Abstract
Structural DNA nanotechnology consists of constructing objects, lattices and devices from branched DNA molecules. Branched DNA molecules open the way for the construction of a variety of N-connected motifs. These motifs can be joined by cohesive interactions to produce larger constructs in a bottom-up approach to nanoconstruction. The first objects produced by this approach were stick polyhedra and topological targets, such as knots and Borromean rings. These were followed by periodic arrays with programmable patterns. It is possible to exploit DNA structural transitions and sequence-specific binding to produce a variety of DNA nanomechanical devices, which include a bipedal walker and a machine that emulates the translational capabilities of the ribosome. Much of the promise of this methodology involves the use of DNA to scaffold other materials, such as biological macromolecules, nanoelectronic components, and polymers. These systems are designed to lead to improvements in crystallography, computation and the production of diverse and exotic materials.
Keywords: Unusual DNA Motifs, Bottom-Up Nanoscale Construction, DNA Sequence Design, Nanoscale DNA Objects, Nanorobotics, Nanoscale Pattern Design
INTRODUCTION
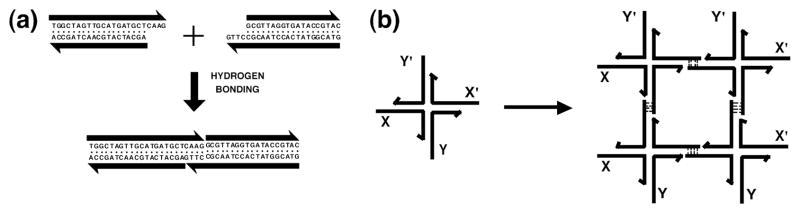
The double helical structure of DNA is well-known. It is the repository of genetic information for all organisms. It is able to function in this role because of the complementarity nature of the pairing between the bases: Adenine (A) pairs specifically with thymine (T) and guanine (G) pairs specifically with cytosine (C). The biological consequence of complementarity is that the cellular machinery canreplicate the information contained in each strand via a semi-conservative mechanism, leading to the production of daughter cells containing the same genetic complement. Pairing between short intermolecular overhangs, called ‘sticky ends’ has been used by genetic engineers for over 30 years to organize DNA fragments. This is a unique intermolecular interaction in chemistry, not only because it is programmable (the base sequences of sticky ends can be changed to a wide variety of different complements), but because the local product structure is known to be similar to the classic B-DNA structure that is seen everywhere as a cultural icon throughout the world.
Of course, from the perspective of nanotechnology, genetic engineering does not provide the structural consequences that one would want from a bottom-up approach to the control of the structure of matter. Although sticky-ended cohesion can be used to order a series of DNA fragments, say on a plasmid, the end product is simply a double helical molecule that flanks a linear helix axis, although that helix may be cyclic, knotted or catenated. The missing element to make DNA an interesting nanotechnological building block is a branch point. Branches enable the construction of N-connected objects and lattices. They are also inherent to most applications of DNA to nanomechanical devices. Sticky ends and their application to forming N-connected DNA objects are illustrated in
Here, I will describe how to design branched DNA motifs and how to select sequences for those motifs. I will then describe some of the key steps taken by the field of structural DNA nanotechnology to reach the point where today it is a discipline practiced by a significant number of investigators. I will describe the construction of the earliest N-connected objects, and of periodic arrays with programmable patterns. I will also describe some of the most exciting developments in the area of nanomechanical devices, leading to the development of DNA-based nanorobotics. Introductions to this material can be found in (Seeman, 2003; Seeman, 2004).
MOTIF AND SEQUENCE DESIGN
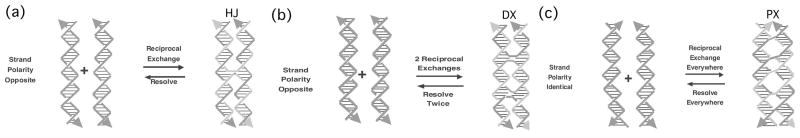
The essential difference between conventional linear DNA and the motifs useful structural DNA nanotechnology is the presence of branching. This notion is thought of most easily as a crossover or strand exchange between two adjacent double helices. Reciprocal exchange is the protocol that can be used to produce branched motifs (Seeman, 2001). Figure 2 illustrates how this method is used to produce a singly-branched Holliday junction (an intermediate in genetic recombination), a stiff double crossover (DX) molecule and a paranemic crossover (PX) molecule.
