Abstract
We have developed an efficient reverse-genetics protocol that uses expedient pooling and hybridization strategies to identify individual transfer-DNA insertion lines from a collection of 6000 independently transformed lines in as few as 36 polymerase chain reactions. We have used this protocol to systematically isolate Arabidopsis lines containing insertional mutations in individual cytochrome P450 genes. In higher plants P450 genes encode enzymes that perform an exceptionally wide range of functions, including the biosynthesis of primary metabolites necessary for normal growth and development, the biosynthesis of secondary products, and the catabolism of xenobiotics. Despite their importance, progress in assigning enzymatic function to individual P450 gene products has been slow. Here we report the isolation of the first 12 such lines, including one (CYP83B1-1) that displays a runt phenotype (small plants with hooked leaves), and three insertions in abundantly expressed genes. The DNAs used in this study are publicly available and can be used to systematically isolate mutants in Arabidopsis.
The isolation and analysis of mutations provides one of the most powerful approaches available for determining the biological function of specific genes. Directed reverse-genetic screens, which require sequence information from the genes of interest and the availability of large numbers of individuals carrying foreign DNA sequences (transposons or T-DNA) randomly inserted within the genome, provide an alternative approach for ascertaining gene-product function. The insertion disrupts and thereby inactivates individual genes. The individual plants containing the disruption can be identified using a PCR primer designed from the gene sequence of interest, combined with a second PCR primer designed from the border sequence of the insertional mutagen, in the strategy described here as T-DNA. Only genomic DNA of lines containing insertional mutations in or near the gene of interest will generate a PCR-amplification product. This method was first applied in Drosophila melanogaster (Ballinger and Benzer, 1989; Kaiser and Goodwin, 1990) and has more recently been successfully applied in other systems, including Caenorhabditis elegans, petunia, maize, and Arabidopsis (Zwaal et al., 1993; Bensen et al., 1995; Koes et al., 1995; McKinney et al., 1995; Krysan et al., 1996; Mena et al., 1996; Frey et al., 1997).
The P450 gene superfamily is a large and ancient gene family (Nelson et al., 1996). More than 500 P450 genes have been identified, and these have been classified into more than 70 gene subfamilies (Nelson and Strobel, 1987; Nelson et al., 1996). Higher eukaryotes typically have a genomic complement of more than 100 P450 genes (Nelson et al., 1996). The P450 gene family has rapidly expanded and diversified during the last 400 million years, partly as a consequence of chemical warfare between the plant and its predators (Gonzalez and Nebert, 1990). P450 gene duplications, conversions, and clustering provide evidence for this rapid evolution (Nelson and Strobel, 1987; Gonzalez and Nebert, 1990). In addition to a limited number of fundamental functions shared by different eukaryotes, such as the 14-α-demethylation of sterols (Bak et al., 1997), P450 genes have evolved to perform numerous diverse functions. In animals these include the metabolism of endogenous signaling molecules, such as steroid hormones (Nebert, 1991), and xenobiotic detoxification (Nelson and Strobel, 1987; Gonzalez and Nebert, 1990; Nebert, 1991). In plants P450 enzymes are involved in more than 50 reactions (Bolwell et al., 1994; Schuler, 1996). Among these reactions are the biosynthesis of structural macromolecules such as lignin, and of plant growth regulators such as jasmonic acid, GAs, and brassinosteroids. P450 enzymes also catalyze the metabolism of xenobiotics, as well as the biosynthesis of numerous secondary products, including alkaloids, terpenoids, and cyanogenic glucosides (Bolwell et al., 1994; Schuler, 1996). The roles of secondary metabolites in plants are largely undefined, but many are hypothesized to play defensive roles.
