A mechanistic understanding of the molecular transactions that govern cellular function requires knowledge of the dynamic organization of the macromolecular machines involved in these processes. Structural biologists employ a variety of biophysical methods to study large macromolecular complexes, but no single technique is likely to provide a complete description of the structure-function relationship of all the constituent components. Since structural studies generally only provide snapshots of these dynamic machines as they accomplish their molecular functions, combining data from many methodologies is crucial to our understanding of molecular function.
Introduction
The intimate relationship between form and function makes structural characterization of macromolecular complexes a powerful tool in understanding the molecular mechanisms that underlie biological function. Visualizing the three-dimensional organization of a biological molecular machine not only helps us conceptualize the assembly’s biochemical properties, but also leads to new mechanistic models that can be further tested. The resolution (spatial detail) and the scope (size of the described sample) of structure-function studies depend largely on the methodology utilized to probe the system. While X-ray crystallography is without question the most popular and successful technique used to analyze molecular structure in atomic detail, the bottleneck of crystallization limits its general applicability. Structure determination by NMR, on the other hand, becomes very difficult for proteins larger than 50 kD. As a result, these “classical” structure techniques may be able to deal only with a subset or small facet of larger complexes.
Structural studies of large macromolecular complexes intractable by X-ray crystallography or NMR have long been the realm of cryo-electron microscopy (cryoEM), which is not limited by size (the bigger the better) nor requires large amounts of sample at high concentration. Developments in both cryoEM technology and image processing software have led to a number of reconstructions solved to better than 4Å resolution, allowing for ab initio chain tracing [1–3]. However, these studies involved exceptionally well-behaved, highly symmetric samples. More generally, the applicability of the cryo-EM methodology, together with the advent of automated data acquisition and more powerful computing resources, has resulted in an exponential growth in the number of cryoEM reconstructions at subnanometer resolutions, including small (<0.5 megadaltons) asymmetric complexes [4–13].
Generally, the structures derived from any of these methods provide only snapshots from the conformational landscape that often characterizes macromolecular function. For this reason, the combination of multiple methodologies holds vast potential in more completely describing the dynamic rearrangements accompanying this landscape. Within this context, hybrid methodology provides informational value greater than the sum of individual techniques. A generally used hybrid approach over the last decade involves rigid-body fitting of high-resolution structures of the constituent fragments into cryoEM reconstructions of full complexes. Here we concentrate on two families of hybrid studies involving cryoEM: those where crystallographic structures are not available for all the components within an assembly, and those dedicated to characterizing the dynamic nature of macromolecular assemblies. In both cases additional information was required beyond EM volumes and X-ray atomic coordinates.
Going Hybrid to Define the Subunit Architecture of Large Assemblies
In particularly favorable cases, atomic-resolution structures are available for most, if not all, of the constitutive components of the assembly being studied by cryoEM. In such cases, subnanometer resolution EM maps not only permit delineation of subunit and domain boundaries, but discernable secondary structure elements allow for unambiguous positioning of atomic structures with an accuracy that far exceeds the resolution of the reconstruction itself. The resulting pseudo-atomic model of the entire macromolecular complex defines the precise location of functional elements, informs on protein-protein interfaces, and provides unique functional information about the complex as a whole.
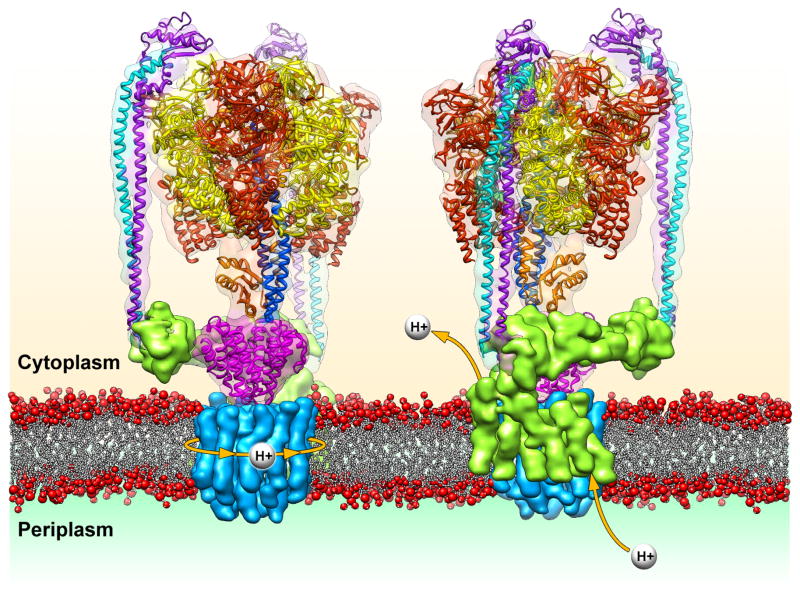
Due to less stringent experimental requirements, it is common for low-resolution cryoEM reconstructions of macromolecular complexes to be solved before all (or even any) atomic-resolution structures of its assembly components are available. In these cases, the EM density may still provide valuable information about the arrangement of proteins within the complex. Docking of the available atomic structures into the cryoEM density can itself shed light on the position and role of the other portions of the complex. This is exemplified in the recent work of Lau and Rubinstein on the Thermus thermophilus ATP synthase [14,15]. ATP synthases consist of a membrane-embedded region, so far intractable by crystallography, that is connected to an extramembranous catalytic subcomplex by both a central stalk and one or more peripheral stalks. Lau and Rubinstein’s single particle analysis by cryoEM revealed the subnanometer structure of an intact ATP synthase, and docking of crystal structures provided key insights into the protein interactions within the catalytic cytoplasmic domain, as well as how these components are structurally coupled to the membrane-embedded ring subcomplex [16–18].