Fig. 2.
Reciprocal Exchange leads to branched structures. (a) A single exchange leads to a Holliday junction. (b) Two exchanges lead to the DX molecule. (c) Exchange at every possible position generates the PX molecule. Note that the exchanges in (b) are between strands of the opposite polarity, but those in (c) are between strands of the same polarity. The exchange in (a) is shown to be between strands of opposite polarity, but the polarity of the strands does not matter in this case.
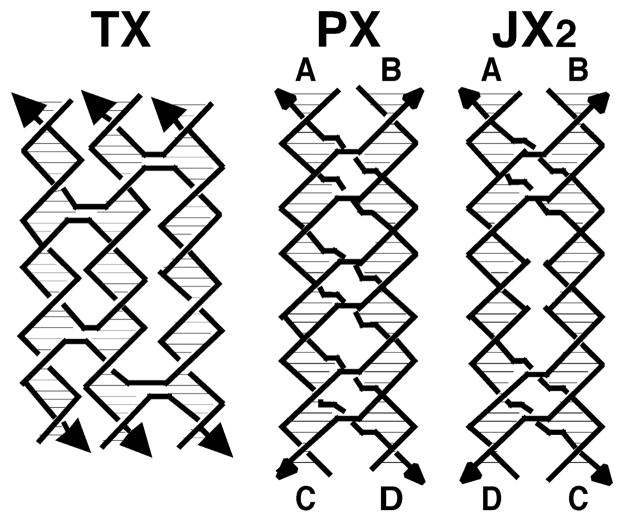
A number of other useful motifs generated in this way are illustrated in Figure 3. These include the TX molecule and the JX2 molecule, a topoisomer of the PX molecule whose relationships are illustrated by the letters flanking their helices. All of these motifs have found application in structural DNA nanotechnology.
Fig. 3.
Branched DNA Motifs. On the left is a TX molecule generated by adding a helical domain to a DX by two reciprocal exchanges between opposite polarity stands. The JX2 molecule on the right can be compared readily to its topoisomer, the PX molecule. It contains the same arrangement of strands, but it lacks two crossovers, leading to a molecule in which the top end is twisted relative to the bottom by a half-turn less than in the PX. This feature is highlighted by the alphabetic labels on the strands.
As noted in Figure 2, it is clear that forming each motif entails fusing DNA double helices. The two strands of the conventional double helical DNA molecule run in opposite directions. Thus, fusion between double helices can occur between strands of the same polarity or of opposite polarity. For the Holliday junction, with a single crossover, this difference has no topological consequences. However, DX molecules can be made with either polarity in its linkages, and the TX molecule can be made with mixed polarities. The DX and TX molecules shown in Figures 2 and 3 have crossovers between strands of opposite polarities. The PX molecule, and its topoisomer, JX2, have crossovers between strands of the same polarity.
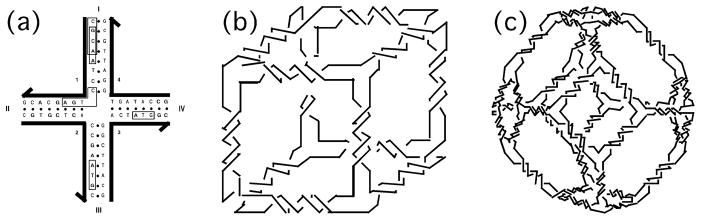
The assignment of sequences to these motifs seeks to maximize the probability that the target structure will form at equilibrium as a consequence of mixing the strands and cooling them. This is done by using the assumption that Watson-Crick base pairing is the preferred form of interaction between strands of DNA. Designing a crossover into the structure introduces a fault into this structure, corresponding to an excited state. The key point is to make sure that the excited state that results is the one sought, rather than one that is easier for the system to accommodate. To ensure that the target is what results, sequence symmetry is minimized throughout the structure (Seeman, 1982). An example of how this is done is shown in Figure 4a, which shows a single-crossover Holliday junction-like molecule in a crossroads representation.
