Abstract
Hematopoietic stem cell transplant (HSCT) therapy is limited by pulmonary infections. Mice with fully reconstituted hematopoietic compartments, including alveolar macrophages (AMs), post-bone marrow transplantation (BMT) have impaired host defense against Gram negative Pseudomonas aeruginosa. Impaired innate immunity is related to increased production of prostaglandin E2 (PGE2) by AMs. Cyclooxygenase (COX)-2 is the rate-limiting enzyme for synthesis of PGE2 from arachidonic acid, and COX-2 expression is elevated in AMs post-BMT. We hypothesized epigenetic mechanisms may be responsible for upregulation of COX-2 in AMs. Using bisulfite sequencing, we observed the 5’-untranslated region and exon 1 of the COX-2 gene is hypomethylated in the AMs of BMT mice compared to control. COX-2 expression was increased in primary AMs and in the AM cell line (MHS) following treatment with 5-aza-2-deoxycytidine (a methyltransferase inhibitor). Methylation by SssI methyltransferase of a 702 bp region of the COX-2 promoter including the beginning of exon 1 driving a luciferase reporter silenced luciferase expression. Because transforming growth factor beta (TGF-β1) is elevated in lungs post-BMT, we tested whether TGF-β1 could promote expression of COX-2 in a hypermethylated COX-2 vector, and observed TGF-β1 induced modest expression of COX-2, suggesting an ability to demethylate the promoter. Finally, BMTs performed with marrow from mice expressing a dominant negative form of the TGF-β receptor on CD11c-expressing cells (which includes AMs) demonstrated improved host defense and AM function. Our findings suggest impaired innate immunity and PGE2 elevation post-BMT are due to hypomethylation of the COX-2 gene which is at least partly regulated by TGF-β1.
Introduction
Hematopoietic stem cell transplantation (HSCT) is commonly used to treat malignant and nonmalignant hematologic disorders (1, 2). Traditionally, a conditioning regimen is implemented prior to intravenous infusion of hematopoietic stem cells (HSC) which may consist of chemotherapy with or without total body irradiation (TBI) (2). TBI is itself myeloablative and immunosuppressive and can affect regions within the body that are not easily accessible by chemotherapeutic agents delivered via the circulation (1–3). Although HSCT has proven to be an effective therapeutic option for malignancy, it is also associated with significant morbidity and mortality (1–5). Following either autologous (i.e. recipient HSCs also serve as donor cells) or allogeneic (i.e. related or unrelated donor provides HSCs) HSCT, transplant recipients are susceptible to developing life-threatening infectious and noninfectious complications (2, 3, 6–9).
The lung is a common target organ post-transplant where pulmonary complications account for significant mortality and morbidity in HSCT recipients (2, 3, 6–9). Such complications develop throughout the timeline of pre-engraftment (0–30 days after transplant), early post-engraftment (30–100 days after transplant) and late post-engraftment (>100 days)(10). Despite full reconstitution or engraftment of donor-derived leukocytes, patients exhibit sustained and enhanced susceptibility to infections post-transplant (6–10). AMs are the resident macrophages in the lung and, together with recruited polymorphonuclear leukocytes (PMNs), play an important role in regulating an immune response in the lung (11–14). Previous studies have reported defective phagocytic and bacterial killing function of human alveolar macrophages within 4 months following HSCT, with some deficiency persisting up to 12 months (15). Thus, impaired innate immune function may explain the prolonged susceptibility to infection observed in post-transplant individuals.
To study the effects of HSCT, our laboratory previously developed a syngeneic bone marrow transplant (BMT) murine model that simulates autologous HSCT in humans and allows for a direct approach to study immune reconstitution and function without the confounding effects of graft-versus-host-disease or immunosuppressive drugs. We have shown that even after full immune reconstitution following syngeneic BMT, donor-derived AMs from BMT mice are defective in phagocytosis and killing compared to mice that did not undergo BMT (11, 16). We discovered this defect is related to decreased cysteinyl leukotrienes (cys LTs) and tumor necrosis factor α (TNFα) production, and increased prostaglandin E2 (PGE2) production (11, 17, 18).
Eicosanoids are lipid mediators derived from arachidonic acid and cells of the myeloid lineage are major producers of both cys LTs and PGE2 (18, 19). Synthesis of prostaglandins is mediated by the cyclooxygenase (COX) enzymes of which there are two isoforms. COX-1 is a constitutive isoform of COX responsible for basal COX expression required for homeostasis, whereas COX-2 is induced primarily by inflammation (19). PGE2 production post-BMT is attributed to the increased activity of COX-2 and PGE2 negatively regulates the innate immune response (11, 20). In our model, PGE2 and COX-2 expression were found to be elevated post-BMT within AMs and PMNs and this caused functional impairments in the innate immune function of both of these cell types (11, 16, 21, 22). However, in our model of P. aeruginosa infection post-BMT, we demonstrated that it was the defect in non-opsonized phagocytosis by AMs post-BMT, rather than PMN function which was responsible for the acute clearance of P. aeruginosa (13); thus, we have focused our current studies on regulation of COX-2 expression in AMs.
PGE2 binds to seven transmembrane spanning E prostanoid (EP) receptors, of which there are 4 discrete EP receptors that couple to G proteins involved in mediating the intracellular signaling in response to PGE2 (20, 23). We have shown that binding of the EP2 receptor by PGE2 results in increased IL-1 receptor associated kinase (IRAK-M) expression in AMs resulting in the inhibition of non-opsonized phagocytosis (14). PGE2 also induces elevation of the enzyme phosphatase and tensin homolog deleted on chromosome 10 (PTEN) which results in the inhibition of serum-opsonized phagocytosis by AMs (21). Thus PGE2 is able to act through distinct signaling pathways to compromise host innate immune defense.
Because COX-2 mRNA and protein expression are upregulated in the AMs of mice post-BMT and remain elevated in cells cultured ex vivo (11, 14), we hypothesized that epigenetic mechanisms may be responsible for the upregulation of COX-2. To test this, we sought to determine the methylation status of the COX-2 gene promoter. Using bisulfite conversion and pyrosequencing, we discovered that the COX-2 gene was significantly hypomethylated in the 5’-untranslated region (UTR) and exon 1 of AMs from mice post-BMT. Using in vitro assays, we determined that COX-2 mRNA expression is regulated by methylation, as treatment of both a murine alveolar macrophage cell line (MHS) and primary AMs with 5-aza-2’-deoxycytidine (a methyl transferase inhibitor) increased COX-2 mRNA levels by RT-PCR and caused demethylation of the COX-2 gene. Similarly, transfections in MHS cells with methylated COX-2 promoter constructs showed reduced luciferase activity. However, COX-2 promoter activity could be enhanced by treatment with TGF-β1, a cytokine known to be elevated in BMT lungs (24). Thus, our data indicate that epigenetic regulation of COX-2 is one mechanism driving the observed elevation of both COX-2 and PGE2 in BMT mice and this alteration is regulated in part by TGF-β1.
Materials and Methods
Animals
C57BL/6 mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Mice expressing dominant-negative TGF-β receptor II (TGF-βRII) under the CD11c promoter (CD11cdnR ) were provided by Dr. Laouar (University of Michigan) (25, 26). Mice were bred and housed under specific pathogen-free conditions and monitored daily by veterinary staff. All mice were euthanized by CO2 asphyxiation. The University of Michigan Committee on the Use and Care of Animals approved these experiments.
Bone marrow transplantation
Recipient mice received 13.5 Gy of total body irradiation (TBI; orthovoltage x-ray source) split in two fractions, 3 h apart. Bone marrow cells were harvested from donor mice and resuspended in serum-free medium (SFM; DMEM, 0.1% BSA, 1% penicillin-streptomycin, 1% l-glutamine, and 0.1% amphotericin B). Bone marrow cells (5 × 106) were administered by tail vein injection into TBI recipient mice. All experiments with BMT mice were performed 5–6 wk post-BMT when mice were fully donor-cell reconstituted. Spleen cells were >94% donor-derived, and AMs were >80% donor-derived at this time point (16).
Harvesting AMs
Resident AMs from mice were obtained via ex vivo lung lavage, using a previously described protocol (11). Briefly, these cells were collected in lavage fluid consisting of complete medium (DMEM, 1% penicillin-streptomycin, 1% l-glutamine, 10% FCS, 0.1% Fungizone) and 5 mM EDTA. The cells were enumerated by counting on a hemocytometer before plating.
Molecular cloning of COX-2 promoter into luciferase expression vector
The full-length (full) murine COX-2 promoter (1203 bp) DNA as defined in (27) and a 498 bp COX-2 promoter (deleted) was amplified using primers to create cloning sites. Supplemental figure 1 shows the sequence of the COX-2 promoter and the location of the created cloning sites (Kpn I, MluI and Bgl II) in grey boxes. Table 1 shows the primers used to amplify the promoter regions by standard PCR from genomic DNA from AMs. The amplified full COX-2 promoter DNA was digested with restriction endonucleases KpnI and BglII (New England BioLabs, Inc.) while the deleted COX-2 promoter was digested with Mlu I and BglII (New England BioLabs, Inc.). pGL3-basic luciferase reporter vector (Promega) was digested with either KpnI and BglII or MluI and BglII. The digested DNA and vectors were ligated with T4 DNA ligase for 24 hours. E. coli strain MC1061 was made competent by CaCl2 and was transformed with ligated DNA. Positive colonies were selected with Ampicillin (100 mg/mL) and correct insert was verified restriction digestion and PCR.
