Abstract
The importance of type I interferons (IFN) in the host response to viral infection is well established, however, their role in bacterial infection is not fully understood. Several bacteria (both Gram-positive and Gram-negative) have been shown to induce IFN-beta (IFN-β) production in myeloid cells, but this IFN-β is not always beneficial to the host. We examined whether Staphylococcus aureus induces IFN-β from myeloid phagocytes, and if so, whether it is helpful or harmful to the host to do so. We found that S. aureus poorly induces IFN-β production compared to other bacteria. S. aureus is highly resistant to degradation in the phagosome because it is resistant to lysozyme. Using a mutant that is more sensitive to lysozyme, we show that phagosomal degradation and release of intracellular ligands is essential for induction of IFN-β and inflammatory chemokines downstream of IFN-β. Further, we found that adding exogenous IFN-β during S. aureus infection (in vitro and in vivo) was protective. Together, the data demonstrate that failure to induce IFN-β production during S. aureus infection contributes to pathogenicity.
Introduction
The importance of type I interferons (IFN) in the host response to viral infection has been known for half a century, however, the role of these cytokines in bacterial infection is still being elucidated. Several bacteria and even some fungi induce type I IFNs in the host, and most reports suggest that this can be beneficial for host defense (1, 2). However, there are some examples in which IFN-beta (IFN-β) induction is detrimental (1). Gram-negative bacteria such as Escherichia coli and Salmonella typhimurium activate type I IFNs through a mechanism that is largely dependent on lipopolysaccharide (LPS) stimulation of TLR4 (3, 4). Type I IFNs can be protective against infection with Gram-negative bacteria such as Salmonella typhimurium (5), although its production exacerbates LPS-induced shock (6). More recently, Gram-positive species have been identified that induce type I IFNs through stimulating additional pattern recognition receptors (7) such as endosomal TLRs (8) and various cytosolic receptors such as DAI (9) and NOD-2 (10). Type I IFN production has been reported to be protective against infection with Gram-positive bacteria such as Group B Streptococcus (Streptococcus agalactiae) (11) and Group A Streptococcus (Streptococcus pyogenes) (12), although its production exacerbates pathology during Listeria monocytogenes infection (13, 14).
Staphylococcus aureus is a Gram-positive bacterium that can be both a commensal and a pathogen. 30-50% of the world’s population is asymptomatically colonized with S. aureus, which mainly inhabits mucosal surfaces (15). However, under certain circumstances it can become a life-threatening pathogen, often causing soft tissue infections (16). The pathophysiology associated with serious infection is an area of intense research, especially in the last 10 years as antibiotic resistant strains (such as methicillin-resistant S. aureus, MRSA) have become increasingly prevalent (16). Little is known about the role of type I IFNs during S. aureus infection. A few papers have suggested a role for production of type I IFNs by airway epithelial cells in host defense during lung infection. Martin et al. determined that type I IFN signaling is detrimental during S. aureus pneumonia in mice (17). In contrast, a recent study has shown that type I IFNs protect against S. aureus alpha toxin in the lungs (18) and another has determined that stimulation of type I IFNs by treatment with CpG DNA can be protective in a model of post-hemorrhage-S. aureus pneumonia (19). However, whether innate immune cells such as macrophages and dendritic cells produce type I IFNs in response to S. aureus and how this might affect host defense during infection remains an important question.
During our investigations into how various Gram-positive bacteria induce IFN-β we noticed that S. aureus induces very little of the cytokine in myeloid phagocytes in vitro or during cutaneous infection in vivo. This seemed unusual to us since the related bacterial species, Group B Streptococcus, readily induced IFN-β production from myeloid phagocytes (11). We found that because S. aureus is resistant to degradation in the phagosome, due to modifications to its cell wall peptidoglycan, it is able to prevent release of immunostimulatory ligands that are necessary to trigger IFN-β production. Further, we show that IFN-β production can stimulate effective host defense against S. aureus and that treatment with IFN-β can promote clearance of S. aureus during cutaneous infection.
Materials and Methods
Bacterial strains and culture
Wild type Staphylococcus aureus 113 (WT-SA) and OatA mutant (ΔOatA-SA) (kindly provided by F. Gotz) (20), S. aureus Pig1 (kindly provided by D. Leung) (21), and S. aureus Newman were grown in Todd Hewitt broth at 37°C with agitation overnight to stationary phase, washed in sterile PBS, then diluted to OD600=0.4 (determined to be 1×108 CFU/ml). We generated a bioluminescent strain of S. aureus Pig1 (CST222). Isogenic strains harboring Tn4001 luxABCDE KmR (22) were constructed using previously established transduction methods, to introduce the plasmid or the alleles into the CST222 background (23). Streptococcus agalactiae (Group B Streptococcus, GBS) was grown in Todd Hewitt broth. Salmonella enterica serovar Typhimurium and Escherichia coli were grown in Luria broth. Listeria monocytogenes was grown in brain-heart infusion broth.
Mice and murine cell culture
Caspase-1−/−, MyD88−/−, and TLR7−/− mice on C57BL/6 background were bred and housed under specific pathogen-free conditions in the Cedars-Sinai Medical Center animal facility. All protocols were reviewed and approved by the Cedars Sinai Institutional Animal Care and Use Committee. C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, Maine). TRIF−/− and TLR9−/− bones were provided by E. Miao (24), UNC93B−/− bones were provided by B. Beutler (25), and MAVS−/− bones were provided by K. Fitzgerald (26). Conventional bone marrow derived dendritic cells (BMDC) were made by culturing bone marrow from femurs and tibias for 7-8 days in complete RPMI (10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine; Mediatech) supplemented with 10 ng/ml recombinant GM-CSF (Peprotech) on day 0 and day 4.
Human cell culture
Monocytes were isolated from fresh human blood. We obtained blood from healthy donors within our hospital, and samples were anonymized before being provided for use and analysis in this study. Blood was layered over lymphocyte separation media (Fisher) and spun at 1000 × g for 25 min with no brake. The interface was removed and washed 3X in PBS. Monocytes were negatively selected using a magnetic bead system (Milltenyi Biotech). Isolated monocytes were allowed to differentiate for seven days into dendritic cells. Growth factors (recombinant human IL-4 and GM-CSF (10 ng/ml each) (Peprotech)) were added on days zero and four.