Fig 4.
Design of Sequences for Branched Junctions and Polyhedral Catenanes. (a) Design of a branched junction. (b) DNA cube and (c) a DNA truncated octahedron.
The strands of the target molecule shown contains sixteen nucleotides. Each strand has been broken up into a series of thirteen overlapping tetramers, such as the CGCA or GCAA that are boxed at the top of strand 1. Sequence symmetry is minimized by insisting that each tetramer be unique. Thus, competition with the target octamers in each arm can come only from trimers, such as the ATG sequences boxed on strands 2 and 3; in principle, they could go to the wrong places, but both the thermodynamics of pairing and the cooperativity of double helix formation work against this eventuality. In addition, an element of negative control is introduced by forbidding the complement of any tetramer that flanks a branch point, such as the CTGA that is boxed at the corner of strand 1; the absence of a TCAG anywhere in the sequence means that the sequence going around the corner does not have the opportunity to form double helical DNA.
STRUCTURAL AND TOPOLOGICAL CONSTRUCTIONS
Given our understanding of branched motif generation and sequence assignment, it is easy to see how DNA can be used to prepare topological and structural targets. Following the notion summarized in Figure 1b, it has been possible to construct DNA polyhedra, molecules whose edges consist of double helical DNA and whose vertices correspond to the branch points of DNA branched junctions. We have reported a cube and a truncated octahedron built from DNA. Schematic versions of these constructs are shown in Figures 4b and 4c. One can think of these polyhedra simply as 3-connected graphs; we have also reported the construction of an irregular graph containing both 3-connected and 4-connected vertices. The connectivity of targets is limited by the number of double helical arms that flank the junctions in the structure. We have made junctions with as many as twelve arms. There are few interesting individual targets with connectivities so high, but lattices built from Platonic and Archimedean solids can have connectivities this high. For example, the stick version of cubic close packing is 12-connected. It is also noteworthy that the plectonemic nature of the DNA double helix makes it an extremely convenient synthon for topological targets; this is true because a half-turn of DNA corresponds to a unit-tangle in a knotted or catenated structure. In this context, the cube is a hexacatenane, and the truncated octahedron is a 14-catenane. In addition, an elusive topo-synthetic target, known as Borromean Rings, was synthesized first from DNA by using this same principle (Seeman, 1998).
Fig. 1.
Basics of Structural DNA Nanotechnology. (a) Sticky-ended cohesion. (b) Self-assembly of branched DNA molecules with complementary sticky end pairs.
The construction of periodic matter is one of the key goals of structural DNA nanotechnology. Periodic matter requires stiff components: The relationship between periodicity and cyclization so prominent in the Fourier analysis of crystals also applies to DNA nanoconstruction; components that can combine to form periodic matter can also form cyclic matter, thereby poisoning the assembly. The best defense against this problem is to use stiff components. Simple branched junctions are somewhat floppy, so they did not seem to be the best molecules to use to construct periodic matter. We found that DX molecules and their relatives, such as TX molecules are very good for this purpose. We have constructed arrays with patterns that can be both programmed and modified from DX molecules. We have also built 2D arrays from parallelograms, and from triangular arrangements of DX molecules (Ding et al., 2004). An example of the latter is shown in Figure 5.
Fig 5.
