in context with the literature: Gene-by-environment interaction
In this report, we present cellular evidence that genetic variation at the CTSL1 locus and an environmental factor (aryl-hydrocarbon) interact to influence gene expression (Fig. 2d). As we also present evidence that CTSL1 polymorphism influences BP (Fig. 3a, b), the results raise the likelihood of a gene-by-environment interaction to regulate BP in the population.
Other reports, both human and experimental, suggest a potential role for the aryl hydrocarbon system in hypertension susceptibility. Indeed, a recent Institute of Medicine report provides epidemiological evidence that exposure to dioxin, both an environmental pollutant and a component of herbicides, may be associated with hypertension [21].
Lund et al. [22] investigated the influence of AHR on BP, using AHR-knockout mice studied at different altitudes, documenting hypoxia-induced systemic hypertension. In the genetically hypertensive rat (SHR), functional variation in the promoter of brain and muscle ARNT-like protein-1 (BMAL1) occurs within a hypertension susceptibility locus, and a subsequent human study found association of BMAL1 haplotypes with BP [23].
Finally, preliminary evidence suggests that a polymorphism in human AHR may influence not only endothelium-dependent vasodilation but also resting BP [24].
Advantages and limitations of this study
We began with systematic polymorphism discovery across the CTSL1 locus in multiple biogeographic ancestry groups (Fig. 1a, b). Computational (Fig. 2b) as well as experimental (Fig. 2d) approaches confirmed that polymorphic disruption of a CTSL1 promoter XRE yielded changes in gene expression, suggesting a novel gene-by-environment interaction, perhaps pertinent to control of BP. We then proceeded to find CTSL1 genetic effects on BP in a sample of individuals selected from the most extreme BP values in the population (Fig. 3a, b); such a sampling strategy maximizes statistical power [7–9], and allows generalization to a larger population. Then extension (supplemental Table 6, http://links.lww.com/HJH/A188) into an independent sample drawn from the same source population confirmed the finding. However, several issues remain unexplored by our studies. We studied the effects of CTSL1 genetic variation on BP in only one population (Fig. 3), and do not know if other human ethnic groups might be susceptible to such variation; promoter C-171A minor allele frequencies of ~7–36% were found across multiple biogeographic ancestries (supplemental Table 2, 3, http://links.lww.com/HJH/A188). We do not yet understand the full spectrum of substrates and products of CTSL1 digestion that might be most important for control of physiological processes such as BP. Finally, the gene-by-environment interaction that we observed in transfected cells (Fig. 2) has not yet been established in vivo, nor do we know precisely which environmental aryl hydrocarbons might participate.
For most (if not all) complex traits, genetic association studies to date have failed to explain a substantial fraction of even the heritable portion of trait variance. For example, BP in twin and family studies typically displays ~40–50% heritability, in which heritability is the fraction of trait variance accounted for by genetic variance; yet the SNPs so far associated with BP in even the largest scale Genome Wide Association Study (GWAS) collaborations account for only a small % of BP variance [25], an issue that has been referred to as the ‘missing heritability problem’; at least some of this deficit may be referable to the stringency of p-values required in the face of genome-wide multiple statistical testing. Indeed, in the most recent large scale meta-analysis published for BP by International Consortium for BP-GWAS (ICBP-GWAS) [25], up to 29 highly significant hypertension susceptibility loci were verified in a multistage study of ~200 000 individuals of European descent, but such loci accounted for only ~0.9% of population BP variance. Although CTSL1 was not discovered by these GWAS methods, the ICBP-GWAS group went on to estimate that an additional ~57–174 such blood pressure risk variants are likely to exist [25]; CTSL1 may be one such gene.
Conclusion and perspectives
We conclude that CTSL1 promoter common polymorphism C-171A is responsible for changes in not only in transcriptional efficiency of the CTSL1 gene, but also BP in the population. This conclusion arises from a convergence of computational and experimental approaches. The effects of C-171A seem to arise from differential actions of specific transcription factors at the CTSL1 promoter: the AHR:ARNT heterodimeric complex acting at an XRE disrupted by C-171A bi-allelic variation. The results raise the potential for novel gene-by-environment interactions in control of BP, thus augmenting our understanding of molecular events underlying inter-individual variations in BP, and the genetic predisposition hypertension, a potent risk factor for cardiovascular disease.
Supplementary Material
Acknowledgments
Support: National Institutes of Health [HL58120; UL1RR031980 (UCSD Clinical and Translational Research Institute); MD000220 (UCSD Comprehensive Research Center in Health Disparities, CRCHD)], Department of Veterans Affairs.