Analysis of natural and induced mutations has been critical in our appreciation of the diversity of P450 gene function. For instance, mutations in the CYP1B1 gene are associated with primary congenital glaucoma in humans (Stoilov et al., 1997), and the cpd (CYP90A) and dwf4 (CYP90B) mutants of Arabidopsis have helped define the role of brassinosteroids in plants (Szekeres et al., 1996; Azpiroz et al., 1998; Choe et al., 1998). However, for the vast majority of plant P450 genes, no information exists regarding their metabolic roles, which means that it is impossible to predict the mutant phenotypes as required by traditional procedures of “forward” genetic screening. By way of explanation, P450 proteins with more than 40% identity are included in the same family, designated with the prefix CYP, e.g. the CYP90 family. Proteins with more than 60% identity are included in the same subfamily, e.g. the CYP90 family contains the CYP90A and CYP90B subfamilies. Individual members of a subfamily are further designated with integers, e.g. CYP90A1 and CYP90A2 (Nelson et al., 1996).
Arabidopsis, with its large EST collection, growing genomic sequence database and T-DNA insertion mutant population (summarized by Azpiroz-Leehan and Feldmann, 1997), and excellent genetics, provides a model system with which to dissect plant P450 gene function. We initiated a systematic screen for mutants of Arabidopsis P450 genes tagged as a consequence of T-DNA insertion. We report here the development of an efficient method of combinatorial screening and the initial characterization, to our knowledge, of the first P450 mutant lines obtained in this manner. This approach can be applied to any single gene or to gene superfamilies.
MATERIALS AND METHODS
Tagged Lines and DNA Pools
The generation of transformed T-DNA lines has been described previously (Forsthoefel et al., 1992). A C58C1Rif Agrobacterium tumefaciens strain carrying the 3850:1003 cointegrate plasmid conferring resistance to kanamycin was used to transform Arabidopsis ecotype Wassilewskija. The pooling strategy used in these experiments is outlined in Figure 1. Seeds from “row” and “column” pools (n = 120) were sown in flats, and plants (50–100 g fresh weight) were harvested at 3 weeks of age and used to prepare DNA (Murray and Thompson, 1980; McKinney et al., 1995). The plants were destarched by placing the flats in the dark for 36 h before they were harvested. DNA concentration was estimated by agarose gel electrophoresis of HindIII-digested samples. DNA pools were adjusted to approximately 0.025 g/L and stored at 4°C. The 120 DNA pools used in this work, and the transformants used to make these pools, are available from ABRC (CD5-7). DNA from “integer” pools (described below) was prepared by a modification of the method of Cocciolone and Cone (1993). Pooled sterile seedlings were frozen in liquid nitrogen and then powdered in 10-mL round-bottom tubes with a glass rod. Extraction buffer (1.0 mL) was added, vortexed for 10 s, and mixed for 10 min, and then 0.5 mL of Cl3CH/phenol was added and the solution was vortexed for 10 s and shaken for 10 min. The preparations were centrifuged for 10 min at the maximum speed in a desktop centrifuge, and the aqueous solution was removed and added to a 1.5-mL centrifuge tube. The DNA was precipitated with one-tenth volume of 3 m sodium acetate and 0.6 volume of isopropanol, washed with 70% ethanol, air dried for 15 min, and dissolved in 50 μL of TE (10 mm Tris [pH 8.0], 1 mm EDTA).
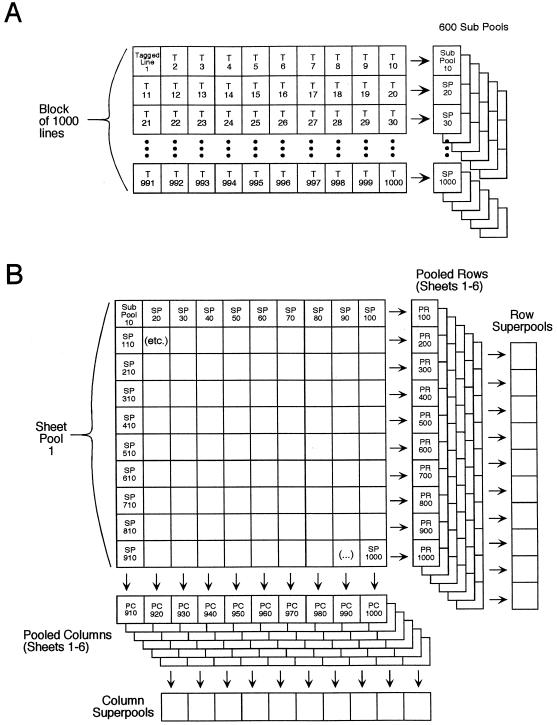
Figure 1.