Peptidoglycan purification
Peptidoglycan was prepared as previously (27). Briefly, overnight cultures of wild-type or ΔoatA S. aureus were heat-killed and washed with acetone. Bacteria were lysed using glass beads and a Turbomix for 10 min at 4°C. Unbroken cells were removed by low speed centrifugation, and cell wall fragments were collected by high speed centrifugation. Washed cells walls were treated with RNase A and DNase I overnight at 37°C followed by overnight treatment with trypsin. The remaining peptidoglycan was extracted for 5 hr at 4°C with 10% trichloroacetic acid and washed with acetone.
S. aureus infection in vitro
BMDC were plated in 24-well plates (3×105 cells/well) for RNA or 96-well plates (1×105 cells/well) for protein measurements 16-24 hours prior to infection. On the day of infection, media was replaced with antibiotic free media 60 minutes before infection. Bacteria were added to cells at indicated multiplicities of infection (MOI), and plates were spun at 450×g for 5 minutes. The infection was allowed to progress for 60 minutes, cells were washed in PBS, and new media containing 400 μg/ml gentamycin was added to cells for 30 minutes to kill extracellular bacteria. After 30 minutes, media was replaced with one containing a lower concentration of gentamycin (100 μg/ml) and cells were incubated at 37°C and 5% CO2 for indicated times. Heat-killed bacteria were heated at 70°C for 90 minutes and washed in PBS 3X. Gentamycin-killed bacteria were incubated with shaking at 37°C for 90 minutes with 100 μg/ml gentamycin and washed in PBS 3X. Paraformaldehyde-fixed bacteria were incubated with 4% PFA for 10 minutes at room temperature and washed in PBS 3X.
Murine whole blood killing assay
Murine peripheral whole blood was harvested aseptically via cardiac puncture using a 22-gauge needle to minimize cell lysis and maintain integrity of blood for the duration of the assay. Blood was mixed with PBS or recombinant murine IFN-β. WT-SA were pelleted, washed, and diluted to 103 CFU/ml in PBS (without Ca2+ and Mg2+), and 25 μl bacteria was immediately mixed with 75 μl blood. 100 μl reactions were performed in sterile heparinized 2 ml round-bottom Eppendorf tubes, in triplicate and incubated at 37°C on a rotary shaker. After three hours of incubation, ten-fold serial dilutions were plated on THB plates to quantify surviving CFU.
Cytokine measurement
IL-1β and TNF-α levels in cell supernatants were measured by ELISA according to the manufacturer’s instructions (BioLegend). IFN-β in cell supernatants was measured via a luciferase L929-ISRE bioassay (kindly provided by B. Beutler) (11). Cell supernatants were harvested and frozen immediately at -80°C, then later incubated with L929-ISRE cells at a dilution of 1:2 for 8 hours. Concentrations of IFN-β were calculated according to a standard curve of luciferase activity from two-fold serial dilutions of IFN-β.
Quantitative RT-PCR
Total RNA was harvested from cells using RNeasy columns (Qiagen) using the manufacturer’s protocol. Total RNA was harvested from skin lesions that were snap frozen in liquid N and then stored at -80° 2 C. Skin was then minced on ice, before being ground using an agate mortar and pestle containing liquid N2. Pulverized samples were put through QIA Shredder columns (Qiagen), incubated with proteinase K, and then run through RNeasy columns (Qiagen) per manufacturer’s protocol. Synthesis of cDNA was completed with MMLV reverse transcriptase (Invitrogen) according to the manufacturer’s recommendations and primed with oligo d(T) (IDT). QRT-PCR was performed on a Realplex Mastercycler (Eppendorf).
Experimental model of in vivo S. aureus infection
Skin infections were performed following an established protocol for generating localized S. aureus subcutaneous infection (21, 28). Briefly, female C57BL/6 mice (7-10 weeks old) underwent hair removal at least 24 hours prior to infection (via shaving and use of Nair). Mice were injected subcutaneously in both flanks with (1×107 CFU). S. aureus strain 113 (WT or ΔOatA) was used for in vivo RT-PCR. S. aureus strain 113 (WT-SA) and S. aureus strain CST222 (bioluminescent Pig1) were used for IFN-β-treated infection studies. Bacterial cultures were washed, diluted, and resuspended in PBS containing 0.5 mg/ml sterile Cytodex beads (GE Healthcare). 50 Units of recombinant murine IFN-β was used in WT-SA infections and 250 Units IFN-β was used in CST222 infections. Mice were anaesthetized with isofluorine and injected with 100 μl on each flank using a 1 ml syringe fitted with a 27 gauge needle. Mice were monitored daily for lesion phenotype and size for WT-SA infections; lesions were harvested after 48 hours of infection and surviving CFU quantified. Mice infected with CST222 were monitored using Xenogen IVIS imaging system (Xenogen Corporation, Alameda, CA) to quantify bioluminescence in the lesion (72 hours post-infection is shown). Images are gray-scale photographs of mice with a color scale overlay that quantifies radiance (photons/sec) within a circular region of interest with Living Image software (Xenogen).