Table I.
Primers and probes for semiquantitative real-time RT-PCR and PCR
| RT-PCR primers and probes | Sequence (5’-3’) |
| β-Actin forward | CCGTGAAAAGATGACCCAGATC |
| β-Actin reverse | CACAGCCTGGAGGCTACGT |
| β-Actin probe | TTTGAGACCTTCAACACCCCAGCCA |
| COX-2 forward | TGACCCCCAAGGCTCAAAT |
| COX-2 reverse | GAACCCAGGTCCTCGCTTATG |
| COX-2 probe | TTTGCCCAGCACTTCACCCACAGT |
| PCR primers for cloning | Sequence (5’-3’) |
| COX-2 forward long | CACAGTAGGAAGGTACCCAACACTATC |
| COX-2 reverse long | CTGCTAGAAAGGAGATCTGAGCCTGAG |
| COX-2 forward short | CCGGTAGCTGTACGCGTGCTCTGAGC |
| COX-2 reverse short | AGATCGCAGATCTGAGCCTGAGG |
| Primers for analysis of CpG methylation on sites 1–6 | Sequence (5’-3’) |
| Forward primer: | AGATGTGGATTTTGATAGAGGATATT |
| Reverse primer: | CTACCCTTAACTACCCCAAATAATAC |
| Sequencing primer: | ATTTTATTAAAAATAGAAGAAA |
In vitro methylation
The COX-2 promoters, both the full and deleted COX-2 versions, inserted into the luciferase reporter plasmid pGL3-basic (Promega) were in vitro methylated with CpG methyltransferase M.SssI (New England BioLabs, Inc.) as specified by the manufacturer for 4 hours. Methylation was confirmed by digestion with methylation-sensitive restriction enzyme SmaI.
In vitro transfections and Dual Luciferase Assay
MHS cells were transfected using Lipofectamine LTX and PLUS Reagent (Invitrogen™) following manufacturer's optimized RAW 264.7 protocol. Briefly, 6.2 ×104 MHS cells were cultured in complete media containing RPMI, 10% fetal calf serum, 1% penicillin-streptomycin, 1% L-glutamine and 0.5 mM 2-mercaptoethanol overnight. Following 24 hours, MHS cells were transfected with COX-2 promoter-driven luciferase reporter plasmid and pRL-SV40 in a 50:1 ratio for a total of 0.3 µg of DNA using lipofectamine LTX and PLUS Reagent (Invitrogen™). Transfections were performed with unmethylated or methylated, long or short COX-2 luciferase reporter plasmids. Where indicated, cells were treated with 10 µg/mL lipopolysaccharide (LPS) or 1 ng/mL porcine TGF-β1.
Real-time RT-PCR
Real-time RT-PCR was performed on an ABI Prism 7000 thermocycler (Applied Biosystems, Foster City, CA). Gene-specific primers and probes were designed using Primer Express software (PE Biosystems, Foster City, CA) as published previously (11, 12). Sequences for all primers and probes used can be found in Table 1. Each AM sample was pooled from two to three mice and was run in duplicate. Average cycle threshold (CT) was determined for each sample and was normalized to β-actin. Relative gene expression was calculated as described previously (28).
DNA methyltransferase or histone deacetylase (HDAC) inhibition
AMs or MHS cells were treated with either varying concentrations of the methyltransferase inhibitor 5-aza-2'-deoxycytidine (Sigma, St.Louis, MO) or the HDAC inhibitor Trichostatin A (Sigma, St.Louis, MO) for 72 hours. Primary AMs were initially stimulated with (1ng/mL) of recombinant murine GM-CSF (R&D, Minneapolis, MN) for 24 hours to promote proliferation prior to receiving the appropriate treatments for the following 72 hours.
ELISA/enzyme immunoassay
MHS cells were cultured at 5 × 105 cells/ml in a 24-well plate for 72 hours. MHS cells were grown in complete media for 48 hours then switched to serum-free media for 24 hours. Supernatants were collected for enzyme immunoassay (EIA). Production of PGE2 and thromboxane B2 (a metabolite of thromboxane A2) was measured by EIA (Cayman Chemical, Ann Arbor, MI), according to the manufacturer’s instructions.
Bisulfite Conversion and Pyrosequencing
DNA was isolated from 5×105 to 1×106 cells using the Dneasy kit (Qiagen, Orange, CA). The Zymo Research EZ DNA Methylation kit (Irvine, CA) was used to bisulfite modify 500 ng of genomic DNA. Bisulfite modification was performed according to manufacturer’s instructions. Briefly, genomic DNA was denatured with Dilution Buffer and further treated with CT Conversion Reagent. Samples were processed following the alternative incubation conditions recommended by manufacturer whereby samples were incubated in a thermocycler under the following conditions: (95°C 30sec; 50°C 60 min) × 16 cycles; 4°C hold. Following bisulfite conversion, the PTGS2 (COX-2) promoter was PCR amplified and sequenced on a Pyrosequencer (Qiagen). The CpG sites analyzed are shown in Supplementary Figure 1 color coded by blue, yellow or pink boxes to denote the regions amplified by different primer pairs for analysis. The CpG sites analyzed are also numbered for clarity. Amplification and sequencing primers for murine PTGS2 were obtained from EpigenDx, Inc, Worcester, MA (assay ADS2001 (sites in yellow) and ADS2002 (sites in pink). Sites in blue were amplified and sequenced using primers we designed in Table 1. The PCR conditions for determining the methylation profile of the DNA using ADS2001 are as follows: 95°C 15 min; 45 × (95°C 30 s; 54°C 30s; 72°C 30s); 72°C 5 min; 4°C hold. PCR conditions using ADS2002: 95°C 15 min; 45 × (95°C 30 s; 60°C 30s; 72°C 30s); 72°C 5 min; 4°C hold.
P. aeruginosa PAO1 preparation and FITC-labeling
P. aeruginosa PAO1 stock was grown in tryptic soy broth (Difco; BD, Sparks, MD), and the culture concentration was determined via absorbance measurements. For FITC-labeling, a P. aeruginosa culture was centrifuged and washed two times by resuspending cell pellet in 1 mL sterile PBS and transferring into a sterile tube. P. aeruginosa was heat-killed by autoclaving for 20 minutes and resuspended at 109 – 1010 CFU/mL in 0.1 M NaHCO3 (pH 9.2). 0.2 mg/mL FITC (Sigma, St. Louis, MO) in DMSO was added to heat-killed P. aeruginosa and allowed to incubate in dark for 1 hour on rocker at room temperature. Following FITC-labeling, heat-killed P. aeurginosa were washed three times and resuspended in 1 mL sterile PBS. Aliquots were prepared and stored at −80°C until use.
In vitro phagocytosis assay
AMs isolated by BAL were plated at 2 × 105 cells per well in 100 µl and cultured overnight in complete media on a 96-well, flat-bottomed, half-area tissue culture plate (Costar, Corning, NY). The following day, FITC-labeled heat-killed P. aeruginosa (prepared as described above) was added at 300:1 MOI. 2 hours following incubation at 37°C in dark, 50 µL of trypan blue (250 µg/mL in 0.09 M citrate buffer solution; Sigma) was added to each well for 1 min to quench fluorescence of non-phagocytosed FITC-labeled bacteria. AM phagocytosis of FITC-labeled bacteria was measured using a microplate fluorimeter and expressed in arbitrary fluorescence intensity units. For possible differences in AM adherence to tissue culture plate, data were normalized for cell number using a LDH Cytotoxicity Detection Kit (Roche Diagnostics, Indianapolis, IN) as previously described (11).
Statistical analysis
Statistical significance was analyzed using the GraphPad Prism 5.0 statistical program (La Jolla, CA). Comparisons between two experimental groups were performed using the Student t test. Comparisons between 3 or more values used an ANOVA analysis with a post-hoc Bonferroni comparison. A value of p < 0.05 was considered statistically significant.