A, To generate the DNAs necessary for this strategy, the seeds were first combined into subpools. Seeds from the first 10 lines (1–10), the next 10 lines (11–20), etc., through 6000 lines were pooled to form 600 subpools. B, For each thousand lines (Sheet Pool), the DNAs from the 100 subpools were mixed in a multiplex array, to give 10 Pooled Column (PC) and 10 Pooled Row (PR) pools. The pooled column and pooled row pools from all of the sheet pools were then amalgamated to give 10 Column Superpools and 10 Row Superpools. When the sheet pool, column superpools, and row superpools containing the positive amplicon are known, it is possible to define the mutant of interest to a subpool of 10 lines.
Identification of P450 Genes in Public Databases
Arabidopsis P450 ESTs were identified by searching the TIGR database (http://www.tigr.org/tdb/at/at.html). The TIGR database analysis contains singleton ESTs and contiguous sequences constructed from Arabidopsis ESTs that have been assigned tentative designations on the basis of BLAST search results (Rounsley et al., 1996). A word search for P450 recovered a total of 81 candidate CYP sequences (release of January 7, 1997), which were reconfirmed by BLASTX analysis. Clones for these P450 genes were obtained from ABRC.
PCR Primers
GSPs were designed with the program PrimerSelect from the LaserGene software package (DNASTAR, Madison, WI). For each sequence, the forward primer was designed to be an average of 100 bp 5′ of the reverse primer to maximize coverage of the gene. When possible the primers were designed to sequences in the middle of the gene or just slightly 5′ of the middle. To reduce the possibility that the primers spanned intron/exon boundaries, primers with sequences most commonly at the exon boundaries were eliminated: AGGT for forward primers and ACCT for reverse primers. All GSPs were designed to anneal at 60°C. Amplification of genomic DNA was used to confirm the suitability of the P450 primer pairs. For five primer pairs, no product was detected by UV visualization of ethidium bromide-stained agarose gels; these may represent cases in which one of the P450 primers of interest is interrupted by an intron. The left border (LB) and right border (RB) primers used in these experiments were: LB102A, GATGCAATCGATATCAGCCAATTTTAGAC, and RB16843, GCTCATGATCAGATTGTCGTTTCCCGCCTT, respectively.
Hybridization Probes
Probes were prepared from both P450 plasmid and genomic sequences. The 50-μL reaction mixture contained 1 ng of pooled P450 plasmid (DNA from all available P450 clones [from ABRC] pooled together), 0.5 unit of Taq DNA polymerase (BRL), 20 mm Tris-HCl (pH 8.4), 50 mm KCl, 50 m deoxyribonucleotide triphosphate, 2.5 m digoxigenin-UTP (Boehringer Mannheim), and 0.5 m forward and reverse primers designed against the LacZ flanking sequence of standard cloning vectors. To prepare probes from genomic DNA, a similar reaction mixture was used, except that 10 ng of genomic DNA as well as forward and reverse P450 GSP primer pairs were used in place of plasmid DNA and the vector primer pair, respectively. All products from plasmid and genomic amplifications were pooled and used in hybridization reactions; this pooled set of probes is referred to as the P450 superprobe.
Control Reactions
The previously described act2-1 mutant line has a T-DNA in the 5′ region of the gene (McKinney et al., 1995) and was used as a positive control in this work to validate the PCR conditions and DNA pooling. Two actin primers were used to amplify sequences; these corresponded to sequences located 1.5 kb 3′ and 600 bp 3′ of the T-DNA insertion, and are available from ABRC (primers sent with CD5-7).