Results
Staphylococcus aureus fails to induce interferon-β in dendritic cells
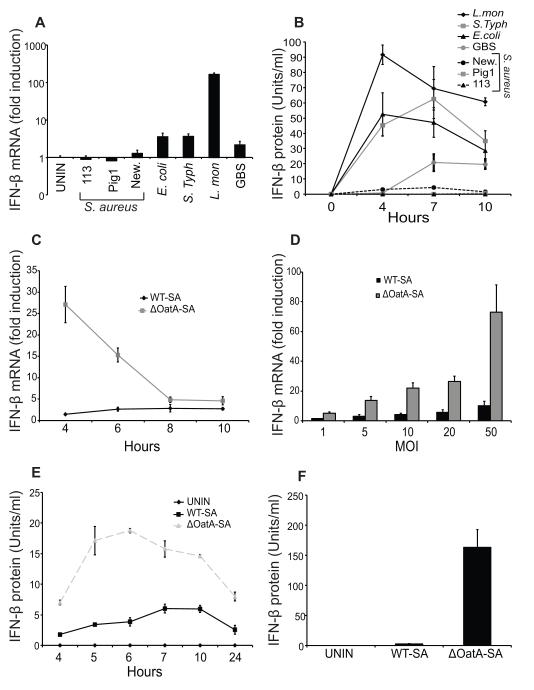
In an attempt to learn more about how Gram-positive bacteria induce type I interferons (IFN) we compared IFN-β production by mouse dendritic cells infected with several different strains of S. aureus or with a panel of bacteria previously reported to induce IFN-β. While control bacteria induced IFN-β mRNA and protein as expected, none of the S. aureus strains did (Fig. 1A, B). Group B Streptococcus (11) is related to S. aureus and readily induced IFN-β production, so it seemed unusual that none of the S. aureus strains did. One major difference between S. aureus and many Gram-positive bacteria is that it is highly resistant to degradation by the lysozyme that is produced in myeloid phagocytes (20). S. aureus makes an enzyme, peptidoglycan O-acetyltransferase, that structurally modifies its cell wall peptidoglycan so that it is not recognized by lysozyme (20). We and others have previously observed that degradation of S. aureus is an important factor in inducing production of certain cytokines by phagocytes (27, 29, 30). We therefore explored whether degradation could lead to induction of IFN-β during S. aureus infection. We infected dendritic cells with a wild type S. aureus, SA113 (WT-SA) or an isogenic mutant lacking peptidoglycan O-acetyltransferase A (ΔOatA-SA) that is sensitive to lysozyme (20). IFN-β mRNA induction was much higher in cells infected with ΔOatA-SA compared to WT-SA over a wide range of infectious doses (Fig. 1C, D). IFN-α did not appear to be induced in response to WT-SA or ΔOatA-SA (data not shown). Infection with ΔOatA-SA also led to greater secretion of IFN-β protein compared to WT-SA (Fig. 1E, F). In contrast, ΔOatA-SA and WT-SA stimulate similar levels of TNF-α production particularly at higher infectious doses, consistent with our previous studies (Supplemental Fig. 1A) (27, 29). This observation is not unique to mouse cells, since we also observed greater production of IFN-β by human dendritic cells infected with ΔOatA-SA compared to WT-SA bacteria (Supplemental Fig. 1B). Taken together the data show that S. aureus poorly induces IFN-β production by human and mouse dendritic cells and that making S. aureus more sensitive to lysozyme enhances IFN-β production.
FIGURE 1. Lysozyme resistance contributes to the failure of Staphylococcus aureus to induce interferon-β (IFN-β) in dendritic cells.
A and B, Bone marrow-derived dendritic cells (BMDC) were infected with Escherichia coli (E. coli), Salmonella enterica serovar Typhimurium (S. Typh), Listeria monocytogenes (L. mon), Streptococcus algalactiae (GBS), and three wild type S. aureus strains, 113, Pig1, and Newman (New.) (MOI 10). RNA was harvested at 4 hours for detection of IFN-β mRNA by RT-PCR (A), and supernatants were harvested at the indicated time points for IFN-β protein bioassay (B). C, BMDC were infected with WT-SA or ΔOatA-SA (MOI 10). RNA was harvested at the indicated time points for detection of IFN-β mRNA by RT-PCR. D, BMDC were infected with WT-SA or ΔOatA-SA (at the indicated multiplicities of infection, MOI). RNA was harvested at 4 hours for detection of IFN-β mRNA by RT-PCR. E, BMDC were infected with WT-SA or ΔOatA-SA (MOI 10). Supernatants were harvested at the indicated time points for IFN-β protein assay. F, BMDC were infected with WT-SA or ΔOatA-SA (MOI 50). Supernatants were harvested at 10 hours for IFN-β protein bioassay. All data are shown as mean ± SD.
Mutant S. aureus induce IFN-β and IL-1β through independent mechanisms
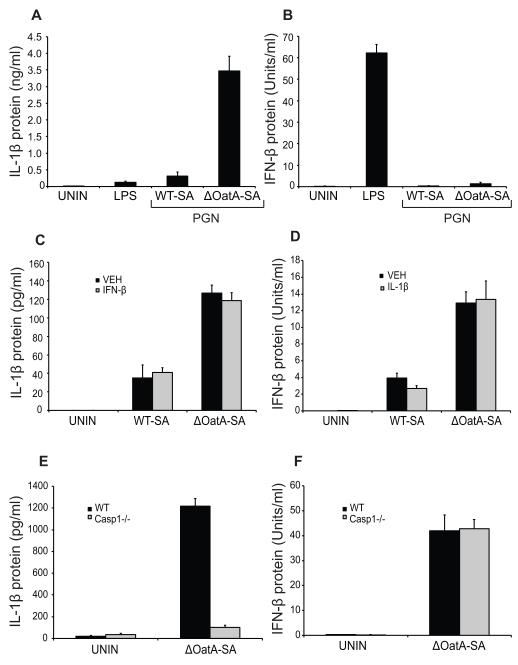
There are a couple publications showing that IL-1β release can be dependent on IFN-β in response to certain intracellular bacteria (31, 32). We have previously shown that S. aureus poorly activates the inflammasome (and subsequent IL-1β release) in myeloid cells, and that this is also mediated by the resistance of the bacteria to lysozyme (Supplemental Fig. 1C) and reference (27). We therefore wondered whether the IL-1β effects we have previously reported were simply caused by the IFN-β changes we have now observed. This does not seem to be the case. First, degradation-sensitive S. aureus peptidoglycan is sufficient to trigger IL-1β production (Fig. 2A) and reference (27), while peptidoglycan is not sufficient to induce IFN-β (Fig. 2B). Second, adding exogenous IFN-β to dendritic cells has no effect on their ability to produce IL-1β in response to WT-SA or ΔOatA-SA infection (Fig. 2C). Therefore, altered IFN-β production was not a cause of our previously reported change in IL-1β. We also do not observe that enhanced IFN-β production during infection with ΔOatA-SA is somehow caused by an increase in IL-1β. First, adding exogenous IL-1β had no effect on the ability of the cells to produce IFN-β in response to WT-SA or ΔOatA-SA infection (Fig. 2D). Second, caspase-1-deficient cells showed no defect in IFN-β production, while IL-1β was blocked in response to ΔOatA-deficient S. aureus (Fig. 2E, F). Taken together, these observations demonstrate that mutant S. aureus induces IFN-β and IL-1β through independent mechanisms.
