Abstract
We have synthesized and investigated properties of new PEI-PEG-based polyplexes containing MC1SP-peptide, a ligand specific for melanocortin receptor-1 (targeted polyplexes), and control polyplexes without this ligand peptide (non-targeted polyplexes). The targeted polyplexes demonstrated receptor-mediated transfection of Cloudman S91 (clone M-3) murine melanoma cells that was more efficient than with the non-targeted ones. Transfection with the targeted polyplexes was inhibited by chlorpromazine, an inhibitor of the clathrin-mediated endocytosis pathway, and, to a lesser extent, by filipin III or nystatin, inhibitors of the lipid-raft endocytosis pathway, whereas transfection with the non-targeted polyplexes was inhibited mainly by nystatin or filipin III. The targeted polyplexes caused significantly higher in vivo transfection of melanoma tumor cells after intratumoral administration compared to the non-targeted control. The targeted polyplexes carrying the HSVtk gene, after ganciclovir administration, more efficiently inhibited melanoma tumor growth and prolonged the lifespan of DBA/2 tumor-bearing mice compared to the non-targeted ones. Packed targeted polyplexes appeared and accumulated in the melanoma cells six hours earlier than the non-targeted ones. The targeted polyplexes enter into the nuclei of the melanoma cells more rapidly than the non-targeted control, and this difference may also be attributed to processes of receptor-mediated endocytosis. We believe that these data may be useful for the optimization of polyplex systems.
Keywords: polyethylenimine, polyethylene glycol, polyplexes, melanocortin receptor-1, intracellular trafficking, transfection
1. Introduction
Mechanisms directing cell penetration, intracellular trafficking, and polyplex unpacking strongly determine transfection efficiency of such non-viral vectors as polyplexes [1,2] – complexes of cationic polymers with DNA. Among the most efficient types of polyplexes are polyethylenimine (PEI)-based ones [3]. Though PEI polymers do not demonstrate obvious cell specificity, chemical modification of the PEI backbone with ligands [4,5] such as sugars, peptides, and proteins, including nanobodies [6], provided cell specificity to the polyplexes made of these targeted PEI polymers. Modification of polyplex polymers with ligands to internalizable receptors helps to achieve at least two goals: cell specificity and receptor-mediated internalization into the target cell [2]. To achieve these goals, one needs to find such receptors on the target cells that are overexpressed on them but only rarely represented on other, non-target cells. Successful attempts to accomplish this approach includes liganding PEI (or PEI-containing block-copolymers) with polypeptide ligands [7] (e.g. transferrin, epidermal growth factor (EGF), transforming growth factor-α, fibroblast growth factor, vascular endothelial growth factor) and with low molecular weight receptor ligands like clenbuterol [8], an agonist of β2-adrenoreceptors. Endocytotic pathways of targeted polyplexes are usually at least partially predetermined by their ligands [5]. In contrast, endocytosis of non-targeted polyplexes sometimes differs from its polymeric part [9,10], which compacts DNA and carries ligands, so it is doubtful whether data about endocytosis pathways of polymeric parts can be used to predict endocytosis of the whole non-targeted polyplex. Endocytotic pathways can be influenced by many factors: e.g. plating mesenchymal stem cells on collagen I-coated surfaces inhibited transfection, while plating them on fibronectin-coated surfaces enhanced transfection, which was due to fibronectin-promoted internalization through clathrin-mediated endocytosis [11]. It should be mentioned that for non-targeted PEI-based polyplexes the pathways may also be different and dependent on the cell type used [12].
Investigations aiming to reveal pathways of polyplex penetration are generally carried out using known inhibitors of different endocytosis pathways: e.g. chlorpromazine, an inhibitor of clathrin-dependent pathway; filipin III, nystatin, and genistein, inhibitors of caveolae-dependent pathway; wortmannin and LY294002, inhibitors of macropinocytosis. Though some investigators used the term “selective” for the above-mentioned inhibitors of endocytosis, one should keep in mind that effects of these inhibitors can be cell-type dependent [3], which is especially important when effects of polyplexes are compared for different cell lines.
Simultaneous characterization of intracellular trafficking and unpacking of polyplexes can be accomplished with polyplexes in which DNA is labeled with quantum dots, whereas their polymeric part is labeled with a fluorescent dye with spectral characteristics permitting Förster resonance energy transfer (FRET) measurements [13–17]. This approach together with mathematical modeling permits estimation of quantitative characteristics of both these processes (e.g. unpacking rate constants, rate constants of entry and exit into/from subcellular compartments, etc.) [16,17].