PCR Reactions and Hybridizations
PCR reactions were set up at 4°C and a hot-start protocol was used, with the last component (either primers or DNA, depending on the screen) being added when the PCR block was at 72°C. The reaction mixture contained 20 mm Tris-HCl (pH 8.4), 50 mm KCl, 1.5 mm MgCl2, 100 m deoxyribonucleotide triphosphate, 50 m of each primer (forward or reverse and right border or left border), and genomic DNA. The PCR program comprised one cycle of 1 min at 95°C, followed by 43 cycles at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 2 min, followed by a 10-min extension cycle at 72°C.
After PCR amplification a 96-well format replicator was used to blot the reactions onto nylon membranes. The DNA was immediately affixed to the filter by either treating with UV light in a Stratalinker (Stratagene) or baking for 1 h at 80°C. The DNA was denatured by immersing the membrane in denaturing solution (0.2 m NaOH, 0.6 m NaCl) for 4 min, then the membranes were transferred into neutralizing solution (0.5 m Tris-HCl [pH 7.5], 1.5 m NaCl) for 4 min, and into sterile water for 1 min. The membranes were either air dried or used directly in hybridizations with the P450 superprobe. DNA-blot hybridizations used digoxigenin-UTP-labeled probes, and were done in a rotary hybridization oven (Robbins Scientific, Sunnyvale, CA) at 65°C in a solution comprising 0.25 m sodium phosphate (pH 7.4), 7% SDS, and 1 mm EDTA. Autoclaved solutions were used in all steps of the hybridizations and posthybridization processing (performed according to the manufacturer's instructions; Boehringer Mannheim).
Reverse Genetic Screening
In the primary screens, 100 to 300 ng of a DNA pool consisting of DNA from each of the 6000 lines was used in each PCR reaction. GSPs were arrayed in a 96-well format and were the last component added to the PCR reactions in the primary screen. An eight-channel pipet was used to add the primers. Primer pairs associated with positively hybridizing spots were tested for random priming by performing PCR without a T-DNA border primer; these were eliminated before the secondary screens.
The secondary screen involved screening DNA stocks pooled in a three-dimensional array with the GSPs that gave positively hybridizing spots in the primary screen in combination with a T-DNA border primer. The pools consisted of: (a) 6 “sheet pools” of 1000 lines (sheet pool 1 consisted of lines 1–1000 pooled together; sheet pool 2 consisted of lines 1001–2000, etc.); (b) 10 “row superpools” of 600 lines each (row superpool 100 consisted of all pooled rows ending in 100 [e.g. PR100, PR1100, PR2100, PR3100, PR4100, and PR5100]; pooled row 200 consisted of all pooled rows ending in 200, etc.); and (c) 10 “column superpools” of 600 lines each (column superpool 10 consisted of all pooled columns ending in 10 [e.g. PC910, PC1910, PC2910, PC3910, PC4910, and PC5910]; column superpool 20 consisted of all pooled columns ending in 20, etc.) (Fig. 1). In the secondary screens, 50 to 100 ng of DNA was used in each PCR reaction. The DNAs for this screening were arrayed in a 96-well format and were added last to the PCR reactions using an eight-channel pipet. For 12 of the GSPs a single positive subpool was identified.
In the tertiary screen, single mutant lines were identified from each of the 12 positive subpools. To avoid extracting DNA from each of the 120 lines that made up the 12 subpools (10 lines per subpool), “integer” pools were generated. Approximately 30 seeds from each of 12 lines ending in 1 were pooled, all 12 lines ending in 2 were pooled, etc. Plantlets from these pools were grown in liquid culture, and total DNA was extracted and arrayed in 96-well plates. In the tertiary screen, DNA was added last to the PCR reactions. PCR products were analyzed by hybridization with the P450 superprobe as described above.