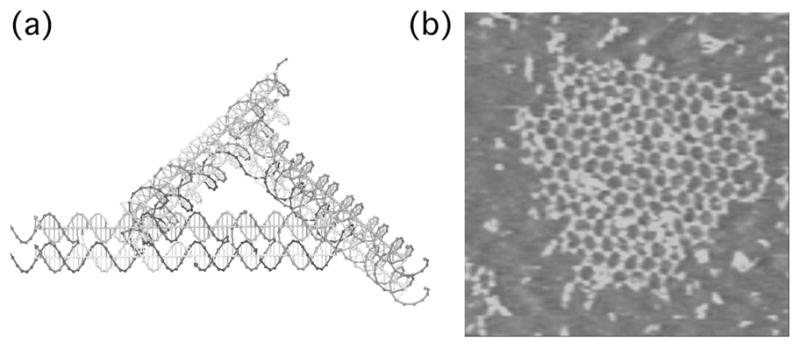
Formation of a Trigonal DNA Array. (a) The robust DX triangle motif. (b) An AFM image of a trigonal array built from two different molecules of this motif.
NANOMECHANICAL DEVICES
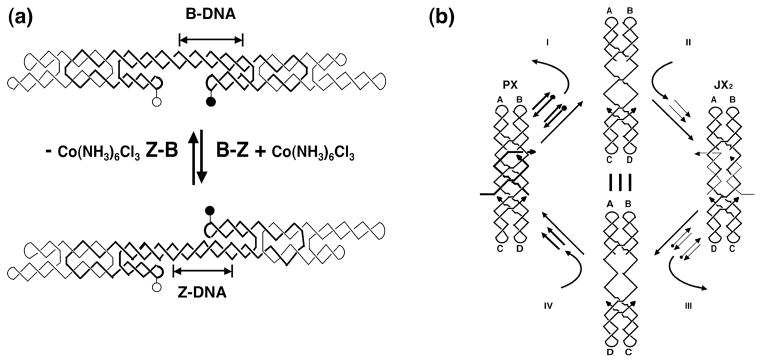
In addition to being an outstanding medium for the construction of objects and arrays, DNA is also convenient for the construction of nanomechanical devices. The first robust device constructed from DNA was based on the B-Z structural transition. Conventional DNA, known as B-DNA is a right-handed double helical molecule whose repeat unit is the nucleotide pair. However, appropriate sequences, typically (CG)n can adopt the Z-DNA conformation which is a left-handed double helix whose repeat unit is a dinucleotide pair. In the absence of Z-DNA-promoting conditions, Z-forming sequences remain in the B-DNA conformation. The device (Figure 6a) consists of two DX molecules connected by a shaft that contains a Z-forming sequence. In the absence of the Z-promoting reagent Co(NH3)63+, both DX molecules have their extra domain on the same side of the shaft; adding Co(NH3)63+ converts the Z-forming sequence to the Z conformation, placing one of the extra domains on the other side of the shaft.
Fig 6.
DNA Nanomechanical Devices. (a) A device based on the B-Z transition of DNA. Addition of Co(NH3)6Cl3 converts a short segment from B-DNA to Z-DNA. (b) The machine cycle of a PX-JX2 device. Removal of PX set strands (I) leaves a naked frame to which JX2 set strands (II) can be added to change state. The PX state can be restored by the same procedures (III) and (IV).
Figure 6b illustrates a robust sequence-dependent device that is capable of using the sequence specificity of DNA, so that a variety of devices can coexist in the same environment, yet be addressed individually. The device can adopt two different states, termed PX and JX2, that differ from each other by a half-turn. The states are set by DNA strands that are termed ‘set strands’. One can make a number of different devices by changing the sequences to which the set strands bind. In the drawing, the set strands for the PX conformation are drawn with thick lines, and the set strands for the JX2 conformation are drawn with thin lines. The set strands contain single-stranded extensions that are not paired with any other strands. Yurke and his colleagues (2000) have shown that when the complete complement to a set strand [termed an ‘unset strand’ or a ‘fuel strand’] is added to solution, it will remove the set strand, leaving a naked device frame. Thus, the machine cycle shown in Figure 6b starts on the left with the PX state, moves clockwise through the addition of unset strands to leave a naked frame. Continuing onwards, the set strands for the JX2 state are then added, and that state is formed. Adding the unset strands for the JX2 state again produces the naked frame, and the addition of the set strands for the PX state completes the cycle.