Results
COX-2 mRNA levels are elevated in BMT AMs
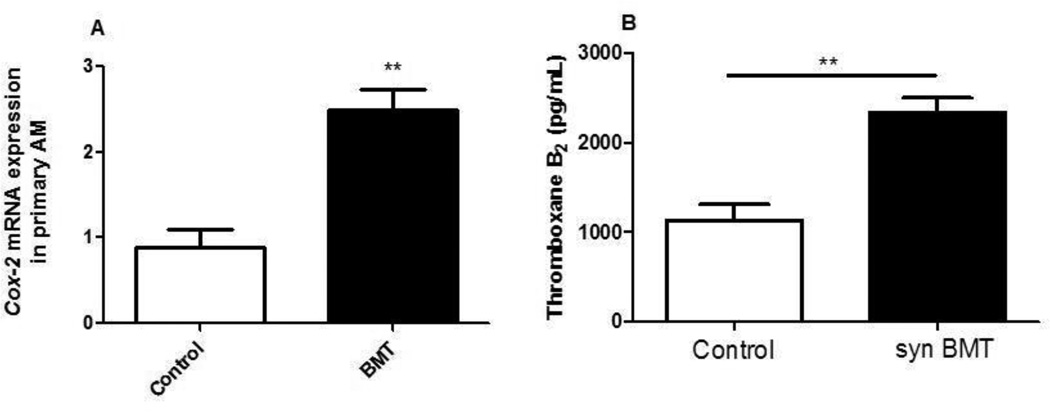
PGE2 levels were previously reported to be elevated in the plasma of patients undergoing autologous HSCT (29). The increased PGE2 detected in these individuals post-transplantation was independent of conditioning regimen (chemotherapy or radiotherapy). Similar results were previously observed in our established syngeneic BMT mouse model, whereby 13 Gy total body irradiation (TBI) or a dual-chemotherapy regimen induced a defective pulmonary immune response associated with elevated levels of PGE2 produced by lung innate immune cells (11, 16). PGE2 production is dependent on the activity of COX enzymes. To determine whether the increase in PGE2 levels are a result of increased COX-2 expression, we measured COX-2 expression levels from control (untransplanted) and BMT AMs harvested by bronchoalveolar lavage (BAL). Following a 24 h culture with serum-free media, RNA was harvested from AMs and COX-2 mRNA expression levels were measured by real-time RT-PCR. These results demonstrate that COX-2 is upregulated in BMT AMs when compared to control AMs (Figure 1A). These results are consistent with our previous observation that COX-2 protein is also elevated in AMs post-BMT(14) and confirm that the BMT-induced alterations which enhance COX-2 expression persist in AMs cultured ex vivo.
Figure 1. COX-2 mRNA and TXB2 are increased in BMT mice.
A: AMs were harvested from control and BMT mice at 5 weeks post-BMT. RNA was isolated and COX-2 was measured by real-time RT-PCR; n=4, representative of two experiments; B: Supernatants were collected from BMT and control AMs cultured in serum-free media for 24 h and measured for TXB2 by ELISA; n=4, representative of two experiments.
COX-2 can catalyze the synthesis of numerous prostaglandins. We previously published that prostacyclin (PGI2) is also elevated in AMs post-BMT (11). To determine if thromboxane A2 was also elevated, we measured the more stable metabolite thromboxane B2 (TXB2), in control and BMT AMs (Fig. 1B). Taken together, these results suggest that BMT causes a global upregulation of prostanoid products.
BMT AMs are hypomethylated in the 5’-UTR and exon 1 of the COX-2 gene
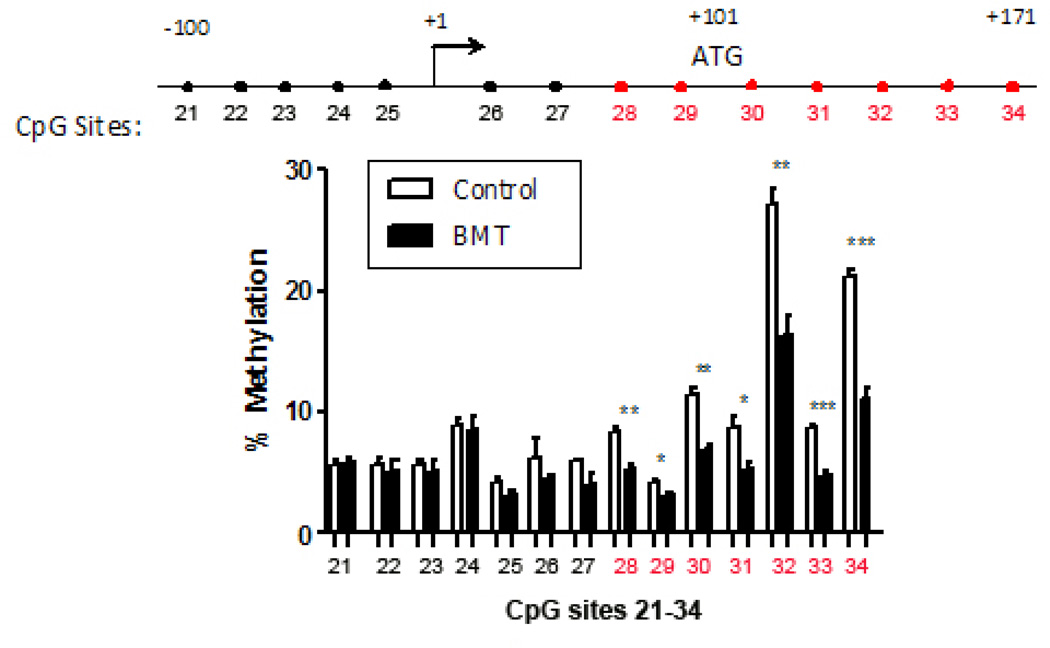
To determine whether epigenetic regulation by DNA methylation was involved in the increased expression of COX-2 mRNA levels observed in Figure 1A, we collected genomic DNA from the AMs of control and BMT mice and performed bisulfite sequencing of the COX-2 promoter. We analyzed methylation at all 34 CpG sites noted in Supplemental Fig. 1. There was no difference in the methylation status of CpG sites 1–27 in control and BMT AMs (data not shown). However, BMT AMs exhibited significant hypomethylation compared to control AMs at CpG loci within the 5’-UTR and exon 1 (sites 28–34; Figure 2).
Figure 2. COX-2 in BMT AMs is hypomethylated around the first exon start site.
At week 5 post-BMT, AMs from control and BMT mice were harvested, DNA was isolated and bisulfite converted. Cytosine to thymine conversion was detected by pyrosequencing to determine methylation patterns. Control: n=4, BMT: n=3; *p < .05, **p<.01, ***p < .001. Representative of 2 experiments showing similar patterns at the same sites. Numbered CpG sites refer to locations specified in Supplemental Fig.1.
5-aza-2'-deoxycytidine increases COX-2 in MHS cells and primary AMs
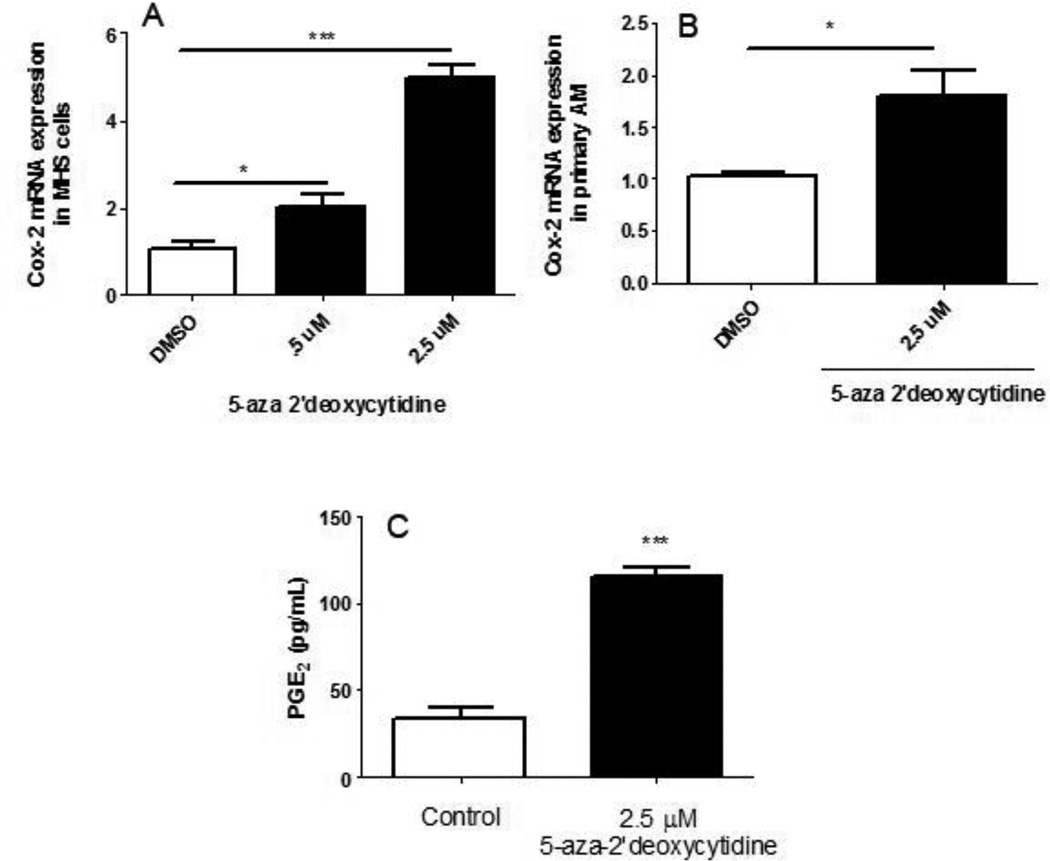
A chemical analog for cytosine, 5-aza-2'-deoxycytidine, inhibits methylation by blocking DNA methyltransferase activity (30). The methylation patterns of COX-2 post-BMT indicate that a decrease in methylation of the COX-2 promoter region may be responsible for the elevation of COX-2 and PGE2 observed post-transplant. To determine whether DNA methylation of COX-2 contributes to its diminished expression in control non-BMT cells, we treated MHS cells, a murine alveolar macrophage cell line, with increasing concentrations of 5-aza-2'-deoxycytidine for 72 hours or DMSO as a vehicle control. RNA was extracted from these cells and COX-2 mRNA levels were measured by real-time RT-PCR. COX-2 expression from MHS cells increased dose-dependently in the presence of the methyltransferase inhibitor (Figure 3A).