Identification of Mutant Phenotypes
Fifty plants from each of the 12 putative mutant lines were grown in soil and screened for visible alterations in phenotype. In the one case in which a line had a visibly altered phenotype and the insertion was located between the GSPs, it was possible to test for linkage of the T-DNA insertion and the phenotypically mutant locus by testing for the presence of wild-type chromosomes in mutant plants. One leaf from each of 12 mutant plants and an equal number of wild-type plants were pooled separately, and DNA was extracted. Reactions were set up using approximately 10 ng of DNA. If the recessive mutant phenotype is caused by T-DNA insertion, no copies of the wild-type allele should be found in the DNA pool from the mutants. Tandem T-DNA insertions, typically 34 to 68 kb in length, preclude amplification of the gene in mutant alleles. For lines lacking a visible alteration in the phenotype, homozygous kanamycin-resistant lines were selected and analyzed for phenotypic alterations and for disruptions in the P450 gene of interest.
Sequencing PCR Products
Products for sequencing were amplified as follows: a 1-min cycle at 95°C, followed by 30 or 35 cycles at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 2 min, followed by a 10-min extension cycle at 72°C. PCR products were separated on agarose gels, reamplified, purified using Promega Wizard columns, and sequenced.
RESULTS AND DISCUSSION
Examination of the TIGR Arabidopsis database (Rounsley, et al., 1996; http://www.tigr.org/tdb/at/at.html) allowed identification of 81 unique P450 ESTs and contigs. This database is estimated to represent about one-half of all of the expressed genes in the genome, suggesting that the final number of P450 genes may exceed 160. We designed forward (F) and reverse (R) GSPs to 70 of these P450 sequences. When full-length Arabidopsis P450 cDNA sequences were in the database, we used those sequences for primer design; these include CYP83A1, CYP86A1, and CYP90A1. Because we expected that within our current population of 6000 Arabidopsis lines a small percentage of P450 genes would carry T-DNA inserts (Krysan et al., 1996), we first determined the detection limit of the screening protocol. These control experiments established that the act2-1 allele, a previously isolated T-DNA mutant (McKinney et al., 1995), could be detected by PCR in a single DNA pool prepared from all 6000 lines (data not shown). This allowed an efficient primary screen to be performed in which all 6000 lines could be screened at one time to determine which of the P450 genes contained mutations.
We arrayed GSPs in a 96-well format and performed a primary screen for each of these 70 P450 genes with the four possible combinations of gene- and T-DNA-specific primers (F/LB, F/RB, R/LB, and R/RB) using the pooled DNA to determine which genes had insertions. PCR products were spotted onto membranes and hybridized with a mixed probe made from the Arabidopsis P450 ESTs. In this primary screen, 29 primer pairs gave products that hybridized to the P450 superprobe. Seven GSPs were subsequently shown to produce positively hybridizing PCR products as a result of single-primer amplification and were eliminated from the screen.
The 22 primer pairs were used in secondary and tertiary screens to identify the individual lines containing the specific mutation. The secondary round of screening involved analyzing sheet pools, column superpools, and row superpools as outlined in Figure 1. This arrangement allows a specific mutation to be defined to a subpool of 10 independent T-DNA lines using 26 PCR reactions (Fig. 1). Based on their three-dimensional address in the array, the PCR products giving positive hybridization signals using the 22 primer pairs described above identified 12 independent subpools containing insertional mutants (Table I). Six of these lines were identified by multiple primer pairs. This indicates redundancy in the EST collection, as would be expected when two nonoverlapping ESTs are derived from the same or closely related genes. Four of the primers did not reproducibly identify the same array addresses in the screen and were discarded.
Table I.