Of particular importance, however, was the ability to define the remaining elements in the structure as the transmembrane helices of both the membrane-bound rotary ring and subunit I, revealing their mode of interaction. Two distinct clusters of helices within subunit I each interact exclusively with specific rotary subunits, one closer to the periplasm and the other closer to the cytoplasm (Figure 1). This organization supports a two half-channel ion-translocating mechanism, in which one helical bundle of subunit I channels protons from the periplasm to the rotary subunits, while the other conducts protons from the rotary subunits to the cytoplasm. The authors propose that the movements of the rotary subunits are transferred to the central rotor by a funnel-shaped connector, linking the transmembrane proton motive force to ATP synthesis by the catalytic domains.
Figure 1.
Organization of the ATP synthase and a model for proton translocation through the membrane. The crystal structures corresponding to the subunits in the extracellular domain are docked into the subnanometer EM reconstruction (EMDB ID: 5335) [15] to show the atomic organization of the catalytic hexamer (red and yellow) as it is held in position by two peripheral stalks (cyan and purple) and the central stalk (orange and dark blue). Motions from the membrane-embedded L-ring rotary motor (blue) are propagated to this central stalk via the cone-shaped C-subunit (magenta). In the model proposed by Lau and Rubinstein, [15] protons would drive the rotation of the L-ring motor as they enter through the periplasmic half-channel of subunit I and exit through the cytoplasmic half-channel.
The 26S proteasome is a classic example of a large macromolecular complex that has been the target of structural studies for several decades, but whose atomic structure remains elusive. The dynamic nature and labile character of the 19S regulatory particle (RP), which controls access to the proteolytic chamber, has significantly hampered efforts to define its structural organization. Atomic-resolution structures of some subunits or fragments had been determined [19–21], but their relative arrangement within the RP could not be decisively defined, limiting our understanding of their contributions to RP function.
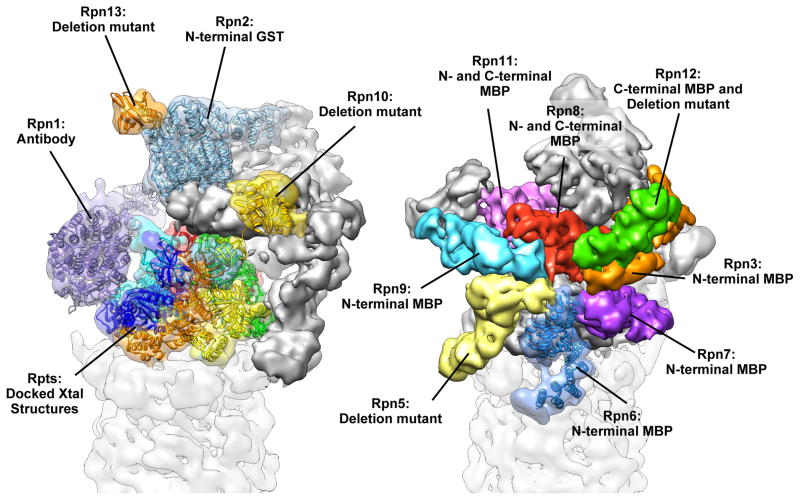
Two recent studies have combined cryoEM reconstructions of the 26S proteasome with biochemical data to provide a much more complete understanding of the RP architecture [22,23]. Although making use of differing methodologies, these studies point to identical models of molecular organization. In the study by Lasker et al., cross-linking/MS experiments [24] were combined with previously determined protein-protein interactions [25–27] and crystal structures, and placed in the context of a subnanometer reconstruction of the 26S to arrive at a description of the RP subunit organization [23]. Martin and colleagues took advantage of a heterologous expression system of the “lid” subcomplex [22], locating components using maltose-binding protein (MBP) fusions and negative stain EM analyses. Combined with antibody and GST-fusion labeling of the RP “base” subcomplex, these studies directly describe the complete architecture of the proteasome RP (Figure 2). This approach could in principle be applied to any new macromolecular complex lacking an extensive history of proteomic studies. More recently, and following determination of additional crystal structures of RP components [28,29] daFonseca et al. [30] have proposed a slightly modified organization for the lid subunits within a cryo-EM reconstruction of the human 26S.