Melanocortin receptor-1 (MC1R), a Gs protein-coupled receptor expressed in melanocytes, determines skin pigmentation by increasing cAMP level and subsequent downstream activation of tyrosinase, a key enzyme for melanogenesis [18]. Mutations of the MC1R gene are often associated with cutaneous melanomas [19]. MC1Rs are often overexpressed on melanoma cells [20–23] as well as on surrounding cells such as keratinocytes [24,25]. The α-melanocyte-stimulating hormone (αMSH), a natural tridecapeptide ligand to these receptors, has the same sequence in humans and mice. Sensitivity of human MC1Rs to αMSH is even higher than that of mouse ones [26]. Here we report results concerning synthesis and properties of new targeted PEI-PEG-based polyplexes containing MC1SP-peptide – a ligand specific for MC1R overexpressed on the cells of many melanomas, both human and murine [20–23].
We believe that our data may be useful for the optimization of polyplex systems.
2. Materials and methods
2.1. Synthesis of polymeric part of polyplexes
Block-copolymer of linear 25 kDa polyethylenimine (PEI), Polysciences (Warrington, PA), and heterobifunctional polyethylene glycol MAL-dPEG24™-NHS ester (PEG), Quanta BioDesign (Powell, OH), was synthesized as previously described [16]. Briefly, PEI solution (2.83 mg/ml) in 0.1 M borate buffer at pH 5.5 was added to bifunctional PEG containing both a maleimide and N-hydroxysuccinimide ester groups (PEG/PEI ratios were 6.8 ± 2.1 (mean ± S.D.)). The activation reaction was carried out for 4 h at room temperature followed by pH adjustment to 7 with NaOH. MC1SP-oligopeptide, CGYGPKKKRKVSGSGSSIISHFRWGKPV (Rusbiolink, Moscow, Russia), was added in 3.2-fold excess and covalently attached to the PEI-PEG block-copolymer. Under these conditions, purification of the reaction mixture from unreacted PEG did not change the coupling of the peptide to the PEI-PEG. The sequence in bold is a specific ligand to MC1Rs [27], and the sequence in italic is the simian virus 40 large T-antigen minimal nuclear localization signal [28]. The dissolved oligopeptide was added to the activated PEI-PEG for the reaction of maleimide residues of the bifunctional PEG with cysteine sulfhydryl groups of MC1SP-peptide. The mixture was stirred for 2 h at room temperature and kept in a refrigerator overnight at 4°C with constant stirring. Unreacted PEG and low molecular weight residues were removed with an Amicon Ultra Ultracel-10k ultrafiltration cell (Millipore, Bedford, MA) equipped with a 10 kDa molecular weight cutoff membrane and 0.1 M borate buffer at pH 7.5 as the eluent. The PEI concentration of the conjugate was measured by a copper assay at 620 nm [29,30]. Measurements of PEI concentration by this method in a mixture of PEI, PEG, and the peptide with known PEI concentrations showed that PEG and the peptide did not interfere with the copper assay. (MC1SP, NLS−) oligopeptide, CGYGPKTKRKVSGSGSSIISHFRWGKPV (Rusbiolink, Moscow, Russia), was similarly attached to the PEI-PEG block-copolymer. The PEG concentration was determined by a method described by Gong et al. [31]. The MC1SP concentration was determined by the increase in amino group concentration [32]. The ratios of MC1SP-peptide/PEI in the conjugates were 0.63 ± 0.22 (mean ± S.D.).
2.2. Preparation of polyplexes
Polyplexes were prepared [16] in sterile isotonic glucose solution at pH 7.4. Briefly, the polymer solution was added rapidly to the DNA and mixed by vortex followed by 20 min incubation at room temperature prior to use.
2.3. Dynamic light scattering and zeta potential determination
Particle sizes of polyplexes were measured using a ZetaPALS instrument (Brookhaven Instruments, Holtsville, NY) in 5% D-glucose, 5 mM HEPES, pH 7.4. Particle sizes of polymers were measured in 0.1 M borate buffer at pH 7.5. The instrument was validated with nanosphere size standards (polystyrene microspheres in water, 92 ± 3.7 nm) from Duke Scientific (Palo Alto, CA). The scattered light was detected at 90° angle. Measurements were performed in 40 μl quartz cuvettes at 25°C with count rates between 10 and 40 kCps in ten or more runs of 240-s duration each and analyzed by multimodal size distribution analysis. Mean hydrodynamic diameter of particles was determined for the fraction with maximum number weight (more than 99% of all particles).