FIGURE 2. IFN-β induced by ΔOatA-SA is independent of IL-1β. A and B, BMDC were stimulated with LPS (1 μg/ml) or peptidoglycan (40 μg/ml) from WT or ΔOatA S. aureus.
Supernatants were harvested at 10 hours for IL-1β ELISA (A) and IFN-β protein bioassay (B). C, Exogenous IFN-β (100 U/ml) was added to WT-SA or ΔOatA-SA infections (MOI 1) in BMDC. Supernatants were harvested at 10 hours for IL-1β ELISA. D, Exogenous IL-1β (500 pg/ml) was added to WT-SA or ΔOatA-SA infections (MOI 1) in BMDC. Supernatants were harvested at 10 hours for IFN-β protein bioassay. E and F, WT and Caspase-1−/− BMDC were infected with ΔOatA-SA (MOI 10). Supernatants were harvested at 10 hours for IL-1β ELISA (E) and IFN-β protein bioassay (F). All data are shown as mean ± SD.
IFN-β induction by mutant S. aureus is MyD88-dependent but TLR-independent
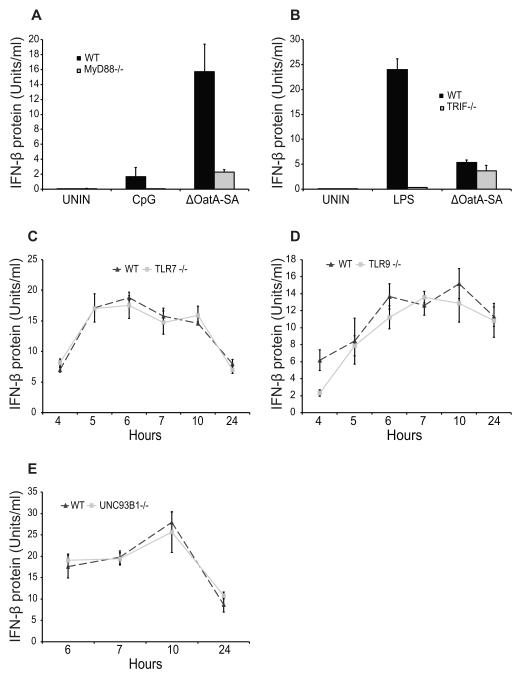
We next set out to define how degradation-sensitive S. aureus induces IFN-β production. Several Toll-like receptors (TLR) are known to induce IFN-β, including TLR3 and TLR4 (which both utilize the adaptor TRIF to signal) and TLR7 and TLR9 (which are dependent on MyD88 as an adaptor molecule) (8). We therefore tested the ability of ΔOatA-SA to induce IFN-β in both MyD88-deficient and TRIF-deficient dendritic cells. MyD88 was required for IFN-β secretion in response to infection with ΔOatA-SA, however TRIF was not (Fig. 3A, B). This suggested that TLR7 and/or TLR9 might be involved. TLR7 recognizes single-stranded RNA within endosomes and requires MyD88 to signal, so we infected TLR7-deficient cells with ΔOatA-deficient S. aureus to see if they were defective for IFN-β production. ΔOatA-SA induced IFN-β was not dependent on TLR7 (Fig. 3C). TLR9 is another endosomal TLR that recognizes CpG DNA and like TLR7 requires MyD88 to signal. TLR9 was also not required for ΔOatA-SA induced IFN-β (Fig. 3D). We therefore hypothesized that ΔOatA-SA could stimulate both receptors and that loss of TLR7 or TLR9 could be compensated for by the other. To test this, we infected UNC93B1-deficient cells with ΔOatA-SA. UNC93B1 is an endoplasmic reticulum membrane protein required for folding and trafficking of all three endosomal TLRs (TLR3, TLR7, and TLR9) and thus UNC93B1-deficient cells fail to express all three receptors (8, 33). Surprisingly, UNC93B1 was not required for IFN-β induced by degradation-sensitive S. aureus (Fig. 3E). We also tested whether IFN-β induced by ΔOatA-SA was TLR2-dependent by infecting TLR2-deficient cells, but also found no difference from wild type cells (data not shown). Together the data show that IFN-β induction in response to ΔOatA-SA is MyD88-dependent yet TLR-independent, a situation that is similar to what has recently been observed for Group A Streptococcus (Streptococcus pyogenes) (34).
FIGURE 3. IFN-β induced by ΔOatA-SA is MyD88-dependent but TLR-independent.
A, WT and MyD88−/− BMDC were stimulated with CpG (5 μg/ml) or infected with ΔOatA-SA (MOI 1). Supernatants were harvested at 10 hours for IFN-β protein bioassay. B, WT and TRIF−/− BMDC were stimulated with LPS (100 ng/ml) or infected with ΔOatA-SA (MOI 1). Supernatants were harvested at 10 hours for IFN-β protein bioassay. C, WT and TLR7−/− BMDC were infected with ΔOatA-SA (MOI 1). Supernatants were harvested at the indicated time points for IFN-β protein bioassay. D, WT and TLR9−/− BMDC were infected with ΔOatA-SA (MOI 1). Supernatants were harvested at the indicated time points for IFN-β protein bioassay. E, WT and UNC93B1−/− BMDC were infected with ΔOatA-SA (MOI 1). Supernatants were harvested at the indicated time points for IFN-β protein bioassay. All data are shown as mean ± SD.