Figure 2.
CryoEM reconstruction of the regulatory particle (RP) of the 26S proteasome (EMDB ID: 1992) [22]. The biochemical marker used to localize each subunit in the study by Martin and colleagues is noted, and atomic coordinates for known or homologous components are shown docked into their corresponding density. The complex is shown from two angles, emphasizing the two subcomponents of the RP. Colored in the left view are the subunits that constitute the base sub-complex and the ubiquitin receptors, and colored on the right are the subunits of the lid sub-complex. The Rpn2 and Rpn6 crystal structures (PDB IDs 4ady and 3txm, respectively [28,29]) were determined after completion of the initial cryoEM study. Docking of these structures illustrates the fidelity of subunit boundary delineation that was provided by the subnanometer reconstructions in the absence of crystal structures. Additionally, docking the Rpn2 crystal structure into the Rpn1 density evidences the high level of structural homology between the two largest components of the proteasome.
Although crystal structures provide precise atomic information, there are frequently unstructured or dynamic regions that do not crystallize and are not visualized in the structure. Such low-complexity regions are frequent sites of post-translational modification and are commonly involved in regulating protein-protein interactions. Often times these unstructured regions become at least partially ordered in the context of a larger assembly. High-resolution cryoEM may allow visualization of these extended segments and insight into their role at interfaces. Extended segments of viral coat proteins are often involved in viral assembly and thus can be described by visualization of fully assembled viruses. One beautiful example is that described by Harrison and coworkers in the study of rotavirus VP7 protein [31]. The authors more recently also visualized the rotavirus penetration protein VP4 in infectious particles [32], revealing an unexpected architecture that resolved many of the perplexing questions regarding rotavirus penetration. Another example, albeit at lower resolution, is the study of microtubules interacting with the kinetochore complex Ndc80. The disordered N-terminal tail of Ndc80 mediates interactions with other Ndc80 molecules, resulting in a self-organization of the complex into clusters along microtubules. Docking of crystal structures revealed a prominent extra density not accounted for by the atomic coordinates, which extended from the N-terminus in a staggered fashion between the globular domains of the complex [33]. Importantly, removal or phosphorylation of this segment abrogates clustering, confirming its involvement in the self-association of Ndc80 complexes.
Subnanometer resolutions like those in the examples mentioned above are not always necessary for accurate positioning of atomic structures into cryoEM density, provided there is sufficient data from other biophysical and biochemical studies. Recent work by Melero et al. reveals the pseudo-atomic architecture of the UPF surveillance complex, a central component of the nonsense-mediated decay pathway, by integrating the results from mass spectrometry, protein and nucleic acid labeling, and biochemical interaction data, into a 16Å-resolution cryoEM reconstruction [34]. The resulting model provides a structural description of how this enzyme is stabilized at an exon junction complex, such that its helicase region of the complex is appropriately situated to remodel the 3′ end of an mRNP.
Localization of specific subunits in complexes purified from endogenous sources is commonly pursued using antibody labeling, but this approach depends on the affinity of the antibody for the epitope in the context of the assembled complex, and often suffers from substoichiometric labeling. When a recombinant expression system exists for the complex, genetic tags are a significant advantage, as demonstrated in the proteasome lid study mentioned previously. In addition to localizing a subunit by tagging one or both of its termini, internal tags can allow the effective “tracing” of the polypeptide path of large subunits. A recent implementation of this idea has been successfully utilized to effectively establish the architecture of the functional domains in human Dicer [35]. By inserting the 15–amino acid AviTag sequence, a substrate for biotin-protein ligase, into surface loops along the structure of this enzyme, followed by biotinylation and tagging with a monovalent form of streptavidin, the protein was visualized by negative stain EM to localize the position of the extra streptavidin density.
An alternative internal tagging method recently implemented for EM labeling purposes takes advantage of the fact that the N- and C-termini of green fluorescent protein (GFP) are in close spatial proximity to one another, such that internal GFP tags, connected by a short loop, can be integrated at desired sites along a main protein chain. This strategy has been used, in combination with isotopic chemical cross-linking and mass spectrometry, to localize all subunit domains within the gene silencing complex PRC2 and generate a detailed map of interactions across the assembly (Claudio Ciferri, G.C.L. and E.N., unpublished results).