Figure 3. 5-aza-2'-deoxycytidine increases COX-2 in MHS cells and primary AMs and PGE2 in MHS cells.
A: 5×105 MHS cells were cultured with 0.5 or 2.5 µM 5-aza 2’-deoxycytidine, or vehicle (DMSO) for 72 hours. RNA was isolated and Cox-2 mRNA levels were analyzed by real-time RT-PCR. Transcripts were normalized to β actin levels (n=4); B: Lungs from untransplanted mice were lavaged and primary AMs were harvested. AMs were initially stimulated with 1 ng/mL GM-CSF for 24 h and further treated with 2.5µM 5-aza-2’-deoxycytidine in the presence of GM-CSF for an additional 72 hours. RNA was isolated and COX-2 mRNA levels calculated by RT-PCR. C: MHS cells (5 × 105) were grown in 10% complete media in the presence or absence of 2.5 µM 5-aza-2’-deoxycytidine for 48 hours. Media was aspirated and fresh serum free media with or without 5-aza-2’-deoxycytidine was added for 24 hours. Supernatants were collected and PGE2 ELISA was performed. (n=3); *p<.05, **p<.01, ***p<.001. All data are representative of at least 2 experiments.
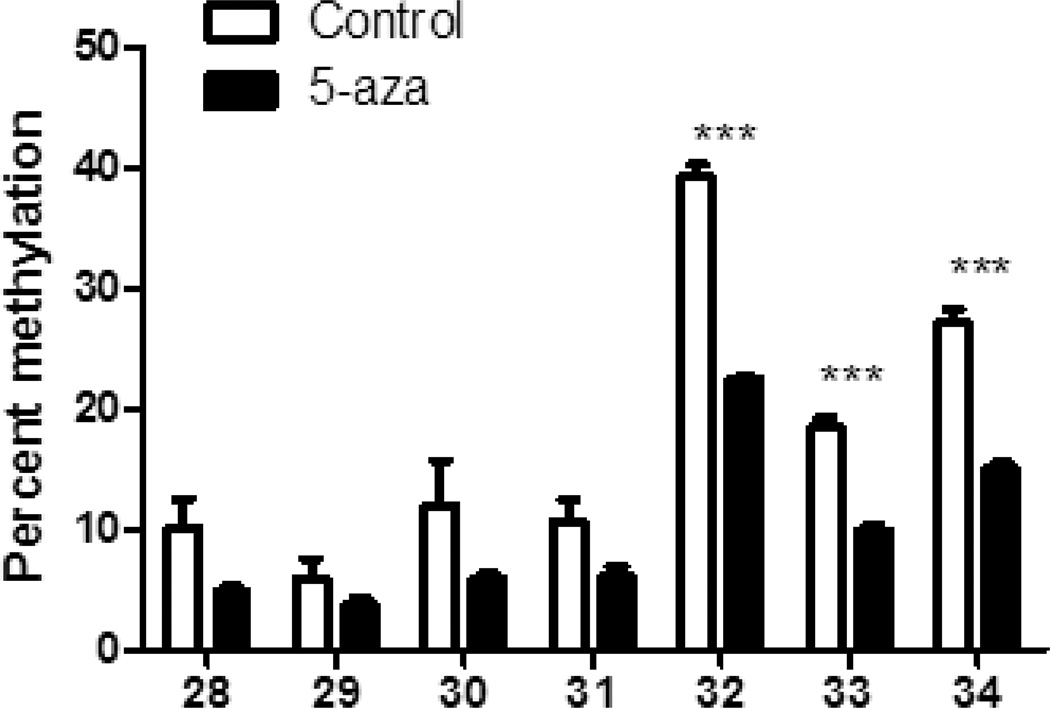
We next sought to verify these results in primary AMs from control mice. We collected AMs by BAL and induced proliferation by initially treating the primary AMs with GM-CSF for 24 h. AMs were then treated with 5-aza-2'deoxycytidine for 72 additional hours in the presence of GM-CSF to maintain proliferation (31) and COX-2 mRNA levels were determined by real-time RT-PCR. Expression patterns of COX-2 transcripts shown in Figure 3B correlate well with those observed in 5-aza-2’-deoxycytidine-treated MHS cells. Furthermore, treatment of MHS cells with 2.5 µM 5-aza-2'deoxycytidine resulted in increased PGE2 levels (Figure 3C). To determine whether the increase in COX-2 mRNA levels in MHS cells treated with the methyltransferase inhibitor were in fact due to demethylation of the promoter in the same region noted post-BMT, DNA was harvested from 5-aza-2’deoxycytidine-treated MHS cells and subjected to methylation analysis. Figure 4 shows an overall decrease in methylation on CpG sites 28–34; the same sites found to be significantly hypomethylated in BMT AMs, supporting the conclusion that the observed effects on COX-2 expression are due to a change in methylation patterns.
Figure 4. 5-aza-2’-deoxycytidine treatment alters COX-2 promoter methylation.
MHS cells (5×105) were treated with either 2.5 µM 5-aza-2’-deoxycytidine or vehicle (DMSO) for 72 h prior to harvesting DNA for methylation analysis by bisulfite conversion and pyrosequencing; n=3, ***p<.001.
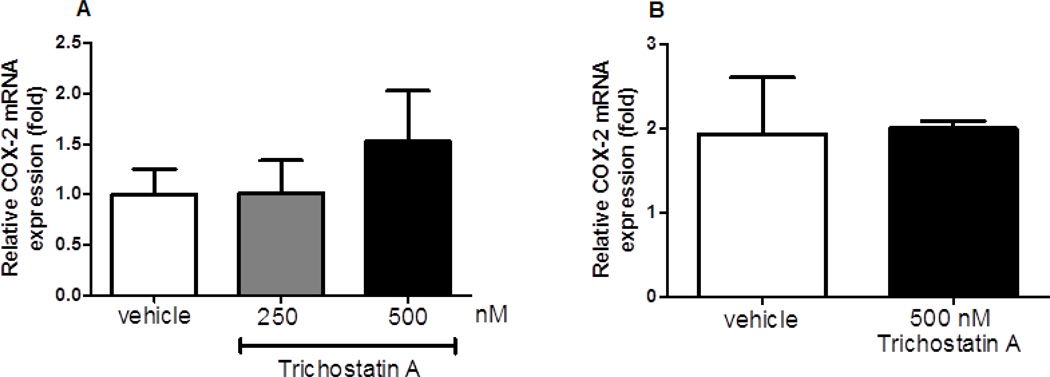
As histone acetylation can affect gene expression, it is possible that COX-2 can also be influenced by other forms of epigenetic regulation. Thus, both MHS and primary AMs were treated with trichostatin A, an HDAC inhibitor. However, MHS (Figure 5A) and primary AMs (Figure 5B) stimulated with trichostatin A for 72 hours did not show a significant difference in COX-2 expression compared to control treatment with DMSO. Taken together, these data indicate that the regulation of COX-2 is primarily influenced at the transcriptional level by DNA methylation.
Figure 5. COX-2 expression is not regulated by histone acetylation.
A: MHS cells (2×105), B: GM-CSF-treated primary AMs (2×105) were incubated with either trichostatin A (200 or 500 nM) or vehicle control (DMSO) for 72 h prior to RNA isolation. mRNA expression was measured by real-time RT-PCR; n=3. No changes were significant compared to vehicle.