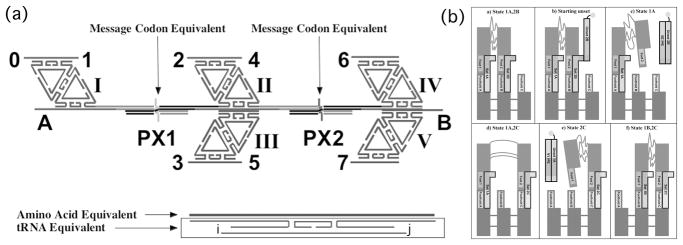
Two of these devices have been used to produce a nanomachine capable of translation (Liao & Seeman, 2004). The devices are used to couple DNA constructs in this machine, shown in Figure 6a. From left to right, the machine contains a DNA diamond, a PX-JX2 device, a DNA double diamond, a distinct PX-JX2 device, and a second DNA double diamond. The numbers represent sticky ends. The gap above the first PX-JX2 device is flanked by 1 and 2, and the gap above the second PX-JX2 device is flanked by 4 and 6. If the state of the left PX-JX2 device is changed from PX to JX2, and the other PX-JX2 device left untouched, the first gap will be flanked by 1 and 3, and the second gap will be flanked by 5 and 7. There are four different states to the machine, and each of them places different sticky end pairs on the two ends of the gaps. The solution contains six DX molecules whose bottom domain is complementary to one of these pairs. Once DX molecules are in place above the gaps, they are ligated to each other and to an initiator DX that binds to site 0. The product is determined by the state of the machine, and there is no transcriptional relationship between the set strands and the produce. The analogy to the components of protein synthesis is indicated in Figure 7a: The DX molecules are analogous to aminoacyl tRNA molecules, their bottom domain to the anticodon, and their top strand to the amino acid. The set strands are analogous to codons.
Fig 7.
Advanced Devices Built from DNA. (a) A translation device. The diamond on the left and the double diamonds in the center and on the right are separated by two PX-JX2 devices. The Arabic numerals indicated sticky ends. The sticky ends at the top will bind a DX molecule complementary to the numbers. As shown, a DX with sticky ends complementary to 1 and 2 and another with sticky ends complementary to 4 and 6 will bind. If the device on the right is switched to the JX2 state, a DX complementary to 4 and 7 will bind. This arrangement allows for positional ligation of the DX molecules bound there. The set strands correspond to an mRNA codon, and the bottom sticky ends (i and j) of the DX are the equivalent of the anticodon. The top strand of the DX is equivalent to the amino acid of an aminoacyl tRNA. (b) A bipedal walker. The parts of a walk are shown. The unset strand removes the set strand of the right foot, and then another set strand fastens it to a new position on the sidewalk. The unset strand of the left foot then frees it and it is fastened by a new set strand to the position where the right foot was bound.
Another sequence-dependent device is illustrated in Figure 7b. This is a bipedal walker that walks on a sidewalk (Sherman & Seeman, 2004). It consists of two double helical domains attached to each other by a flexible linker. The bottom end of each domain is a single-stranded segment. Similarly, the top end of each part of the sidewalk (a TX molecule) is single-stranded. The walker is positioned onto the sidewalk by set strands, similar to those in the PX-JX2 device. Removal of a set strand by adding an unset strand frees a double helical domain of the walker; addition of a set strand for a different position of the sidewalk locks it down to the new position. The drawing shows how the right leg of the walker is freed from the sidewalk, and then it is tied down to a new position. The left leg is then freed from the sidewalk, and is tied down to the position originally occupied by the right leg. Thus, a step has occurred. The device can walk in both directions. By itself this one-dimensional example is only a prototype. However, the same device or a triped can walk in two directions on a two-dimensional lattice. The key point is that the device’s motions is programmable in an interesting fashion by the input strands; a device that responds in this way to external signals can be regarded as a robot. This device could also could be programmed by the output of logical processes, and could be used to keep track of the state to which a system had arrived.