Going Hybrid to See Large Macromolecules in Action
Given that many complexes undergo dramatic rearrangements in order to accomplish a molecular task, the goal of many cryoEM studies is to derive multiple reconstructions describing the different states along the conformational trajectory of a given macromolecular assembly. Ideally this is performed through biochemical selection of specific states and studying them individually. An example is the study of the bacterial CASCADE complex involved in RNA-guided immunity by Wiedenheft et al. [36], in which the subnanometer cryoEM structures of this nucleoprotein complex were solved before and after binding to its nucleic acid target. The subunit organization of the complex was defined using a couple of existing structures of homologues and the known stoichiometry within the complex. Clear non-protein density was assigned to single stranded RNA in the apo complex and to segments of double stranded RNA in the target-bound structure. The complex rearrangements upon target-binding occur along a static backbone that allows for CRISPR RNA protection while maintaining its availability for base-pairing to target nucleic acid. The dramatic conformational change likely functions as a molecular signal for recruitment of an endonuclease that degrades the bound foreign oligonucleotides.
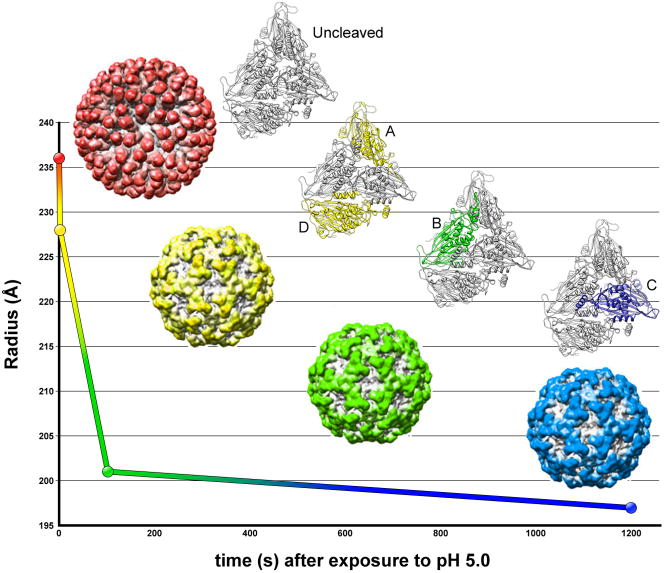
Icosahedral viruses are a particularly favorable sample for cryoEM structure determination, in some cases providing reconstructions at high enough resolutions to allow derivation of atomic models [37–41] as reviewed in [42]. In studying viral maturation, it is essential to first determine the conditions that trigger key conformational rearrangements, and then to trap particular states for detailed characterization. The recent study by Johnson and colleagues of Nudaurelia capensis ω virus maturation as a function of pH required, not only X-ray crystallography and cryoEM structures [43], but the high throughput and time-resolved capabilities of small angle X-ray scattering (SAXS) [44]. SAXS allows the assessment of even subtle changes in viral organization, while monitoring particle homogeneity. Careful control of pH during equilibrium SAXS experiments showed three discrete phases in virus maturation, beginning with a sharp collapse in the diameter of the virus particles, taking place on the order of milliseconds, followed by a slow but continuous decrease in size over 5 seconds, and ending with an even slower final transition that lasts several minutes [44]. CryoEM reconstructions at subnanometer resolution representing each of these important kinetic stages of maturation (Figure 3) showed that the subunits of the virus capsid undergo autocatalytic cleavage of their maturation peptide at different rates, depending on their symmetric position in the virus shell [43]. The slower rates of cleavage were observed at regions of the capsid where the larger molecular rearrangements are necessary for maturation, ensuring proper reorganization of the capsid before solidifying the mature architecture.
Figure 3.
Maturation pathway of the Nudaurelia capensis ω virus (NωV). Initiating maturation by lowering the pH to 5.0, Matsui et al. used time-resolved SAXS experiments to precisely describe three distinct kinetic stages in the maturation pathway of NωV [44]. The maturation, which involve the autocatalytic cleavage of the C-terminal 74 residues, begins with an initial and dramatic reduction in capsid diameter (red to yellow), which occurs on a millisecond time scale, followed by two progressively slower reductions in size (yellow to green, and then green to blue). Further SAXS experiments showed that homogeneous populations of particles representing each of these intermediates could be attained by analyzing a non-cleaving (Glu73Gln) mutant with cryoEM at specific time points [43]. The reconstructions revealed the underlying mechanism responsible for these transitions. Autocatalytic cleavage occurs initially in subunits A and D (colored yellow in the crystal structure), followed by a slower cleavage of subunit B (green), and a final cleavage of C (blue).