Summary of T-DNA insertions detected with primers designed from P450 genes and ESTs
| Mutation No. | Primer from GenBank Accession No. | P450 Family | Notes |
|---|---|---|---|
| 1 | U61231 | CYP89A2 | |
| 2 | N38590 | CYP71A | |
| 3 | T04134 | CYP72 | Identified by multiple primers from nonoverlapping ESTs and by degenerate priming from a related EST |
| 4 | T14112 | CYP71B | Identified by multiple primers from nonoverlapping ESTs |
| 5 | T75944 | CYP83B1 | Runt phenotype; identified by multiple primers from nonoverlapping ESTs |
| 6 | Z33963 | CYP71B | |
| 7 | T20987 | CYP71B | Identified by degenerate priming from a related EST; in a linked CYP71B gene in the same line as mutation 4 |
| 8 | X90458 | CYP86A1 | |
| 9 | X90458 | CYP86A1 | |
| 10 | T04172 | CYP86A2 | Identified by multiple primers from nonoverlapping ESTs |
| 11 | H76866 | CYP71A | |
| 12 | H37250 | CYP76B | Identified by multiple primers from nonoverlapping ESTs |
| 13 | X92510 | CYP74A |
The mutation numbers are used as abbreviations in Figure 2. All insertions have left-border T-DNA at the T-DNA junction, except for mutation 8, which has a right-border junction.
Each subpool consists of 10 lines and these 12 positive subpools can be screened efficiently by pooling lines in a fourth dimension, which we refer to as integer pools. To identify single mutant lines from these 12 subpools, approximately 30 seeds from each line were pooled in the fourth dimension, i.e. all lines ending in 1 were pooled, all lines ending in 2, etc., for a total of 10 pools. Plantlets from these 10 pools were grown in liquid culture and total DNA was extracted. PCR reactions were performed with the appropriate primers, blotted, and analyzed by hybridization to the P450 superprobe. In this tertiary round of screening, 12 lines were identified that contained separate T-DNA insertions.
Direct sequencing of the PCR products was used to establish both the veracity of the insertions and the positions of the T-DNA insertions in these mutant lines (Fig. 2). Two independent primers designed for two different CYP72 genes identified the same insertion, a consequence of a high degree of sequence similarity between these two particular genes (Table I).
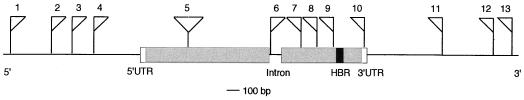
Figure 2.
Summary of the approximate positions of the T-DNA insertions in the P450 genes. A prototypical P450 gene with just one intron represented is shown (UTR, untranslated region; HBR, conserved heme-binding region). The insertions, numbered from 1 to 13, are described in Table I. The directions of the flags indicate the orientations of the border primers relative to the GSPs used to identify the insertion.
The insertions were found in various locations in these genes, further confirming the randomness of T-DNA insertion at the gene level. Four occurred in 5′ regions, four were in exons, one was in an intron, one was in the 3′ untranslated region of the corresponding mRNA (Fig. 2), and three others were found at various distances 3′ to the transcribed region. The five insertions in either exons or introns are almost certain to result in null mutations (Azpiroz-Leehan and Feldmann, 1997). The insertional events in the 5′ and 3′ regions will require further analysis to determine whether gene expression has been altered. In other work, it has been found that T-DNA insertions approximately 1 to 2 kb 5′ or 3′ of the transcribed region can result in modified gene expression and mutant phenotypes (Oppenheimer et al., 1991; Klucher et al., 1996).
The situation for mutations 4 and 7 is particularly complex. These mutations were identified, in a single T-DNA line, by six independent primers designed to noncontiguous ESTs in the CYP71B subfamily (see Table I). Sequencing of the amplification products established that two primers from two related genes amplified the same mutant CYP71B gene by degenerate priming and that the third and fourth primers amplified a second CYP71B gene. Only one kanamycin-resistant linkage group was detected in this line, consistent with this line having only one T-DNA insertion (data not shown). A simple explanation would be that some members of the CYP71B family form a tandemly oriented cluster. Because this line has one insertion within a CYP71B exon, and one insertion 5′ of a CYP71B gene, there could be two T-DNA insertions that have occurred in close proximity to linked P450 genes. A related set of CYP71 genes, the CYP71C gene family of maize, has also been observed to be clustered (Frey et al., 1995). Further analysis will be required to determine the structure of these insertions.