IFN-β induction by mutant S. aureus requires internalization and degradation of bacteria and intact RNA species, but is independent of cytosolic RNA sensors RIG-I and Mda5
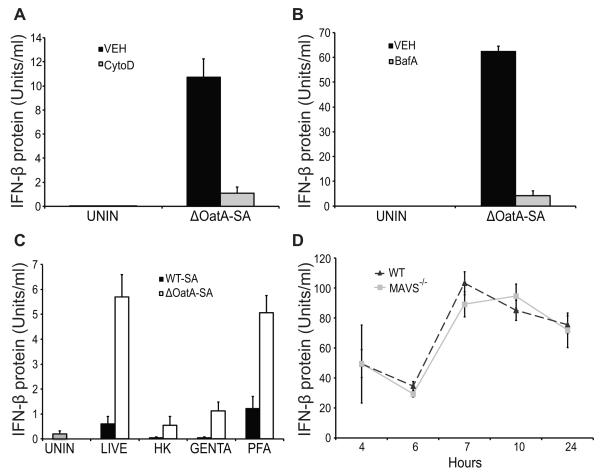
Having observed that TLRs are not involved in IFN-β induction by ΔOatA-SA, we next explored other processes that might be required. In our previous work with these bacteria, we noted that phagocytosis and degradation within phagolysosomes is important for induction of certain inflammatory responses (27, 29). We therefore examined whether these cellular processes are important for induction of IFN-β. When we treated cells with cytochalasin D to block phagocytosis, ΔOatA-SA-induced secretion of IFN-β was blocked (Fig. 4A). Also, when we treated cells with bafilomycin A to prevent acidification of lysosomes, IFN-β production was blocked (Fig. 4B). Together, these results suggested involvement of an internal receptor recognizing ligands exposed in the process of bacterial degradation, but the ligand and the response were different from any we had previously looked at.
FIGURE 4. IFN-β induced by ΔOatA-SA requires phagocytosis, lysosomal acidification, and intact RNA.
A, BMDC were pre-treated with cytochalasin D (CytoD) for 30 minutes and infected with ΔOatA-SA (MOI 1). Supernatants were harvested at 10 hours for IFN-β protein bioassay. B, BMDC were pre-treated with bafilomycin A (BafA) for 30 minutes and infected with ΔOatA-SA (MOI 10). Supernatants were harvested at 10 hours for IFN-β protein bioassay. C, BMDC were infected with live, heat-killed (HK), gentamycin-killed (Genta.), or paraformaldehyde-killed (PFA) WT-SA or ΔOatA-SA (MOI 1). Supernatants were harvested at 18 hours for IFN-β protein bioassay. D, WT and MAVS−/− BMDC were infected with ΔOatA-SA (MOI 10). Supernatants were harvested at the indicated time points for IFN-β protein bioassay. All data are shown as mean ± SD.
Recent work by Sander et al demonstrated the importance of “vita-PAMPs”, pattern associated molecular patterns that are found in live but not dead bacteria (35). The study showed that E. coli RNA released during phagocytosis could stimulate IFN-β production. The authors showed that RNA vita-PAMPs are present in live bacteria and formaldehyde-preserved bacteria, but are lost in heat- and antibiotic-killed bacteria. Whether a similar mechanism is at work for discrimination between live and dead Gram-positive bacteria is not known. We therefore tested whether IFN-β induced by ΔOatA-deficient S. aureus was dependent on the presence of vita-PAMPs by exposing dendritic cells to live, heat-killed, gentamycin-killed, or paraformaldehyde-fixed WT-SA and ΔOatA-SA. Killing bacteria with heat or gentamycin, conditions that promote degradation of bacterial RNAs (35), abrogated IFN-β production. In contrast, killing bacteria with formaldehyde, a condition that preserves bacterial RNAs, maintained IFN-β production (Fig. 4C). Taken together, the data demonstrate that wild type S. aureus avoids inducing IFN-β by making itself resistant to degradation within phagosomes and preventing the release of vita-PAMPs. In an effort to identify the receptor that ΔOatA-SA RNA stimulates to induce IFN-β, we explored whether the cytosolic receptors RIG-I or MDA5 might play a role. These receptors recognize RNA in the cytosol leading to IFN-β production and thus seemed plausible candidates. We infected MAVS-deficient cells (an adaptor required by both RIG-I and MDA5 for signaling) with ΔOatA-SA and compared the amount of IFN-β protein produced to similarly infected wild-type cells, however we saw no decrease in the MAVS-deficient cells (Fig 4D). RIG-I and MDA5 do not appear to play a role in IFN-β induction by ΔOatA-SA.
IFN-β stimulates host responses that are protective against S. aureus
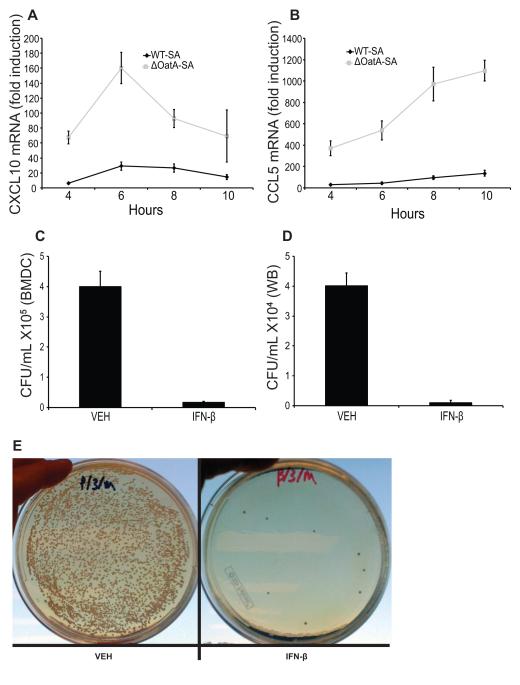
In different bacterial infection models, IFN-β can either be protective or detrimental to the host (1). Since wild type S. aureus is able to avoid production of IFN-β in myeloid cells through synthesis of degradation-resistant peptidoglycan and reduced release of vita-PAMPs, we wondered whether reducing IFN-β production would be beneficial to the bacteria. IFN-β is a potent cytokine known to turn on hundreds of downstream genes (36, 37). We measured two such genes in dendritic cells infected with ΔOatA-SA compared to WT-SA. Both CXCL10 and CCL5 were highly up regulated in cells infected with ΔOatA-SA (Fig. 5A, B). Thus, reduced IFN-β secretion in response to wild type S. aureus also reduces induction of downstream genes, many of which are chemokines important for orchestrating local inflammatory responses including neutrophil recruitment (38).