CONCLUSIONS, APPLICATIONS AND CHALLENGES
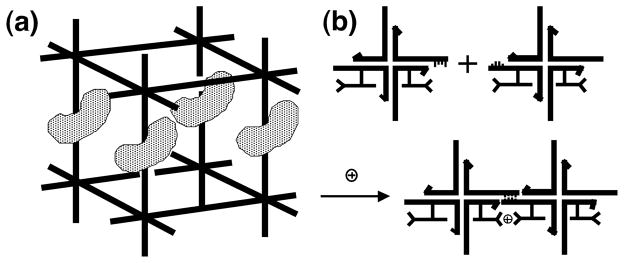
We have demonstrated the facility and versatility of DNA as a bottom-up construction medium on the nanoscale. Objects, arrays and devices are all readily within the scope of this technology. Where can this approach be utilized? The system was originally devised to facilitate the crystallization of biological macromolecules. The idea is to use the DNA lattice as a macromolecular-scale host for macromolecular guests; if all components are well ordered, crystallographic analysis can be performed on both the host and the guest (Seeman, 1982). An example is shown in Figure 8a. However, if one can conceive of organizing biological systems, one also can imagine that nanoelectronic components can be organized. Thus, by using the superb architectural properties of DNA described above, we may be able to organize molecular electronics in an efficient self-assembled fashion (Robinson & Seeman, 1987). This notion is shown in Figure 8b. We expect that the various devices noted above will be of use in producing materials that previously were inaccessible.
Fig 8.
Applications of Structural DNA Nanotechnology. (a) A crystallographic application. A DNA box is shown with sticky ends in each direction. The sticky ends can associate to make a host lattice. Macromolecular guests are shown ordered within the host lattice, enabling their structure determination. (b) Ordering nanoelectronic components. Two branched junctions are shown, and their cohesion organizes molecular wires that are pendent from them.
Many technical challenges remain to be overcome before all of the potential of this approach can be realized. We must extend our 2D capabilities to 3D, with high order. We must be able to incorporate both devices and hetero-species into these lattices. Both hierarchical and functional lattices remain to be developed, and sophistication of the chemistry must be achieved to make this a biomimetic, rather than a biokleptic system. Nevertheless, we are at the very beginning of structural DNA nanotechnology, and the future appears to be boundless. For those interested in controlling the structure of matter, this is a very exciting time!
Acknowledgments
This paper is dedicated to Bengt Norden on the occasion of his 60th birthday. Rresearch has been supported by grants GM-29554 from the NIGMS, grants DMI-0210844, EIA-0086015, CCF-0432009, CCF-05323290 and CTS-0103002 from the NSF, 48681-EL from ARO and an award from Nanoscience Technologies, Inc.
References
- DING B, SHA R, SEEMAN NC. Pseudohexagonal 2D DNA crystals from double crossover cohesion. Journal of the American Chemical Society. 2004;126:10230–10231. doi: 10.1021/ja047486u. [DOI] [PubMed] [Google Scholar]
- LIAO S, SEEMAN NC. Translation of DNA signals into polymer assembly instructions. Science. 2004;306:2072–2074. doi: 10.1126/science.1104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON BH, SEEMAN NC. The Design of a Biochip: A self-assembling molecular-scale memory device. Protein Engineering. 1987;1:295–300. doi: 10.1093/protein/1.4.295. [DOI] [PubMed] [Google Scholar]
- SEEMAN NC. Nucleic acid junctions and lattices. Journal of Theoretical Biology. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- SEEMAN NC. Nucleic acid nanostructures and topology. Angewandte Chemie International Edition. 1998;37:3220–3238. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3220::AID-ANIE3220>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- SEEMAN NC. DNA nicks and nodes and nanotechnology. NanoLetters. 2001;1:22–26. [Google Scholar]
- SEEMAN NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- SEEMAN NC. Nanotechnology and the double helix. Scientific American. 2004;290 (6):64–75. doi: 10.1038/scientificamerican0604-64. [DOI] [PubMed] [Google Scholar]
- SHERMAN WB, SEEMAN NC. A precisely controlled DNA bipedal walking device. NanoLetters. 2004;4:1203–1207. [Google Scholar]
- YURKE B, TURBERFIELD AJ, MILLS AP, Jr, SIMMEL FC, NEWMANN JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]