The ζ-potentials of polyplexes were measured in the standard electrophoresis cell of the ZetaPALS instrument in 5% D-glucose, 5 mM HEPES, pH 7.4. The instrument was validated with ζ-potential reference material (BI-ZR3, −53 ± 4 mV) from Brookhaven Instruments (Holtsville, NY). Measurements were performed in 4 ml glass cuvettes (with 1.6 ml of sample solution added) at 25°C in ten or more runs of 20 cycles duration each. The results are the mean values of indicated number of (usually, 10–40) runs with S.E.M. For ζ-potential measurements, the following parameters were used: viscosity, 1.078 cP; refractive index, 1.338; dielectric constant, 78.54.
2.4. Plasmids
pEGFP-N3 (Clontech, Mountain View, CA), pGL3-CMV, and pCMV-HSVtk encoding enhanced green fluorescent protein, firefly luciferase, and HSVtk gene respectively were propagated in Escherichia coli (DH5α), purified by EndoFree Plasmid Maxi Kit (Qiagen, Hilden, Germany), and kept at −40 °C. We used for all these genes the cytomegalovirus immediate early promoter (CMV) that is often employed for PEI-mediated transfection [33–35]. For experiments with plasmid DNA labeling with QD605 quantum dots, we used the phTERT-TK plasmid encoding HSVtk under human telomerase reverse transcriptase promoter as described earlier [16].
2.5. Cell culture
Cloudman S91 mouse melanoma cells (clone M-3) and Cloudman S91 mouse melanoma cells (clone M-3) stably expressing the CopGFP gene, hereafter called M-3 and M-3 CopGFP cells, as well as HEK293 cells (human embryonic kidney cells) were cultured in DMEM/F12 medium supplemented with 10% FBS. All cultured cells were grown at 37°C in a humidified 5% CO2 atmosphere.
2.6. In vitro gene delivery
Two cell lines were used: M-3 and HEK293 cells. The cells were grown in DMEM/F12 medium supplemented with fetal calf serum (both Gibco Invitrogen, Eugene, OR) at 37°C and 5% CO2 in air. For transfection, the cells were seeded onto 48-well plates, 12,000 cells per well, and incubated in serum-containing medium for 24 h. After replacement of the cultural serum-containing medium, polyplex solutions were added to the cells to final DNA concentration of 0.5 μg/ml. For details for transfections with inhibitors of endocytosis, see Supplementary Materials and Methods.
Transfection efficacy was estimated either as a percentage of cells expressing EGFP or as firefly luciferase activity (relative light units (RLU) per mg protein). In the first case, the cells were photographed 48 h after the start of incubation with polyplexes under a Zeiss LSM 510 META NLO (Carl Zeiss, Oberkochen, Germany) confocal laser scanning microscope, with ×10 lens. Then the number of cells expressing EGFP as well as the total number of the cells in the monolayer were counted, and the percentage of EGFP-expressing cells was calculated. For description of measuring of firefly luciferase activity, see Supplementary Materials and Methods.
2.7. Cellular uptake of polyplexes
To assay polyplex internalization, M-3 cells were seeded in 24-well plates at density 3×104 cells per well 48 h prior to transfection. They were preincubated with chlorpromazine, nystatin, or filipin III in fresh serum-free culture medium for 45 min (see details in Supplementary Materials and Methods). Subsequently, polyplexes containing QD605-labeled DNA were added to the cells at DNA concentration 0.5 μg/ml. After 4.5 h, the cells were washed once and treated with 100 IU/ml heparin for 15 min at 37 °C to remove extracellularly bound complexes. The cells were then harvested by treatment with trypsin (0.25 %) in Versene solution (PanEko, Moscow, Russia), precipitated by centrifugation at 1000×g for 5 min, and dissolved in Versene solution. Analysis was performed using an Epics Altra Flow Cytometer (Beckman Coulter, Miami, FL). The QD605 fluorophore was excited at 488 nm, and emission was detected at 610 nm. To discriminate between vital and apoptotic cells, cells showing low level in forward-angle light scatter and high level in side-angle light scatter as a measure for the cell diameter and the conformation of inner cellular structures were excluded from further analysis [36]. Per sample, 1×104 gated events were collected. To study the influence of the inhibitors on cellular uptake of polyplexes, we analyzed mean fluorescence intensity of all cells in the sample. Control cells were pre-chilled on ice for 4.5 h with polyplexes before commencing the study. Control cells were used to account for cell autofluorescence and exclude fluorescence from extracellularly bound complexes.