Cloned COX-2 luciferase vector shows normal transcriptional regulation
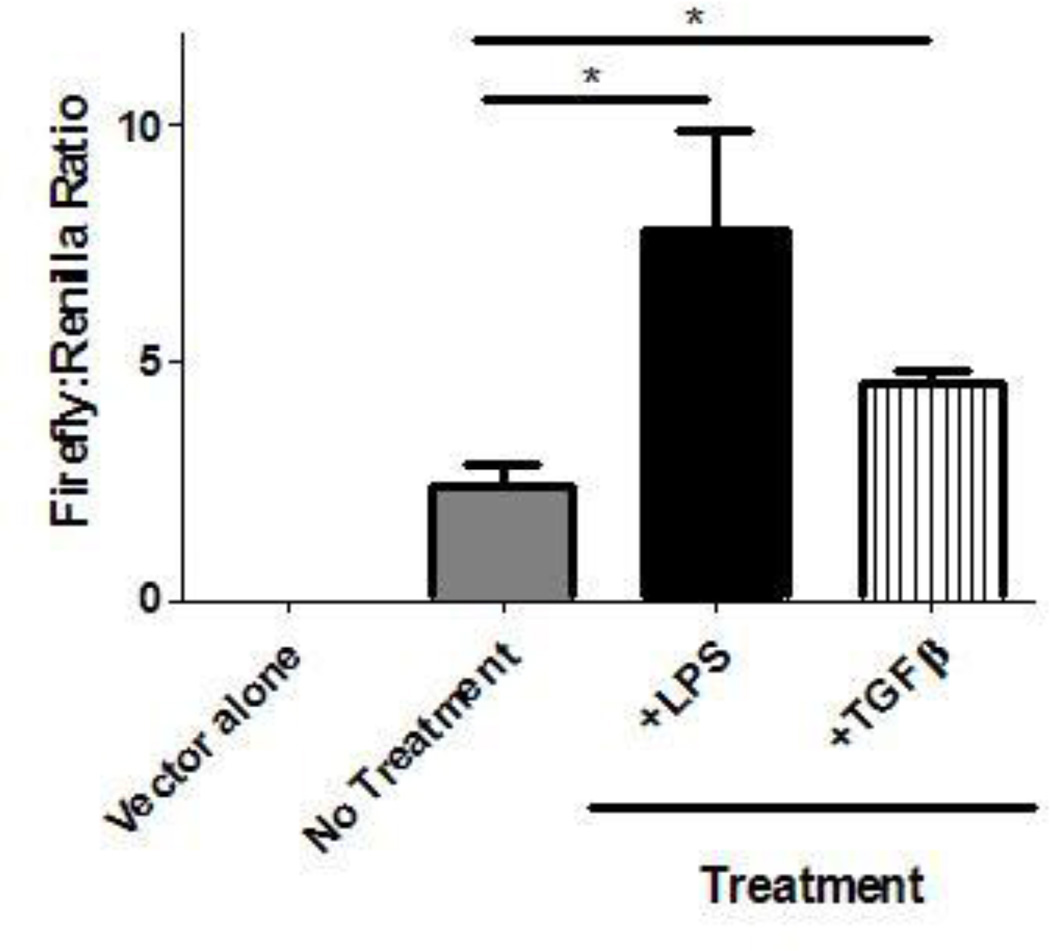
To further study the regulation of COX-2, the full COX-2 promoter region (from the KpnI to the Bgl II site) was cloned into a pGL3-basic firefly luciferase expression plasmid. MHS cells were then transfected with the cloned COX-2 promoter-driven firefly luciferase vector and a control SV-40 promoter-driven renilla luciferase vector via lipofectamine. As COX-2 expression is induced by inflammatory stimuli, MHS cells were cultured in the presence or absence of proinflammatory mediators LPS or TGF-β1 for 24 hours. Treatment with both LPS and TGF-β1 showed a significant increase in firefly luciferase over untreated MHS cells, validating the luciferase assay as an assay of regulatable COX-2 promoter activity (Figure 6). Transfections of the COX-2 promoter-driven expression vector and control expression were attempted in primary AMs of wild-type mice but despite using high concentrations of plasmid DNA and trying both lipofectamine and electroporation protocols, we were unable to achieve significant luciferase expression (even of the control vector) in primary AMs.
Figure 6. Transfected COX-2 promoter-driven luciferase is induced by LPS and TGF-β compared to untreated MHS cells.
MHS cells (6.2×104) were cultured overnight in the presence or absence of either LPS (10 µg/mL), or porcine TGFβ (1 ng/mL). MHS cells were transfected the next day with either luciferase vector alone without COX-2 promoter insert or the long COX-2 promoter driven-luciferase vector, and pRL-SV40 (control vector) in a 50:1 ratio using lipofectamine LTX and PLUS Reagent. Firefly and renilla luciferase activities were measured and are shown as a ratio. (n=3, representative of 2 experiments; *p<.05).
In vitro methylation suppresses COX-2 promoter activity in MHS cells
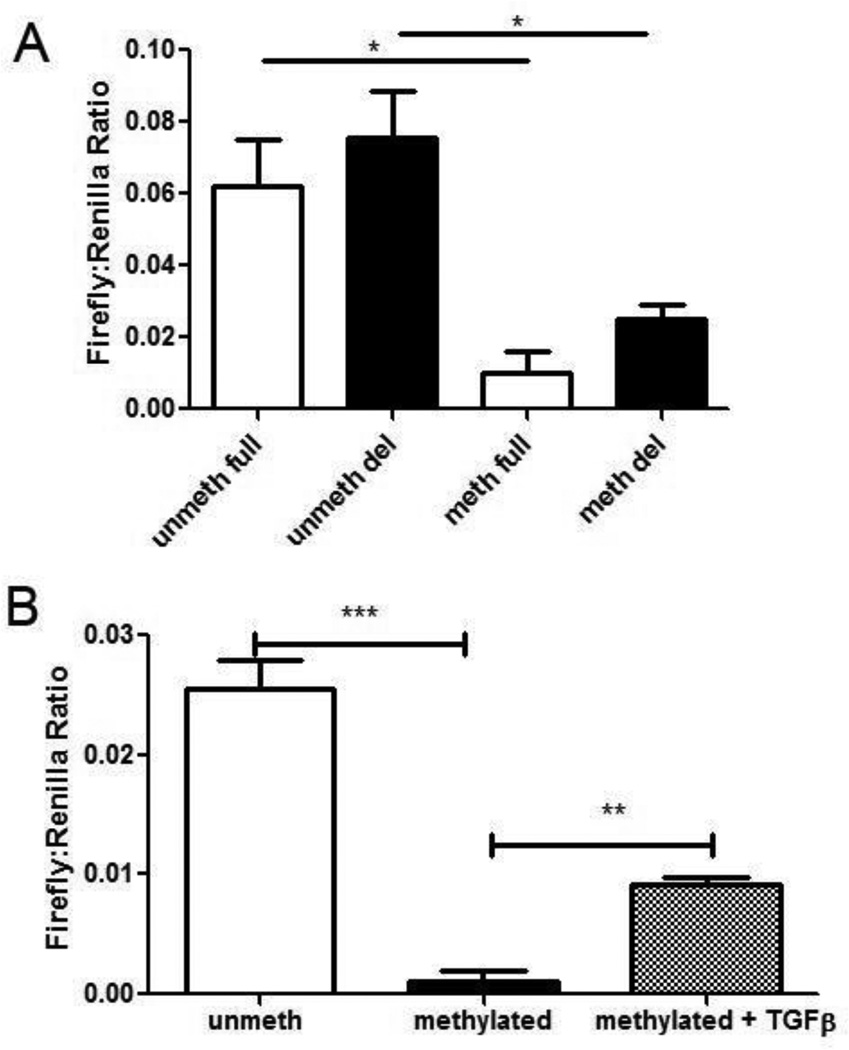
Both full (KpnI to Bgl II) and deleted (MluI to Bgl II) COX-2 promoter constructs were cloned into pGL3-basic firefly luciferase expression plasmids. The deleted COX-2 promoter contains all CpG sites analyzed in primary AMs by bisulfite conversion and pyrosequencing whereas the full COX-2 construct contains additional upstream CpG sites outside the range of our current bisulfite analysis. To determine whether methylation of the COX-2 promoter-driven expression vectors decreases COX-2 expression, the full-length and deleted COX-2 promoter expression plasmids were methylated in vitro with a CpG methyltransferase, M.SssI. Methylation of the construct was verified by treating the COX-2-driven luciferase plasmids with a methylation-sensitive restriction endonuclease, SmaI (data not shown). MHS cells were then transfected with methylated or unmethylated COX-2 firefly luciferase reporter plasmids. The control vector (pRL-SV40) was not treated with the methyltransferase prior to transfection. As shown in Figure 7A, when unmethylated, the deleted COX-2 promoter was able to stimulate firefly luciferase at levels similar to the full-length construct. This suggests that the deleted COX-2 promoter construct contains all relevant sequences for ensuring transcription in MHS cells. Furthermore, when methylated, expression driven by either promoter construct was significantly impaired. Taken together, these results demonstrate that methylation of the deleted promoter region is sufficient to impair transcriptional activity and thus indicate that the relevant CpG sites for analysis lie within the deleted promoter region which was previously analyzed in Figure 2.
Figure 7. COX-2 promoter-driven luciferase expression is decreased following treatment with methyltransferase, but expression is regulated by TGF-β1.
A: Transfections of MHS cells (6.2 × 104) were performed with a 50:1 ratio of either methylated or unmethylated, full-length or deleted (del) COX-2 promoter-driven firefly luciferase plasmid to pRL-SV40 vector using lipofectamine. Firefly:Renilla luciferase activities were quantified by a dual luciferase assay system. (n = 6; *p < .05). B: MHS were cultured overnight in the presence or absence of porcine TGF-β1 (1 ng/mL) and transfected with methylated or unmethylated short COX-2 reporter plasmids and pRL-SV40 and assayed as above (n=3, **p<0.001, ***p<0.001).
TGF-β1 can induce expression of COX-2 from methylated constructs
We have previously shown that BMT mice exhibit increased levels of TGF-β1 in the lung (24), and TGFβ1 can induce expression of COX-2 (Figure 6). To determine whether TGF-β1 could stimulate the transcription of COX-2 from a methylated promoter, we transfected MHS cells as above with the deleted COX-2 promoter-driven firefly luciferase construct that had either been methylated in vitro by M. Sss1, or left unmethylated. Cells were co-transfected with the SV40-driven renilla luciferase vector as a control. Cells transfected with the methylated construct were also treated with 1 ng/ml TGF-β1 or vehicle control. As expected, luciferase expression from the methylated COX-2 construct was impaired when compared to the expression from unmethylated constructs (Fig 7B). However, cells transfected with the methylated constructs exhibited increased luciferase expression when treated with TGF-β1. These data suggest that TGF-β1 may be able to promote the demethylation of the COX-2 promoter, or may promote the activity of the transcriptional machinery on methylated regions of DNA. Interestingly, LPS was not able to increase the transcriptional activity of the methylated COX-2 promoter (data not shown), suggesting that this effect may be specific to TGF-β1 signaling.