FIGURE 5. IFN-β induces chemokines and enhances phagocyte killing of S. aureus.
A and B, BMDC were infected with WT-SA or ΔOatA-SA (MOI 10) and RNA was harvested at the indicated time points. CXCL10 (A) and CCL5 (B) mRNAs were measured by RT-PCR. C, BMDC were infected (MOI 10) with WT-SA plus 100 Units/ml recombinant murine IFN-β or vehicle control (-). Cells were lysed after 24 hours of infection and surviving intracellular CFU were plated and counted. D and E, Murine whole blood was treated with PBS (-) or recombinant murine IFN-β (final concentration 103 Units/ml) and mixed with WT-SA (final concentration 1×103 CFU/ml). Blood was incubated for 3 hours, and surviving bacteria were plated and counted (D). Representative images of PBS or IFN-β treated infected whole blood (E).
We also examined whether the presence of IFN-β could directly affect the ability of cells to kill S. aureus. Addition of exogenous IFN-β to dendritic cells infected with WT-SA substantially aided in clearance of bacteria after 24 hours (Fig. 5C). Of course, IFN-β acts on many cell types other than dendritic cells, and we therefore wanted to examine the broader effects of IFN-β on cellular responses to S. aureus. We treated fresh murine whole blood with IFN-β and infected with WT-SA. IFN-β-treated whole blood had fewer surviving bacteria after 3 hours of infection compared to blood treated with vehicle (Fig. 5D, E). Thus, the presence of IFN-β during infection with wild type S. aureus directly activates phagocytes to kill bacteria.
IFN-β production is suppressed during in vivo S. aureus infection and adding exogenous IFN-β enhances protection
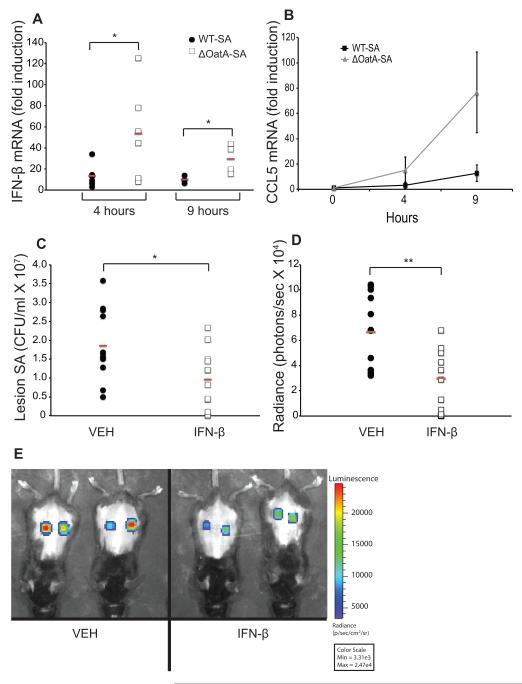
Having demonstrated that the presence of IFN-β could enhance bacterial killing in vitro, we next wanted to investigate the role of this cytokine in vivo. Using a subcutaneous model of S. aureus infection we measured the induction of IFN-β mRNA in lesions infected with WT-SA or ΔOatA-SA at four and nine hours post-infection. ΔOatA-SA lesions had significantly increased IFN-β expression over WT-SA at both time points (Fig. 6A). In addition, CCL5, an important chemokine induced by IFN-β, was also expressed at higher levels in lesions infected with degradation-sensitive S. aureus (Fig. 6B). These data agree with the in vitro studies and confirm that S. aureus is a poor inducer of IFN-β during in vivo infection.
FIGURE 6. IFN-β production is suppressed during in vivo S. aureus infection, and adding exogenous IFN-β enhances protection.
A and B, Mice were injected subcutaneously with 1×107 CFU/ml WT-SA or ΔOatA-SA (3 mice/group, 2 lesions/mouse, n=6 lesions/group). Lesions were harvested and RNA was extracted at the indicated time points for detection of IFN-β (A) and CCL5 (B) mRNA by RT-PCR. C, Mice were injected subcutaneously with 1×107 CFU WT-SA with a vehicle control or with 50 Units recombinant murine IFN-β (6 mice/group, 2 lesions/mouse, n=12 lesions/group). Skin lesions were harvested at 48 hours post infection, and surviving bacteria were plated and counted. D, Mice were injected subcutaneously with 1×107 CFU of the clinical S. aureus isolate Pig1 (bioluminescent strain CST222) with a vehicle control or with 250 Units recombinant murine IFN-β (6 mice/group, 2 lesions/mouse, n=12 lesions/group). Bioluminescence was quantified at 72 hours post infection. E, Representative Xenogen images of skin lesions at 72 hours. Red bars indicate means. * p < 0.05, ** p < 0.01 (unpaired two-tailed t test).
Since S. aureus is a poor inducer of IFN-β during infection, we next explored whether providing exogenous IFN-β could be a viable strategy for enhancing host defense against S. aureus. We infected mice subcutaneously with WT-SA alone or together with 50 Units of IFN-β and assessed CFU in the lesion and lesion size after 48 hours. IFN-β treatment significantly reduced the number of S. aureus recovered from lesions after 48 hours indicating that the cytokine is protective in vivo (Fig. 6C). Enhanced bacterial clearance in IFN-β-treated mice was also associated with accelerated formation of necrotic lesions 24 and 48 hours after infection (Supplementary Fig. 2A, B), consistent with a more aggressive inflammatory response to the bacteria. WT-SA is a commonly used laboratory strain (SA113) and we therefore wondered whether IFN-β could be used to suppress growth of a clinically important strain as well. We generated a bioluminescent form of S. aureus strain Pig1 (a highly pigmented strain isolated from the skin of a child with atopic dermatitis) that we have confirmed fails to stimulate IFN-β production from immune cells (Fig. 1A, B). We infected mice subcutaneously with S. aureus Pig1 alone or together with 250 Units of IFN-β and assessed bacterial load in the lesion by Xenogen imaging after 72 hours. IFN-β treatment significantly reduced the number of S. aureus in lesions indicating that the cytokine is protective against a clinical strain of bacteria as well (Fig. 6D, E). Together, these data show that S. aureus reduces degradation-induced IFN-β production by phagocytes, resulting in less local inflammatory cell recruitment and enhanced bacterial survival.