To define the roles of the P450 genes identified in the reverse-genetic screen, the segregating families were screened for visible alterations in phenotype. Fifty plants from each of the 12 lines were grown in soil and screened on a weekly basis. One line was observed to segregate a clearly visible phenotype. This CYP83B1-1 mutant line segregated small plants having hooked leaves (Fig. 3), with a frequency appropriate for a single recessive mutation. To establish that the CYP83B1-1 mutant phenotype was caused by T-DNA insertion, we examined the linkage between the mutant phenotype and the T-DNA insertion in a pool of 12 homozygotes. No wild-type alleles were detected in this pool, indicating linkage between the CYP83B1-1 insertion and the mutant phenotype, and suggesting that the mutant phenotype is caused by the T-DNA. Restoration of function by transformation using the wild-type CYP83B1-1 gene or isolation and characterization of additional mutant alleles will be required to confirm the CYP83B1 mutant phenotype. The phenotype of the CYP83B1-1 mutant indicates that the CYP83B1 gene is essential for normal growth.
Figure 3.
The phenotypes of 10-d-old soil-grown plants: left, wild type; right, homozygote containing the CYP83B1-1 mutant allele.
We did not observe an obvious visible phenotype for 11 other lines. Several of the P450 subfamilies of Arabidopsis, including CYP71B, CYP72, and CYP89A, have multiple closely related members, and therefore may be genetically redundant. All mutant lines are currently being analyzed for possible biochemical alterations as a consequence of blocks in primary or secondary metabolism.
The systematic identification of P450 mutants in Arabidopsis provides a methodical approach for dissecting the complex roles of plant P450 genes in plant growth and development, secondary metabolism, and plant defense. In particular, the roles of plant defensive compounds in interactions with insects and pathogens are probably best isolated by reverse-genetic strategies rather than by the direct selection for mutants. The P450 genes for which we have isolated mutants are of an unknown function. Because single amino acid changes can drastically alter P450 substrate specificity, it is not possible to predict enzymatic reactions from sequence information. However, the availability of a complete collection of P450 knockout mutations should provide important information relevant to the study of the functional evolution of this large gene family. It should also lead to new targets for engineering herbicide and host-plant resistance, and to potential catalysts for green chemistry.
To recover mutations in the remaining P450 genes or any gene sequence, the following considerations are important. First, the screening strategy appears efficient. Because the sequence data from our 12 tagged lines showed that one-third of the noncontiguous ESTs were redundant in our screen (6 of 18), we estimate that our screen targeted approximately 46 P450 genes. Larger populations of T-DNA-tagged lines will increase the probability of obtaining insertional mutants in the P450 gene family; it is estimated that 105,000 lines are required to have a 95% chance of uncovering an insertion within an average Arabidopsis gene of 2.3 kb. Second, the screening strategy is not restricted to EST-based information, but can be successful with degenerate primers for the P450 heme-binding region (we have independently isolated the CYP83B1-1 mutation in this manner). Furthermore, as more complete P450 sequences are released in the Arabidopsis genomic sequencing project, it will be possible to target these genes more efficiently by designing primers to the expected 5′ and 3′ regions of the P450 gene to ensure complete coverage of the predicted gene by the primers. Additionally, the knowledge of the complete genomic sequence of the target gene will allow the position of the insertion within the gene to be confirmed on the basis of the size of PCR product(s). The methods we have described can also be applied to any Arabidopsis gene or gene family. In particular, as the amount of Arabidopsis sequence information in the public databases grows, the number of genes that can be targeted will grow accordingly. Furthermore, as more T-DNA lines are made available the probability of successfully targeting the gene of interest grows; in this regard, the 6000 lines described here, as well as 6000 more lines donated by T. Jack (Dartmouth), are available from ABRC as seeds and pooled DNAs.
ACKNOWLEDGMENTS
We thank Frans Tax and Richard Schneeberger for critical review of the manuscript.