As advances in cryoEM continue to improve the resolution of maps beyond the subnanometer mark, computational algorithms have emerged that introduce biomolecular flexibility during the docking of a crystal structure [45–56]. This is generally achieved through compartmentalization of atomic coordinates into secondary structural elements that are treated as rigid bodies, or through molecular dynamics techniques that apply a force field to atomic coordinates while constraining atomic movements to the envelope offered by cryoEM electron density. Application of these flexible fitting methods is still relatively new to the field of cryoEM, but there are many examples where this technique has provided valuable insight into the dynamics of a molecular assembly [57–61]. It is important to note, however, that great care should be exercised in performing such analyses, especially in cases where resolution of the EM map is not consistent throughout. Within a particular region of the EM reconstruction, the size of the structural element being docked as an independent unit should not be smaller than the true local resolution of the map. The movements permitted by a given fitting algorithm must be limited to the local structural details present in the map, and failure to account for poorly resolved regions of density might result in inaccurate results. Validation criteria for the models produced by these techniques are under development within the molecular dynamics community, and worldwide modeling exercises, such as the “cryoEM modeling challenge” (see editorial by Ludtke et al. [62]), will likely play a crucial role in establishing such criteria in addition to improving this technique.
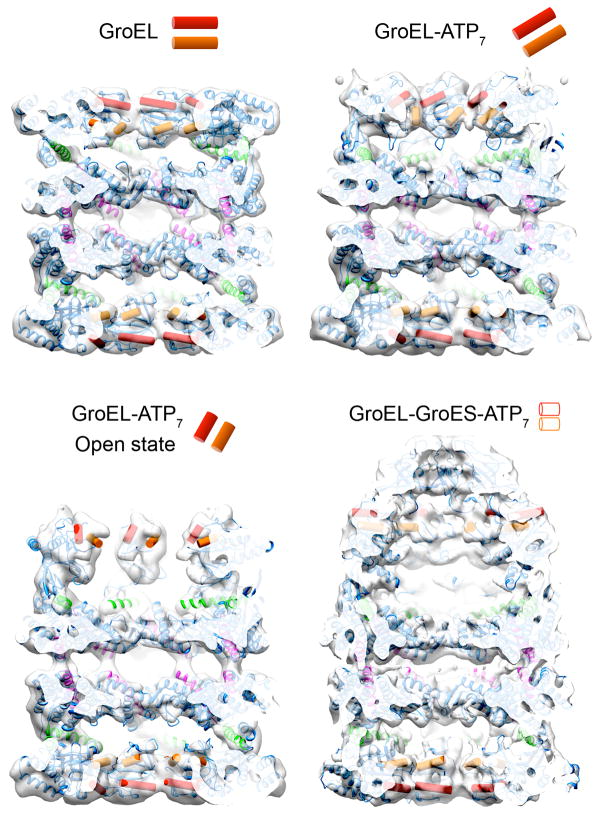
A recent study of GroEL dynamics by Clare et al. describes a true marriage of flexible fitting and cryoEM reconstruction [63]. Extensive studies have shown that the molecular chaperone GroEL binds and encapsulates non-native polypeptides to facilitate their proper folding. Rapid binding of ATP induces a series of concerted motions of the GroEL subunits that trigger binding of GroES, potentially exerting force on the unfolded substrate and culminating in its seclusion and folding inside the newly formed hydrophilic folding chamber. Although the crystal structures of the initial and final states have long been known, the precise atomic trajectory of GroEL subunits as they interact with and encapsulate substrates has not been determined.
Applying extensive computational analysis to a large cryoEM dataset of a GroEL ATPase mutant, Clare et al. were able to determine six distinct three-dimensional reconstructions representing different GroEL-ATP states. With the reconstructed densities at subnanometer resolution, flexible fitting and energy minimization of GroEL crystal structures into the electron density resulted in a series of pseudo-atomic models describing the trajectory of subunit motions as GroEL binds ATP. The motions can be divided into two phases, the first involving coordinated subunit tilting and elevation that is able to maintain substrate binding, while at the same time generating the appropriate docking site for the GroES cap. The second phase is described as the “power stroke”, involving a 100° twist of the GroEL subunits that ejects the substrate from the hydrophobic binding patches, releasing it into the hydrophilic folding chamber. This study perfectly exemplifies the incredibly dynamic nature of macromolecular complexes, and how these dynamic motions can be examined quantitatively and in exquisite detail by properly combining multiple biophysical methodologies.