Improved phagocytosis in AMs from BMT mice following reconstitution with donor HSCs from CD11cdnR mice
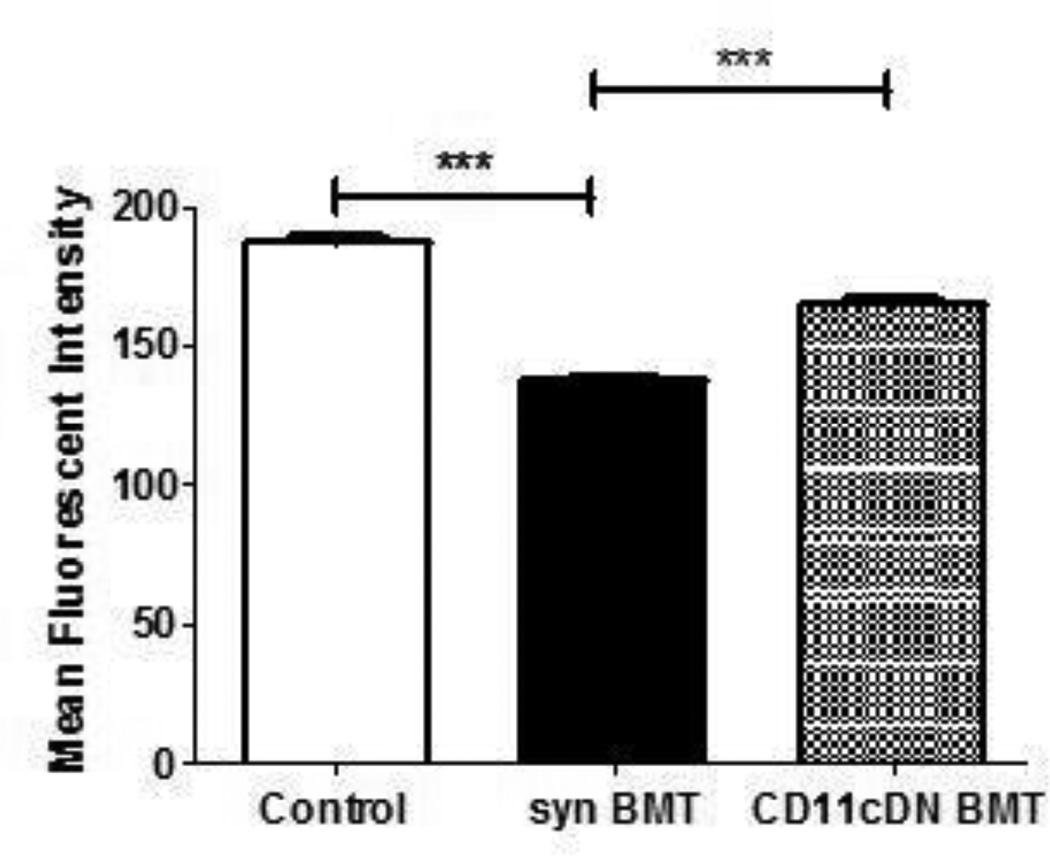
TGF-β1 can play an important role in both the activation and suppression of monocytes and macrophages, however, its role in tissue macrophages is primarily immunosuppressive (32). To determine how TGF-β1 may contribute to the defect in BMT AMs, we developed bone marrow chimeras using HSCs from CD11cdnR donor mice. These mice express a dominant-negative TGF-β1 receptor II under the control of the CD11c promoter which results in the generation of TGF-β1-resistant myeloid cells including dendritic cells, macrophages and natural killer cells (25). Five weeks post-BMT, AMs were harvested by BAL and phagocytosis was measured and compared to WT BMT and control mice. As expected, AMs from WT BMT mice displayed defective phagocytosis of P. aeruginosa when compared to cells from control mice. However, AMs collected from CD11cdnR BMT mice showed enhanced phagocytosis activity compared to WT BMTs (Figure 8). To determine whether this enhancement in AM function is mediated by a change in COX-2 gene methylation, DNA from CD11cdnR BMT AMs was harvested and analyzed for extent of methylation. Interestingly, the methylation of the COX-2 promoter from CD11cdnR BMT AMs reverted to a pattern more similar to control AMs than WT BMT AMs at some, but not all sites previously found to be demethylated in WT BMT AMs (Figure 9).
Figure 8. Improved AM phagocytosis with CD11cdnR bone marrow chimeras.
AMs were harvested by BAL 5 weeks post-transplant from BMT mice reconstituted with either WT or CD11cdnR donor marrow. Phagocytosis of heat-killed FITC-P. aeuroginosa was then assessed and compared to AMs from control mice (n=5; ***p<.001).
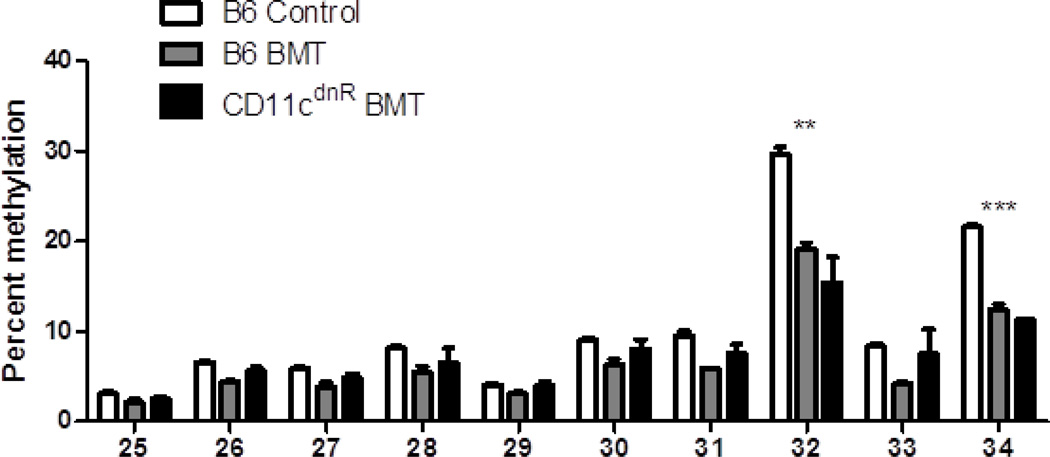
Figure 9. CD11cdnR BMT COX-2 promoter methylation is partially rescued.
Following DNA isolation from primary AMs from control, WT BMT and CD11cdnR BMT mice, DNA was subjected to bisulfite conversion and pyrosequencing for methylation analysis; n=5. WT BMT patterns were hypermethylated compared to control AMs at all sites. However, AMs from CD11cdnR BMT mice were not significantly different from control AMs except at CpG sites 32 and 34.
Discussion
We previously showed that AMs post-BMT are defective in phagocytosis and killing of P. aeuroginosa, a clinically relevant nosocomial pathogen which can afflict patients following hematopoietic stem cell transplantation (11, 13, 14, 16, 28). Defective AM function post-BMT is not exclusive to mice receiving TBI, but is also seen in mice receiving chemotherapy conditioning regimens (16), and persists despite immune reconstitution. We have previously shown that both PGE2 and TGF-β1 are elevated in lungs of BMT mice following immune reconstitution and both can have immunosuppressive effects (11, 24). We now show that irradiated mice receiving CD11c+ donor cells that are unable to respond to TGF-β1 exhibit improved AM function (Figure 8), indicating that TGF-β1 is contributing to the defect in AMs following transplantation. As the dominant negative TGFβ1 receptor II is under the CD11c promoter, it is possible that the incomplete rescue of AM function is due to decreased CD11c expression post-transplant (33). The methylation pattern of COX-2 in CD11cdnR BMT AMs supports this possibility as we observed an incomplete restoration of methylation to control patterns (Figure 9).
Overproduction of PGE2 in the lung has been reported following hematopoietic reconstitution and is intimately linked with a defective lung innate immune system (11). Here, we show that COX-2 transcription in AMs is elevated in mice post-BMT relative to control AMs. Additionally, previously published data showed upregulation of PGE2 specific synthase enzymes post-BMT as well (14). These results correlate with the elevated PGE2 levels reported previously in both mice and patients that have undergone HSCT (11, 13, 14, 16, 29). As other prostanoids [e.g. PGI2 and TXA2 (Figure 1b)] are also elevated after transplant these data indicate that increased COX-2 has a broad effect on downstream effectors. PGI2 signaling through its IP receptor on macrophages and neutrophils may play a similar role as PGE2 in inhibiting proper cell function as IP is a Gs–coupled receptor that signals through cAMP (23, 34). While TXA2 is also increased post-BMT, how it affects AM function is unclear. It is possible however that upregulation of TXA2 may have deleterious effects on platelet activation or myocardial ischemia [reviewed in (34)].