Abbreviations:
- ABRC
Arabidopsis Biological Resource Center (Ohio State University, Columbus)
- EST
expressed sequence tag
- GSP
gene-specific primer
- T-DNA
transfer DNA
Footnotes
This work was supported by the National Science Foundation-Department of Energy-U.S. Department of Agriculture (USDA) Joint Program of Collaborative Research in Plant Biology (92-20332), by USDA grant no. 9701472 to K.A.F., R.F., and D.W.G., and by the University of Arizona Agricultural Experiment Station.
LITERATURE CITED
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Bak S, Kahn RA, Olsen CE, Halkier BA. Cloning and expression in Escherichia coli of the obtusifoliol 14 alpha-demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14 alpha-demethylases (CYP51) from fungi and mammals. Plant J. 1997;11:191–201. doi: 10.1046/j.1365-313x.1997.11020191.x. [DOI] [PubMed] [Google Scholar]
- Ballinger DG, Benzer S. Targeted gene mutations in Drosophila. Proc Natl Acad Sci USA. 1989;86:9402–9406. doi: 10.1073/pnas.86.23.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB. Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed]
- Bolwell GP, Bozak K, Zimmerlin A. Plant cytochrome P450. Phytochemistry. 1994;37:1491–1506. doi: 10.1016/s0031-9422(00)89567-9. [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatuso S, Sakurai A, Feldmann KA. Arabidopsis DWF4 encodes a cytochrome P450 that mediates multiple steps of 22α-hydroxylation in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone SM, Cone KC. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics. 1993;135:575–588. doi: 10.1093/genetics/135.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel NR, Wu Y, Schulz B, Bennett MJ, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: prospects and perspectives. Aust J Plant Physiol. 1992;19:353–366. [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP and others. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- Frey MR, Kliem, Saedler H, Gierl A. Expression of a cytochrome P450 gene family in maize. Mol Gen Genet. 1995;246:100–109. doi: 10.1007/BF00290138. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Nebert DW. Evolution of the P450 gene superfamily: animal-plant ‘warfare,’ molecular drive and human genetic differences in drug oxidation. Trends Genet. 1990;6:182–186. doi: 10.1016/0168-9525(90)90174-5. [DOI] [PubMed] [Google Scholar]
- Kaiser K, Goodwin SF. “Site-selected” transposon mutagenesis of Drosophila. Proc Natl Acad Sci USA. 1990;87:1686–1690. doi: 10.1073/pnas.87.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Souer E, van Houwelingen A, Mur L, Spelt C, Quattrocchio F, Wing J, Opperdijk B, Ahmed S, Maes T and others. Targeted gene inactivation in petunia by PCR-based selection of transposon insertion mutants. Proc Natl Acad Sci USA. 1995;92:8149–8153. doi: 10.1073/pnas.92.18.8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, McDowell JM, Meagher RB. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2-1 and act4-1. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. Diversification of C-function activity in maize flower development. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW. Proposed role of drug-metabolizing enzymes: regulation of steady state levels of the ligands that affect growth, homeostasis, differentiation, and neuroendocrine functions. Mol Endocrinol. 1991;5:1203–1214. doi: 10.1210/mend-5-9-1203. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW and others. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Strobel HW. Evolution of cytochrome P-450 proteins. Mol Biol Evol. 1987;4:572–593. doi: 10.1093/oxfordjournals.molbev.a040471. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- Rounsley SD, Glodek A, Sutton G, Adams MD, Somerville CR, Venter JC, Kerlavage AR. The construction of Arabidopsis expressed sequence tag assemblies. A new resource to facilitate gene identification. Plant Physiol. 1996;112:1177–1183. doi: 10.1104/pp.112.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA. Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci. 1996;15:235–284. [Google Scholar]
- Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997;6:641–647. doi: 10.1093/hmg/6.4.641. [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP090, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Broeks A, van Meurs J, Groenen JTM, Plasterk RHA. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc Natl Acad Sci USA. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]