Conclusions
Proteomics initiatives continue to identify new molecular ensembles involved in vital cellular functions. Defining the architecture of these complexes has benefited tremendously from the combination of cryoEM structures of full complexes with available atomic structures of components from X-ray crystallography and NMR studies. In cases where few or none of the structures are available, EM labeling schemes and additional data from biochemical and biophysical approaches become indispensable. As cryoEM technology continues to improve, atomic-resolution reconstructions are likely to become more common for even small asymmetric complexes. However, these reconstructions will probably be of highly rigid, stable macromolecules that, much like crystallography, will provide only a single snapshot of the complex. The more useful developments in cryoEM will involve the sorting of complex heterogeneity that coexists for a given set of biochemical parameters. We believe that it will soon be possible to obtain as many subnanometer reconstructions as required to describe the full conformational ensemble present in a single dataset, even in the case of small and asymmetric macromolecules. It is these more dynamic, and therefore troublesome, complexes that will benefit the most from a blending of cryoEM, atomic-resolution studies and additional biochemical and biophysical methods.
Figure 4.
Four major conformations of the chaperonin GroEL in the binding and folding of substrate proteins. The structures are shown as cut-away cryo EM maps with docked atomic structures of the subunit domains. The red and orange cylinders, also shown schematically, delineate the layout of hydrophobic binding sites for non-native substrate proteins. Top left: unliganded state of GroEL in which both rings are lined by a continuous band of hydrophobic sites (EMD 1997). Top right: GroEL-ATP7, a conformation triggered by ATP binding to the upper ring (EMD 1998). The binding sites tilt but still form a continuous lining to the end cavity. Bottom left: GroEL-ATP7 open state, an expanded ATP state in which the binding sites detach and elevate, so that the hydrophobic lining becomes discontinuous (EMD 2000). The elevated binding sites are in position to bind the GroES lid. Bottom right: GroEL- GroES-ATP7, the final folding chamber formed by a further 100o twist of the substrate-binding domains, capped by GroES (EMD 1180). In this structure, the hydrophobic sites are occluded by contacts between subunits or with GroES, to form a hydrophilic chamber. The trajectory between the GroEL-ATP open state and the GroEL-GroES chamber is proposed to provide the power stroke of chaperonin action by ejecting the non-native polypeptide from its hydrophobic binding sites, allowing it to fold in the chamber [63].
HIGHLIGHTS.
The combination of multiple structural methodologies is required to describe dynamic macromolecular rearrangements underlying function.
Structural analysis of macromolecular complexes by cryo-EM is greatly aided by labeling and biochemical data when component atomic structures are lacking.
Time-resolved methods like SAXS can identify structural transitions in a conformational pathway, guiding the pursuit of key structures by higher resolution methodologies.
Elucidation of dynamic pathways is aided by molecular dynamics techniques that apply a force field to atomic coordinates while constraining movements to the cryoEM envelope.
Acknowledgments
G.C.L. acknowledges support from the Damon Runyon Cancer Research Foundation. Work was funded by NIGMS R01 GM63072 (E.N.) and the Human Frontiers in Science program (E.N.). H.R.S. acknowledges support from the Wellcome Trust. E.N. is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Jin L, Zhou ZH. 3. 88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 5.Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, Aebersold R, Forster F, Baumeister W, Nickell S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci U S A. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Q, Yuan Y, Xu Y, Feng B, Liu L, Chen K, Sun M, Yang Z, Lei J, Gao N. Structural basis for the function of a small GTPase RsgA on the 30S ribosomal subunit maturation revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A. 2011;108:13100–13105. doi: 10.1073/pnas.1104645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludtke SJ, Tran TP, Ngo QT, Moiseenkova-Bell VY, Chiu W, Serysheva Flexible architecture of IP3R1 by Cryo-EM. Structure. 2011;19:1192–1199. doi: 10.1016/j.str.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schraidt O, Marlovits TC. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Ma B, DiMaio F, Douglas NR, Joachimiak LA, Baker D, Frydman J, Levitt M, Chiu W. Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure. Structure. 2011;19:633–639. doi: 10.1016/j.str.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 13.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs Recognize a Nucleotide-Dependent Structural Cap at Growing Microtubule Ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau WC, Rubinstein JL. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound V(O) motor. Proc Natl Acad Sci U S A. 2010;107:1367–1372. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Lau WC, Rubinstein JL. Subnanometre-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2012;481:214–218. doi: 10.1038/nature10699. The subnanometer resolution structure of an archaeal ATPase presented here defines the architecture of the membrane-embedded and cytoplasmic catalytic domains, leading to a model of how the transmembrane proton motive force is directly linked to ATP synthesis. [DOI] [PubMed] [Google Scholar]