Upregulation of COX-2 expression and PGE2 production has been associated with a wide range of disease processes, of which cancer is most commonly reported (35). In these circumstances, PGE2 may promote cancer progression and metastasis as well as immune suppression. Furthermore, it was recently reported that chronic influenza infection was associated with upregulation of miR29b (36). The consequence of miR29b upregulation was the destabilization of DNA methyltransferases and the upregulation of COX-2 gene expression secondary to DNA demethylation (36). It is interesting that we have also observed upregulation of miR29b in BMT AMs (data not shown). Thus, it is possible that the process of BMT regulates COX-2 demethylation via miR29b expression.
To further understand the regulation of COX-2 expression in our syngeneic murine model for studying innate immune cells post-BMT, we examined methylation patterns of the COX-2 promoter of transplanted and untransplanted mice. Interestingly, we found that the COX-2 promoter region analyzed from AMs of transplanted mice was significantly hypomethylated, particularly around the transcription start site and into the first exon, compared to control untransplanted mice. The fact that the deleted COX-2 promoter drove expression in MHS cells and the fact that no CpG sites located upstream of the transcription start site showed methylation differences, suggests that the region of interest is located near the exon 1 border. Methylation changes of this magnitude have previously been shown to alter EP2 expression (37) and methylation around the transcription start site and in gene exons has been previously described [e.g. in (36, 38–40)]. It is interesting that in human studies related to Helicobacter pylori-induced COX-2 expression and influenza-induced COX-2 expression, demethylation occurs at similar sites as those identified in our murine study (36, 40). However, there are examples of regulation at different sites as well (e.g. hepatitis B demethylates COX-2 in the NFAT sites upstream of the transcription start site (41) and silencing of the COX-2 gene in human gastric cancers involves hypermethylation of promoter regions upstream of exon 1(42)). When we further explored this epigenetic mechanism of regulation and treated a murine AM cell line with a methyltransferase inhibitor, 5-aza-2'-deoxycytidine, an increase in COX-2 mRNA levels was detected and this induced expression was dose-dependent with increasing concentrations of 5-aza-2'-deoxycytidine (Figure 3A). A similar response was observed in primary AMs of control untransplanted mice that were treated with either 5-aza-2'-deoxycytidine or vehicle (DMSO, Figure 3B). Furthermore, when methylation patterns were analyzed following 5-aza-2’-deoxycytidine treatment, there was a significant decrease in methylation on CpG sites found to be hypomethylated in BMT AMs (Figure. 4). In contrast, HDAC acetylation did not appear to regulate COX-2 expression (Figure 5). Overall, these results suggest methylation of the promoter and/or beginning of exon 1 is an important mechanism for regulating COX-2 mRNA levels.
To establish a correlation between elevated levels of mRNA and COX-2 activity in response to methylating or demethylating conditions, the COX-2 promoter region was cloned into a luciferase vector whereby firefly luciferase activity could be driven by the COX-2 promoter and serve as a measure of COX-2 expression. MHS cells were successfully transfected, measured by pRL-SV40-driven activity, and luciferase activity of the COX-2 plasmid was increased in the presence of proinflammatory LPS and TGF-β1 (Figure 6). These data suggest that the reporter vector is regulated similarly to the native gene in MHS cells. Despite numerous attempts, primary AMs were unable to be transfected by either lipofectamine or electroporation methods. We next determined that a deleted COX-2 promoter construct (which contained the CpG sites we had analyzed for methylation patterns) was sufficient to induce COX-2 expression in MHS cells. We also studied the effects of methylation on the activity of this construct. In vitro methylation of the COX-2-driven luciferase vector resulted in decreased COX-2 promoter activity when transfected into the MHS cells. These results verify that methylation inhibits expression of COX-2, and suggest that the methylation of the COX-2 gene which is noted in AMs collected from untransplanted mice likely plays a role in the limited expression of COX-2 and the limited production of PGE2 in these cells under homeostatic conditions.
In contrast, treatment of MHS cells transfected with methylated COX-2 reporter plasmids with TGF-β1 was able to induce modest expression of COX-2 (Figure 7B). These results suggested that the increased levels of TGF-β1 which have been previously reported in BMT mice (24), may serve to induce COX-2 demethylation or increase transcription from the methylated promoter in the AMs of BMT mice. Interestingly, this activity was not noted with LPS, a molecule which is able to stimulate transcription off of unmethylated COX-2 promoters (Figure 6). Thus, these results suggest that the ability to stimulate transcription off of a methylated COX-2 promoter may be a unique action of TGF-β1 signaling. One caveat to these studies is that we do not know for certain whether the level of methylation achieved by in vitro reactions is similar to the levels noted in vivo in BMT mice. However, our finding of improved host defense in the CD11cdnR BMT mice which are unresponsive to TGF-β1 signaling in AMs is highly supportive, and our in vivo methylation patterns of CD11cdnR BMT mice are more similar to control AMs than to WT BMT AMs (Figure 9).
Our results suggest that TGF-β1 may be one mechanism to regulate the increased expression of COX-2 and thus PGE2 which occurs in AMs post-BMT. However, it is likely that other signals also occur secondary to conditioning regimen-induced alterations. This is supported by our observation that the phenotype of CD11cdnR BMT mice is only partially restored to control AM levels regarding phagocytosis or methylation patterns. The production of reactive oxygen species has been observed previously following ionizing radiation and has been implicated in compromised lung immunity (43). Reactive nitrogen species, particularly nitric oxide, have been suggested to induce changes in COX within airway epithelium during inflammation (44). In these studies, increasing concentrations of nitric oxide was able to induce the production of PGE2 in alveolar epithelial cells. Nitric oxide and PGE2 expression can be induced by proinflammatory cytokines (44). Therefore, it is possible that the formation of reactive species caused by irradiation or a chemotherapy regimen may also be playing a role in the induction of COX-2 transcription and expression.
Because irradiation can only directly affect cells that have been irradiated, the fact that unirradiated donor AMs are altered suggests that this impaired AM function may be due to an alteration in the lung microenvironment. Therefore, it is likely that the changes are induced by soluble factors that cause a change in cell behavior or function. Previous data from our lab has indicated that alveolar epithelial cells may influence AM function through the secretion of soluble factors like GM-CSF (28).While we have ruled out the contribution of GM-CSF to the induction of COX-2 in this model (28), it is possible that other soluble factors, in addition to TGF-β1 may trigger the epigenetic changes. This will be an area for future study.
Supplementary Material
Acknowledgments
The authors would like to thank Carly Smith for early help with the methylation studies.
Footnotes
Supported by NIH grants AI065543 (BBM) and T32 AI007413 (R D-G); HL094657 (SKH) ; National Multiple Sclerosis Society RF 4106A1/3 (YL).
References
- 1.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354:1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 2.Wingard JR, Hsu J, Hiemenz JW. Hematopoietic stem cell transplantation: an overview of infection risks and epidemiology. Infect Dis Clin North Am. 2010;24:257–272. doi: 10.1016/j.idc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Mir MA, Battiwalla M. Immune deficits in allogeneic hematopoietic stem cell transplant (HSCT) recipients. Mycopathologia. 2009;168:271–282. doi: 10.1007/s11046-009-9181-0. [DOI] [PubMed] [Google Scholar]
- 4.Guillaume T, Rubinstein DB, Symann M. Immune reconstitution and immunotherapy after autologous hematopoietic stem cell transplantation. Blood. 1998;92:1471–1490. [PubMed] [Google Scholar]
- 5.Auletta JJ, Lazarus HM. Immune restoration following hematopoietic stem cell transplantation: an evolving target. Bone Marrow Transplant. 2005;35:835–857. doi: 10.1038/sj.bmt.1704966. [DOI] [PubMed] [Google Scholar]
- 6.Afessa B, Abdulai RM, Kremers WK, Hogan WJ, Litzow MR, Peters SG. Risk factors and outcome of pulmonary complications after autologous hematopoietic stem cell transplant. Chest. 2012;141:442–450. doi: 10.1378/chest.10-2889. [DOI] [PubMed] [Google Scholar]
- 7.Chen CS, Boeckh M, Seidel K, Clark JG, Kansu E, Madtes DK, Wagner JL, Witherspoon RP, Anasetti C, Appelbaum FR, Bensinger WI, Deeg HJ, Martin PJ, Sanders JE, Storb R, Storek J, Wade J, Siadak M, Flowers ME, Sullivan KM. Incidence, risk factors, and mortality from pneumonia developing late after hematopoietic stem cell transplantation. Bone Marrow Transplant. 2003;32:515–522. doi: 10.1038/sj.bmt.1704162. [DOI] [PubMed] [Google Scholar]