Discussion
In this study, we have shown that S. aureus is a poor inducer of IFN-β production by host immune cells in vitro and in vivo and that the failure to stimulate IFN-β production is beneficial to the bacteria. S. aureus avoids host activation of IFN-β in phagocytes through modifications to its cell wall that make it resistant to degradation. This blocks release of molecules from live bacteria that would otherwise be released during killing and degradation in phagosomes and would stimulate IFN-β production. Using a mouse model of S. aureus skin infection, we show that treatment with IFN-β can promote clearance of bacteria.
The lack of IFN-β induced in myeloid cells by S. aureus is notable with respect to other Gram-positive bacterial species that induce large amounts of IFN-β, including Group B Streptococcus (Streptococcus agalactiae) (11), Group A Streptococcus (Streptococcus pyogenes) (12), and Streptococcus pneumonia (34). Consistent with previous reports, we observed that Group B Streptococcus stimulates IFN-β production in myeloid cells. An important difference between Group B Streptococcus and S. aureus is that it does not modify its cell walls to make it resistant to phagosomal degradation the way that S. aureus does. We hypothesize that this is the reason that Group B Streptococcus stimulates IFN-β production, but S. aureus does not.
We have previously shown that degradation of S. aureus is important for activation of the inflammasome and secretion of IL-1β (27, 27, 27, 27), although our current study shows that the mechanisms by which degradation promotes induction of IL-1β and IFN-β are different. Purified, degradation-sensitive peptidoglycan (from ΔOatA-SA) induces IL-1β production, while wild type peptidoglycan does not (27). This suggests that the inflammasome is activated by degradation products of peptidoglycan. In contrast, we have observed here that IFN-β production is not stimulated by purified peptidoglycans from either wild type or mutant S. aureus, suggesting that the activating ligands for this response are not part of peptidoglycan.
Some recent reports have suggested that IL-1β production and IFN-β production are interrelated (31, 32), however the two responses appear completely independent of each other in our study of S. aureus. Exogenous addition of either cytokine does not affect the production of the other in response to infection with S. aureus, and IFN-β production is not affected in caspase-1 deficient cells. These findings are consistent with the study by Henry et al. in which IFN-β and IL-1β production are independent of each other in response to extracellular/vacuolar bacteria such as Salmonella, but dependent on each other in response to infection with intracellular bacteria, such as Francisella tularensis and Listeria monocytogenes (31).
We have previously observed that degradation of S. aureus in phagosomes can lead to exposure of bacterial DNA that triggers TLR9-mediated enhancement of production of classical inflammatory cytokines such as IL-6 and TNF-α (29), and we were thus surprised to discover that TLR9 and other endosomal TLRs do not make significant contributions to induction of IFN-β. Instead, we found that IFN-β production is dependent largely on release of vita-PAMPs as defined by Sander et al. (35). These authors demonstrated that mRNA from live E. coli within phagosomes is able to leak out and leads to activation of inflammatory responses (including IFN-β production). In contrast, mRNAs are rapidly degraded in dead bacteria and thus are not released when these bacteria are phagocytosed. This led the authors to coin the term “vita-PAMPs” to describe inflammatory ligands associated with live, but not dead microbes. Our data are consistent with this concept in that dead S. aureus (even degradation-sensitive S. aureus) do not stimulate IFN-β production unless they have been killed in a manner that preserves mRNA (i.e. fixed with paraformaldehyde). Thus we have extended the previous work showing that vita-PAMPs from Gram-negative bacteria can influence inflammatory signaling to now include vita-PAMPs from a Gram-positive organism. Further, we document a strategy by which a live bacterium can prevent exposure of vita-PAMPs and the resulting immune responses. One way in which recognition of E. coli and S. aureus vita-PAMPs appear to be different is in their requirement for MyD88. Most inflammatory cytokines induced by live E. coli are MyD88-independent, while we have observed that IFN-β induction by live degradation-sensitive S. aureus is MyD88-dependent, even though this response is TLR-independent. Recent work by Gratz et al. has also described a MyD88-dependent, TLR-independent type of IFN-β induction for Group A Streptococcus (34).
We have demonstrated that the ability of S. aureus to avoid inducing IFN-β during infection in vivo is significant for its survival. Upon infection with a degradation-sensitive S. aureus, IFN-β induction lead to activation of downstream chemokines, including CXCL10 and CCL5, that are important for orchestration of local inflammatory responses. Further, treating dendritic cells, whole blood, and mice with exogenous IFN-β increased protection against infection with S. aureus. The protective effects of IFN-β during skin infection were seen for two different strains of S. aureus, including a medically relevant clinical isolate, Pig1. The beneficial role of IFN-β during S. aureus infection was previously unrecognized and adds to our understanding of how this important human pathogen is able to subvert the host’s immune defenses.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 GM085796 to D.M.U., T32 AI089553-01 to A.J.W., and R01 AI074832 to G. Y. L.).