- 16.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2. 8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 17.Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, Makyio H, Hankamer B, Barber J, Yoshida M, et al. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc Natl Acad Sci U S A. 2004;101:59–64. doi: 10.1073/pnas.0305165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee LK, Stewart AG, Donohoe M, Bernal RA, Stock D. The structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase. Nat Struct Mol Biol. 2010;17:373–378. doi: 10.1038/nsmb.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riedinger C, Boehringer J, Trempe JF, Lowe ED, Brown NR, Gehring K, Noble ME, Gordon C, Endicott JA. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J Biol Chem. 2010;285:33992–34003. doi: 10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanches M, Alves BS, Zanchin NI, Guimaraes BG. The crystal structure of the human Mov34 MPN domain reveals a metal-free dimer. J Mol Biol. 2007;370:846–855. doi: 10.1016/j.jmb.2007.04.084. [DOI] [PubMed] [Google Scholar]
- 21.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. A novel heterologous expression system of the yeast “lid” proteasome subcomplex was used to visualize MBP labels on each subunit by negative stain EM. Combined with antibody and GST-fusion labeling of subunits in the “base” subcomplex, these studies directly describe the complete architecture of the proteasome regulatory particle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23**.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. In this integrative study, crystal structures, cross-linking/MS experiments, and other data from protein-protein interactions studies were combined with computational techniques and a subnanometer reconstruction of the pombe 26S proteasome to describe the subunit organization in the regulatory particle subcomplex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid. insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davy A, Bello P, Thierry-Mieg N, Vaglio P, Hitti J, Doucette-Stamm L, Thierry-Mieg D, Reboul J, Boulton S, Walhout AJ, et al. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2001;2:821–828. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forster F, Lasker K, Nickell S, Sali A, Baumeister W. Toward an integrated structural model of the 26S proteasome. Mol Cell Proteomics. 2010;9:1666–1677. doi: 10.1074/mcp.R000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Korner R, Nickell S, Lasker K, Sali A, Tamura T, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A. 2012;109:149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He J, Kulkarni K, da Fonseca PC, Krutauz D, Glickman MH, Barford D, Morris EP. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure. 2012;20:513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 30.da Fonseca PC, He J, Morris EP. Molecular Model of the Human 26S Proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 31.Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–10648. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO J. 2011;30:408–416. doi: 10.1038/emboj.2010.322. A 4.3Å structure of the infectious rotavirus particle allowed the authors of this study to generate a nearly complete atomic model for the VP4. Combining this model with existing crystal structures of other portions, a complete atomic model of rotavirus is described, including a mechanism for rotavirus infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. This paper describes the subnanometer cryoEM structure of the Ndc80 kinetochore complex bound to microtubules, showing the presence of extra density not accounted for by the atomic coordinates. This novel density is interpreted as the N-terminal tail of the Ndc80 protein, which is involved in cooperative binding of Ndc80 molecules to microtubules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melero R, Buchwald G, Castaño R, Raabe M, Gil D, Lázaro M, Urlaub H, Conti E, Llorca O. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised towards the RNA 3′ end. Nat Struct Mol Biol. doi: 10.1038/nsmb.2287. in press. By integrating modest resolution cryo-EM maps with the results of mass spectrometry, protein and nucleic acid labeling, and biochemical interaction data, the authors describe in this paper the full architecture of the UPF surveillance complex and its control of nonsense-mediated decay. [DOI] [PubMed] [Google Scholar]
- 35.Lau PW, Guiley KZ, De N, Potter CS, Carragher B, Macrae IJ. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19:436–440. doi: 10.1038/nsmb.2268. This study establishes the architecture of the human Dicer’s functional domains using negative stain EM labeling studies. A novel internal tagging method was utilized, which involved the insertion of a 15–amino acid AviTag sequence into protein surface loops, followed by biotinylation and tagging with monovalent streptavidin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. The E. coli CASCADE complex, involved in RNA-guided immunity, is here examined before and after binding to its nucleic acid target by cryoEM. Known subunit stoichiometry and homologous crystal structures were used to describe the architecture and conformational changes upon target binding, which may signal for degradation of foreign oligonucleotides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci U S A. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. At 3.3 Angstroms, the highest reported resolution for any structure solved by single-particle cryoEM, the density was sufficient to build a complete atomic model of the aquareovirus infectious subvirion. The structure provides important details about the mechanism of nonenveloped virus entry into host cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng L, Sun J, Zhang K, Mou Z, Huang X, Ji G, Sun F, Zhang J, Zhu P. Atomic model of a cypovirus built from cryo-EM structure provides insight into the mechanism of mRNA capping. Proc Natl Acad Sci U S A. 2011;108:1373–1378. doi: 10.1073/pnas.1014995108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr Opin Struct Biol. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Matsui T, Lander GC, Khayat R, Johnson JE. Subunits fold at position-dependent rates during maturation of a eukaryotic RNA virus. Proc Natl Acad Sci U S A. 2010;107:14111–14115. doi: 10.1073/pnas.1004221107. Based on the results of time-resolved SAXS experiments, three discrete kinetic phases in the pH-dependent maturation of Nudaurelia capensis ω virus are here described by subnanometer resolution CryoEM. The reconstructions reveal how autocatalytic cleavage occurs at different rates depending on the symmetric position of the peptide in the virus shell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsui T, Tsuruta H, Johnson JE. Balanced electrostatic and structural forces guide the large conformational change associated with maturation of T = 4 virus. Biophys J. 2010;98:1337–1343. doi: 10.1016/j.bpj.2009.12.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JZ, Furst J, Chapman MS, Grigorieff N. Low-resolution structure refinement in electron microscopy. J Struct Biol. 2003;144:144–151. doi: 10.1016/j.jsb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Chan KY, Gumbart J, McGreevy R, Watermeyer JM, Sewell BT, Schulten K. Symmetry-restrained flexible fitting for symmetric EM maps. Structure. 2011;19:1211–1218. doi: 10.1016/j.str.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmed A, Whitford PC, Sanbonmatsu KY, Tama F. Consensus among flexible fitting approaches improves the interpretation of cryo-EM data. J Struct Biol. 2012;177:561–570. doi: 10.1016/j.jsb.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng W. Accurate flexible fitting of high-resolution protein structures into cryo-electron microscopy maps using coarse-grained pseudo-energy minimization. Biophys J. 2011;100:478–488. doi: 10.1016/j.bpj.2010.12.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suhre K, Navaza J, Sanejouand YH. NORMA: a tool for flexible fitting of high-resolution protein structures into low-resolution electron-microscopy-derived density maps. Acta Crystallogr D Biol Crystallogr. 2006;62:1098–1100. doi: 10.1107/S090744490602244X. [DOI] [PubMed] [Google Scholar]
- 51.Fabiola F, Chapman MS. Fitting of high-resolution structures into electron microscopy reconstruction images. Structure. 2005;13:389–400. doi: 10.1016/j.str.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Grubisic I, Shokhirev MN, Orzechowski M, Miyashita O, Tama F. Biased coarse-grained molecular dynamics simulation approach for flexible fitting of X-ray structure into cryo electron microscopy maps. J Struct Biol. 2010;169:95–105. doi: 10.1016/j.jsb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandurangan AP, Topf M. Finding rigid bodies in protein structures: Application to flexible fitting into cryoEM maps. J Struct Biol. 2012;177:520–531. doi: 10.1016/j.jsb.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 55.Schroder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Falke S, Tama F, Brooks CL, 3rd, Gogol EP, Fisher MT. The 13 angstroms structure of a chaperonin GroEL-protein substrate complex by cryo-electron microscopy. J Mol Biol. 2005;348:219–230. doi: 10.1016/j.jmb.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 58.Tama F, Ren G, Brooks CL, 3rd, Mitra AK. Model of the toxic complex of anthrax. responsive conformational changes in both the lethal factor and the protective antigen heptamer. Protein Sci. 2006;15:2190–2200. doi: 10.1110/ps.062293906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsin J, Gumbart J, Trabuco LG, Villa E, Qian P, Hunter CN, Schulten K. Protein-induced membrane curvature investigated through molecular dynamics flexible fitting. Biophys J. 2009;97:321–329. doi: 10.1016/j.bpj.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci U S A. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chappie JS, Mears JA, Fang S, Leonard M, Schmid SL, Milligan RA, Hinshaw JE, Dyda F. A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell. 2011;147:209–222. doi: 10.1016/j.cell.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ludtke SJ, Lawson CL, Kleywegt GJ, Berman H, Chiu W. The 2010 cryo-em modeling challenge. Biopolymers. 2012;97:651–654. doi: 10.1002/bip.22081. [DOI] [PubMed] [Google Scholar]
- 63**.Clare DK, Vasishtan D, Stagg S, Quispe J, Farr GW, Topf M, Horwich AL, Saibil HR. ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the GroEL Chaperonin. Cell. 2012;149:113–123. doi: 10.1016/j.cell.2012.02.047. Extensive computational analysis of a large GroEL ATPase mutant cryoEM dataset reveals the subnanometer structures of six distinct states for GroEL-ATP. Flexible fitting and energy minimization of docked GroEL crystal structures led to a series of pseudo-atomic models, describing the trajectory of subunit motions as GroEL binds ATP. [DOI] [PMC free article] [PubMed] [Google Scholar]