Abbreviations
- AHR
aryl hydrocarbon receptor
- ARNT
aryl hydrocarbon nuclear translocator
- BP
blood pressure
- CTSL1
cathepsin L1
- LD
linkage disequilibrium
- SNP
single nucleotide polymorphism
- XRE
xenobiotic response element
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Funkelstein L, Toneff T, Hwang SR, Reinheckel T, Peters C, Hook V. Cathepsin L participates in the production of neuropeptide Y in secretory vesicles, demonstrated by protease gene knockout and expression. J Neurochem. 2008;106:384–391. doi: 10.1111/j.1471-4159.2008.05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas N, Rodriguez-Flores JL, Courel M, Gayen JR, Vaingankar SM, Mahata M, et al. Cathepsin L colocalizes with chromogranin a in chromaffin vesicles to generate active peptides. Endocrinology. 2009;150:3547–3557. doi: 10.1210/en.2008-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stypmann J, Glaser K, Roth W, Tobin DJ, Petermann I, Matthias R, et al. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc Natl Acad Sci U S A. 2002;99:6234–6239. doi: 10.1073/pnas.092637699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth W, Deussing J, Botchkarev VA, Pauly-Evers M, Saftig P, Hafner A, et al. Cathepsin L deficiency as molecular defect of furless: Hyper-proliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 2000;14:2075–2086. doi: 10.1096/fj.99-0970com. [DOI] [PubMed] [Google Scholar]
- 5.Wen G, Mahata SK, Cadman P, Mahata M, Ghosh S, Mahapatra NR, et al. Both rare and common polymorphisms contribute functional variation at CHGA, a regulator of catecholamine physiology. Am J Hum Genet. 2004;74:197–207. doi: 10.1086/381399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozen S, Skaletsky H. Primer3 on the www for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 7.Rana BK, Insel PA, Payne SH, Abel K, Beutler E, Ziegler MG, et al. Population-based sample reveals gene-gender interactions in blood pressure in white Americans. Hypertension. 2007;49:96–106. doi: 10.1161/01.HYP.0000252029.35106.67. [DOI] [PubMed] [Google Scholar]
- 8.Schork NJ, Nath SK, Fallin D, Chakravarti A. Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am J Hum Genet. 2000;67:1208–1218. doi: 10.1086/321201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenesa A, Visscher PM, Carothers AD, Knott SA. Mapping quantitative trait loci using linkage disequilibrium: Marker- versus trait-based methods. Behav Genet. 2005;35:219–228. doi: 10.1007/s10519-004-0811-5. [DOI] [PubMed] [Google Scholar]
- 10.Cui JS, Hopper JL, Harrap SB. Antihypertensive treatments obscure familial contributions to blood pressure variation. Hypertension. 2003;41:207–210. doi: 10.1161/01.hyp.0000044938.94050.e3. [DOI] [PubMed] [Google Scholar]
- 11.Jideus L, Ericson M, Stridsberg M, Nilsson L, Blomstrom P, Blomstrom-Lundqvist C. Diminished circadian variation in heart rate variability before surgery in patients developing postoperative atrial fibrillation. Scand Cardiovasc J. 2001;35:238–244. doi: 10.1080/14017430152581341. [DOI] [PubMed] [Google Scholar]
- 12.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 15.D’Agostino RBJ, Sr, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB. General cardiovascular risk profile for use in primary care: the Framingham heart study. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 16.Probst MR, Reisz-Porszasz S, Agbunag RV, Ong MS, Hankinson O. Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol Pharmacol. 1993;44:511–518. [PubMed] [Google Scholar]
- 17.Lattanzi W, Bernardini C, Gangitano C, Michetti F. Hypoxia-like transcriptional activation in tmt-induced degeneration: Microarray expression analysis on PC12 cells. J Neurochem. 2007;100:1688–1702. doi: 10.1111/j.1471-4159.2006.04331.x. [DOI] [PubMed] [Google Scholar]
- 18.Nowroozi N, Raffioni S, Wang T, Apostol BL, Bradshaw RA, Thompson LM. Sustained ERK1/2 but not STAT1 or 3 activation is required for thanatophoric dysplasia phenotypes in PC12 cells. Hum Mol Genet. 2005;14:1529–1538. doi: 10.1093/hmg/ddi161. [DOI] [PubMed] [Google Scholar]
- 19.Apostol BL, Illes K, Pallos J, Bodai L, Wu J, Strand A, et al. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum Mol Genet. 2006;15:273–285. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- 20.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 21.Institute of Medicine. Veterans and agent orange: Update 2006. Institute of Medicine. 2007 [Google Scholar]
- 22.Lund AK, Agbor LN, Zhang N, Baker A, Zhao H, Fink GD, et al. Loss of the aryl hydrocarbon receptor induces hypoxemia, endothelin-1, and systemic hypertension at modest altitude. Hypertension. 2008;51:803–809. doi: 10.1161/HYPERTENSIONAHA.107.100586. [DOI] [PubMed] [Google Scholar]
- 23.Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, Farrall M, Gauguier D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A. 2007;104:14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pia Monica Lind EI, Syvänen Ann-Christine, Lind Lars. A polymorphism in the AH-receptor gene is related to hypertension and endothelium-dependent vasodilation (abstract) Toxicol Lett. 2009;189:S100–S101. [Google Scholar]
- 25.Ehret GB, Munroe PB, Rice KM, Bochud VM, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.