- 8.Lossos IS, Breuer R, Or R, Strauss N, Elishoov H, Naparstek E, Aker M, Nagler A, Moses AE, Shapiro M, et al. Bacterial pneumonia in recipients of bone marrow transplantation. A five-year prospective study. Transplantation. 1995;60:672–678. doi: 10.1097/00007890-199510150-00010. [DOI] [PubMed] [Google Scholar]
- 9.Youssef S, Rodriguez G, Rolston KV, Champlin RE, Raad, Safdar A. Streptococcus pneumoniae infections in 47 hematopoietic stem cell transplantation recipients: clinical characteristics of infections and vaccine-breakthrough infections, 1989–2005. Medicine (Baltimore) 2007;86:69–77. doi: 10.1097/md.0b013e31803eb176. [DOI] [PubMed] [Google Scholar]
- 10.Afessa B, Peters SG. Major complications following hematopoietic stem cell transplantation. Semin Respir Crit Care Med. 2006;27:297–309. doi: 10.1055/s-2006-945530. [DOI] [PubMed] [Google Scholar]
- 11.Ballinger MN, Aronoff DM, McMillan TR, Cooke KR, Okiewicz K, Toews GB, Peters-Golden M, Moore BB. Critical Role of Prostaglandin E2 Overproduction in Impaired Pulmonary Host Response Following Bone Marrow Transplantation. J. Immunol. 2006;177:5499–5508. doi: 10.4049/jimmunol.177.8.5499. [DOI] [PubMed] [Google Scholar]
- 12.Deng JC, Cheng G, Newstead MW, Zeng X, Kobayashi K, Flavell RA, Standiford TJ. Sepsis-induced suppression of lung innate immunity is mediated by IRAK-M. J Clin Invest. 2006;116:2532–2542. doi: 10.1172/JCI28054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard L, Wilke C, White E, Moore B. PTEN Limits Alveolar Macrophage Function Against Pseudomonas aeruginosa Following Bone Marrow Transplantation. Am. J. Respir. Cell Mol. Biol. 2011;45:1050–1058. doi: 10.1165/rcmb.2011-0079OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard LL, Ballinger MN, Thomas PE, Wilke CA, Standiford TJ, Kobayashi KS, Flavell RA, Moore BB. A Role for IL-1 Receptor-Associated Kinase-M in Prostaglandin E2-Induced Immunosuppression Post-Bone Marrow Transplantation. J Immunol. 2010;184:6299–6308. doi: 10.4049/jimmunol.0902828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winston DJ, Territo MC, Ho WG, Miller MJ, Gale RP, Golde DW. Alveolar macrophage dysfunction in human bone marrow transplant recipients. Am J Med. 1982;73:859–866. doi: 10.1016/0002-9343(82)90777-x. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard LL, Ballinger MN, Wilke CA, Moore BB. Comparison of conditioning regimens for alveolar macrophage reconstitution and innate immune function post bone marrow transplant. Exp Lung Res. 2008;34:263–275. doi: 10.1080/01902140802022518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballinger MN, McMillan TR, Moore BB. Eicosanoid regulation of pulmonary innate immunity post-hematopoietic stem cell transplantation. Arch Immunol Ther Exp (Warsz) 2007;55:1–12. doi: 10.1007/s00005-007-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coomes S, Hubbard L, Moore B. Impaired pulmonary immunity post-bone marrow transplant. Immunologic Research. 2011;50:78–86. doi: 10.1007/s12026-010-8200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabral GA. Lipids as bioeffectors in the immune system. Life Sci. 2005;77:1699–1710. doi: 10.1016/j.lfs.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Aronoff DM, Canetti C, Peters-Golden M. Prostaglandin E2 inhibits alveolar macrophage phagocytosis through an E-prostanoid 2 receptor-mediated increase in intracellular cyclic AMP. J Immunol. 2004;173:559–565. doi: 10.4049/jimmunol.173.1.559. [DOI] [PubMed] [Google Scholar]
- 21.Hubbard LLN, Wilke CA, White ES, Moore BB. PTEN Limits Alveolar Macrophage Function against Pseudomonas aeruginosa after Bone Marrow Transplantation. American Journal of Respiratory Cell and Molecular Biology. 2011;45:1050–1058. doi: 10.1165/rcmb.2011-0079OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbard LL, Ballinger MN, Thomas PE, Wilke CA, Standiford TJ, Kobayashi KS, Flavell RA, Moore BB. A Role for IL-1 Receptor-Associated Kinase-M in Prostaglandin E2-Induced Immunosuppression Post-Bone Marrow Transplantation. J Immunol. doi: 10.4049/jimmunol.0902828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narumiya S, Sugimoto Y, Ushikubi F. Prostanoid Receptors: Structures, Properties, and Functions. Physiol Rev. 1999;79:1193–1226. doi: 10.1152/physrev.1999.79.4.1193. [DOI] [PubMed] [Google Scholar]
- 24.Coomes SM, Wilke CA, Moore TA, Moore BB. Induction of TGF-β1, Not Regulatory T Cells, Impairs Antiviral Immunity in the Lung following Bone Marrow Transplant. The Journal of Immunology. 2010;184:5130–5140. doi: 10.4049/jimmunol.0901871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-[beta] controls T helper type 1 cell development through regulation of natural killer cell interferon-[gamma] Nat Immunol. 2005;6:600–607. doi: 10.1038/ni1197. [DOI] [PubMed] [Google Scholar]
- 26.Laouar Y, Town T, Jeng D, Tran E, Wan Y, Kuchroo VK, Flavell RA. TGF-beta signaling in dendritic cells is a prerequisite for the control of autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2008;105:10865–10870. doi: 10.1073/pnas.0805058105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraemer SA, Arthur KA, Denison MS, Smith WL, DeWitt DL. Regulation of prostaglandin endoperoxide H synthase-2 expression by 2,3,7,8,-tetrachlorodibenzo-p-dioxin. Arch Biochem Biophys. 1996;330:319–328. doi: 10.1006/abbi.1996.0259. [DOI] [PubMed] [Google Scholar]
- 28.Ballinger MN, Hubbard LL, McMillan TR, Toews GB, Peters-Golden M, Paine R, 3rd, Moore BB. Paradoxical role of alveolar macrophage-derived granulocyte-macrophage colony-stimulating factor in pulmonary host defense post-bone marrow transplantation. Am J Physiol Lung Cell Mol Physiol. 2008;295:L114–L122. doi: 10.1152/ajplung.00309.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cayeux SJ, Beverly PCL, Schulz R, Dorken B. Elevated plasma prostaglandin E2 levels found in 14 patients undergoing autologous or stem cell transplantation. Bone Marrow Transplant. 1993;12:603–608. [PubMed] [Google Scholar]
- 30.Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 31.Chen GH, Curtis JL, Mody CH, Christensen PJ, Armstrong LR, Toews GB. Effect of granulocyte-macrophage colony-stimulating factor on rat alveolar macrophage anticryptococcal activity in vitro. J Immunol. 1994;152:724–734. [PubMed] [Google Scholar]
- 32.Letterio JJ, Roberts AB. Regulation of immune responses by TGF-β*. Annual Review of Immunology. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 33.Ojielo C, Cooke KR, Mancuso P, Standiford TJ, Olkiewicz KM, Cloutheir S, Corrion L, Ballinger MN, Toews GB, Paine R, Moore BB. Defective phagocytosis and clearance of Pseudomonas aeruginosa in the Lung Following Bone Marrow Transplantation. J. Immunol. 2003;171:4416–4424. doi: 10.4049/jimmunol.171.8.4416. [DOI] [PubMed] [Google Scholar]
- 34.Hata AN, Breyer RM. Pharmacology and signaling of prostaglandin receptors: multiple roles in inflammation and immune modulation. Pharmacol Ther. 2004;103:147–166. doi: 10.1016/j.pharmthera.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Dubois RN, Abramson SB, Crofford L, Gupta RA, Simon LS, Van De Putte LBA, Lipsky PE. Cyclooxygenase in biology and disease. The FASEB Journal. 1998;12:1063–1073. [PubMed] [Google Scholar]
- 36.Fang J, Hao Q, Liu L, Li Y, Wu J, Huo X, Zhu Y. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J Virol. 2012;86:1010–1020. doi: 10.1128/JVI.06169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, Peters-Golden M. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am. 2010;177:2245–2255. doi: 10.2353/ajpath.2010.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics. 2010;11:137. doi: 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pero R, Peluso S, Angrisano T, Tuccillo C, Sacchetti S, Keller S, Tomaiuolo R, Bruni CB, Lembo F, Chiariotti L. Chromatin and DNA methylation dynamics of Helicobacter pylori-induced COX-2 activation. Int J Med Microbiol. 2011;301:140–149. doi: 10.1016/j.ijmm.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Yue X, Yang F, Yang Y, Mu Y, Sun W, Li W, Xu D, Wu J, Zhu Y. Induction of cyclooxygenase-2 expression by hepatitis B virus depends on demethylation-associated recruitment of transcription factors to the promoter. Virol J. 2011;8:118. doi: 10.1186/1743-422X-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hur K, Song SH, Lee HS, Ho Kim W, Bang YJ, Yang HK. Aberrant methylation of the specific CpG island portion regulates cyclooxygenase-2 gene expression in human gastric carcinomas. Biochem Biophys Res Commun. 2003;310:844–851. doi: 10.1016/j.bbrc.2003.09.095. [DOI] [PubMed] [Google Scholar]
- 43.Yang HJ, Youn H, Seong KM, Yun YJ, Kim W, Kim YH, Lee JY, Kim CS, Jin Y-W, Youn B. Psoralidin, a dual inhibitor of COX-2 and 5-LOX, regulates ionizing radiation (IR)-induced pulmonary inflammation. Biochemical Pharmacology. 2011;82:524–534. doi: 10.1016/j.bcp.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 44.Neil Watkins D, Garlepp MJ, Thompson PJ. Regulation of the inducible cyclo-oxygenase pathway in human cultured airway epithelial (A549) cells by nitric oxide. British Journal of Pharmacology. 1997;121:1482–1488. doi: 10.1038/sj.bjp.0701283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.