References
- 1.Decker T, Müller M, Stockinger S. The Yin and Yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 2.Bourgeois C, Majer O, Frohner IE, Lesiak-Markowicz I, Hildering KS, Glaser W, Stockinger S, Decker T, Akira S, Muller M, Kuchler K. Conventional Dendritic Cells Mount a Type I IFN Response against Candida spp. Requiring Novel Phagosomal TLR7-Mediated IFN-Signaling. J. Immunol. 2011;186:3104–3112. doi: 10.4049/jimmunol.1002599. [DOI] [PubMed] [Google Scholar]
- 3.Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, Halaas O, Nilsen NJ, Stenmark H, Latz E, Lien E. The Rab11a GTPase Controls Toll-like Receptor 4-Induced Activation of Interferon Regulatory Factor-3 on Phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sing A, Merlin T, Knopf HP, Nielsen PJ, Loppnow H, Galanos C, Freudenberg MA. Bacterial induction of beta interferon in mice is a function of the lipopolysaccharide component. Infect. Immun. 2000;68:1600–1607. doi: 10.1128/iai.68.3.1600-1607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freudenberg MA, Merlin T, Kalis C, Chvatchko Y, Stübig H, Galanos C. Cutting edge: a murine, IL-12-independent pathway of IFN-gamma induction by gram-negative bacteria based on STAT4 activation by Type I IFN and IL-18 signaling. J. Immunol. 2002;169:1665–1668. doi: 10.4049/jimmunol.169.4.1665. [DOI] [PubMed] [Google Scholar]
- 6.Karaghiosoff M, Steinborn R, Kovarik P, Kriegshäuser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, Levy D, Decker T, Müller M. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 7.Monroe KM, Monroe KM, McWhirter SM, McWhirter SM, Vance RE, Vance RE. Induction of type I interferons by bacteria. Cellular Microbiology. 2010;12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasius AL, Beutler B. Intracellular Toll-like Receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Parker D, Martin FJ, Soong G, Harfenist BS, Aguilar JL, Ratner AJ, Fitzgerald KA, Schindler C, Prince A. Streptococcus pneumoniae DNA Initiates Type I Interferon Signaling in the Respiratory Tract. mBio. 2011;2:e00016–11. doi: 10.1128/mBio.00016-11. e00016–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao N, Eidenschenk C, Krebs P, Brandl K, Blasius AL, Xia Y, Khovananth K, Smart NG, Beutler B. The Tpl2 Mutation Sluggish Impairs Type I IFN Production and Increases Susceptibility to Group B Streptococcal Disease. J. Immunol. 2009;183:7975–7983. doi: 10.4049/jimmunol.0902718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gratz N, Siller M, Schaljo B, Pirzada ZA, Gattermeier I, Vojtek I, Kirschning CJ, Wagner H, Akira S, Charpentier E, Kovarik P. Group A Streptococcus Activates Type I Interferon Production and MyD88-dependent Signaling without Involvement of TLR2, TLR4, and TLR9. Journal of Biological Chemistry. 2008;283:19879–19887. doi: 10.1074/jbc.M802848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertheim H, Melles D, Vos M, van Leeuwen W, van Belkum A, Verbrugh H, Nouwen J. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 16.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin FJ, Gomez MI, Wetzel DM, Memmi G, O’Seaghdha M, Soong G, Schindler C, Prince A. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J. Clin. Invest. 2009;119:1931–1939. doi: 10.1172/JCI35879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lizak M, Yarovinsky TO. Phospholipid Scramblase 1 Mediates Type I Interferon-Induced Protection against Staphylococcal α-Toxin. Cell Host Microbe. 2012;11:70–80. doi: 10.1016/j.chom.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roquilly A, Gautreau L, Segain JP, de Coppet P, Sebille V, Jacqueline C, Caillon J, Potel G, Lejus C, Josien R, Asehnoune K. CpG-ODN and MPLA Prevent Mortality in a Murine Model of Post-Hemorrhage-Staphyloccocus aureus Pneumonia. PLoS ONE. 2010;5:e13228. doi: 10.1371/journal.pone.0013228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bera A, Herbert S, Jakob A, Vollmer W, Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu GY. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. Journal of Experimental Medicine. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francis KP, Joh D, Bellinger-Kawahara C, Hawkinson MJ, Purchio TF, Contag PR. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 2000;68:3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng CW, Stewart GC. Rot repression of enterotoxin B expression in Staphylococcus aureus. J. Bacteriol. 2005;187:5301–5309. doi: 10.1128/JB.187.15.5301-5309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Paolo NC, Miao EA, Iwakura Y, Murali-Krishna K, Aderem A, Flavell RA, Papayannopoulou T, Shayakhmetov DM. Virus binding to a plasma membrane receptor triggers interleukin-1 alpha-mediated proinflammatory macrophage response in vivo. Immunity. 2009;31:110–121. doi: 10.1016/j.immuni.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim Y-M. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. The Journal of Cell Biology. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Götz F, Liu GY, Underhill DM. Staphylococcus aureus Evades Lysozyme-Based Peptidoglycan Digestion that Links Phagocytosis, Inflammasome Activation, and IL-1β Secretion. Cell Host Microbe. 2010;7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 29.Wolf AJ, Arruda A, Reyes CN, Kaplan AT, Shimada T, Shimada K, Arditi M, Liu G, Underhill DM. Phagosomal degradation increases TLR access to bacterial ligands and enhances macrophage sensitivity to bacteria. J. Immunol. 2011;187:6002–6010. doi: 10.4049/jimmunol.1100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ip WKE, Sokolovska A, Charriere GM, Boyer L, Dejardin S, Cappillino MP, Yantosca LM, Takahashi K, Moore KJ, Lacy-Hulbert A, Stuart LM. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J. Immunol. 2010;184:7071–7081. doi: 10.4049/jimmunol.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. Journal of Experimental Medicine. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, Farlik M, Decker T, Du Pasquier RA, Romero P, Tschopp J. Type I Interferon Inhibits Interleukin-1 Production and Inflammasome Activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi G-P, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, Drobits B, Li X-D, Knapp S, Kovarik P. Type I Interferon Production Induced by Streptococcus pyogenes-Derived Nucleic Acids Is Required for Host Protection. PLoS Pathog. 2011;7:e1001345. doi: 10.1371/journal.ppat.1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Müller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platanias LC, Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 37.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly-Scumpia KM, Scumpia PO, Delano MJ, Weinstein JS, Cuenca AG, Wynn JL, Moldawer LL. Type I interferon signaling in hematopoietic cells is required for survival in mouse polymicrobial sepsis by regulating CXCL10. Journal of Experimental Medicine. 2010;207:319–326. doi: 10.1084/jem.20091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.