2.8. Establishment of melanoma tumors
M-3 or M-3 CopGFP mouse melanoma tumors were established in female DBA/2 mice by subcutaneous injection of 2×106 M-3 cells suspended in 20 μl DMEM/F12 media into the flank region. Polyplexes were locally injected into the tumor mass when the tumor size was 400–600 mm3 for mRNA and luciferase expression studies or 250 mm3 for gene therapy study. Tumor volume was calculated according to ellipsoid formula [37]. Tumor-bearing mice were euthanized when their tumors achieved 1500 mm3 volume. The animals were maintained under specific pathogen-free conditions with access to mouse chow and water ad libitum. The experimental protocol was approved by the Institute Commission for Animals.
2.9. Gene therapy study in vivo
Polyplexes with pCMV-HSVtk DNA concentration of 80 μg/ml and PEI nitrogen to DNA phosphate ratio (N/P) equal to 30 were used for treatment of tumors. DBA/2 mice bearing experimental subcutaneous M-3 melanoma tumors were intratumorally (10 μg DNA/tumor) injected with 125 μl of polyplexes on the 18th day after inoculation. Seven intraperitoneal injections of ganciclovir (25 μg/g, Hoffmann-La Roche Ltd., Switzerland) were made twice per day. One day after the last ganciclovir injection, the polyplexes were injected the second time with subsequent seven ganciclovir administrations as earlier. The control group was intraperitoneally injected with ganciclovir only.
2.10. Fluorescent labeling of polyplex components and formation of fluorescently labeled polyplexes
Plasmid DNA labeling with QD605 quantum dots, PEI-based block-copolymer labeling with Alexa Fluor647 dye, and formation of fluorescently labeled polyplexes were performed as described earlier [16].
2.11. Transfection of living cells with labeled polyplexes and laser scanning microscopy
M-3 cells were seeded into POCmini chambers (Cell Cultivation System: Open Cultivation, PeCon, Erbach, Germany), 20,000–50,000 cells per chamber in 1 ml of DMEM/F12 medium with 10% bovine fetal serum, and cultivated at 37°C under 5% CO2. After a specified time, the cells were repeatedly washed with Improved MEM Zinc Option medium without indicator supplemented with 10% bovine fetal serum, 8 mM HEPES, and 0.15 % NaHCO3 (IMEM) and transfected with the labeled polyplexes (0.5 μg/ml DNA) in the same medium. Then 50–60 min before imaging, 5 μg/ml Hoechst 33258 was added to the transfected cells to visualize the cell nuclei. Endocytosis processes in the living cells were inhibited by cooling the chambers to 10–12°C for 10–15 min. Then the medium was replaced with IMEM containing 5 μg/ml wheat germ agglutinin labeled with Cy3 to visualize plasma membranes, and the cells were incubated for 10 min more at 10–12°C on a cooled microscope stage. The cells were then washed two times with IMEM, and the initial culture medium containing polyplexes was added to them.
At times 2, 4, 6, and 8 h after transfection, the cells were examined using the LSM-510 Meta NLO multiphoton laser scanning microscope equipped with Plan-Apochromat ×63/1.4 Oil DIC lens and cooled stage (10–12°C). For each time point, at least 150–200 cells were randomly selected. Cell images were obtained using multi-track Z-stack mode (not less than 25 optical sections for every stack). Hoechst 33258 was registered using 2-photon excitation at 760 nm and 435–485 nm pass band for emission. FRET measurements for the QD605–Alexa Fluor647 pair were made: (1) at 458 nm excitation and 650–710 pass band for emission (AlexaFluor647 emission, energy transfer, if any) and (2) at 458 nm excitation and 590–612 nm pass band for emission (QD605, absence of energy transfer).
3. Results
3.1. Characterization and transfection properties of targeted and non-targeted polyplexes
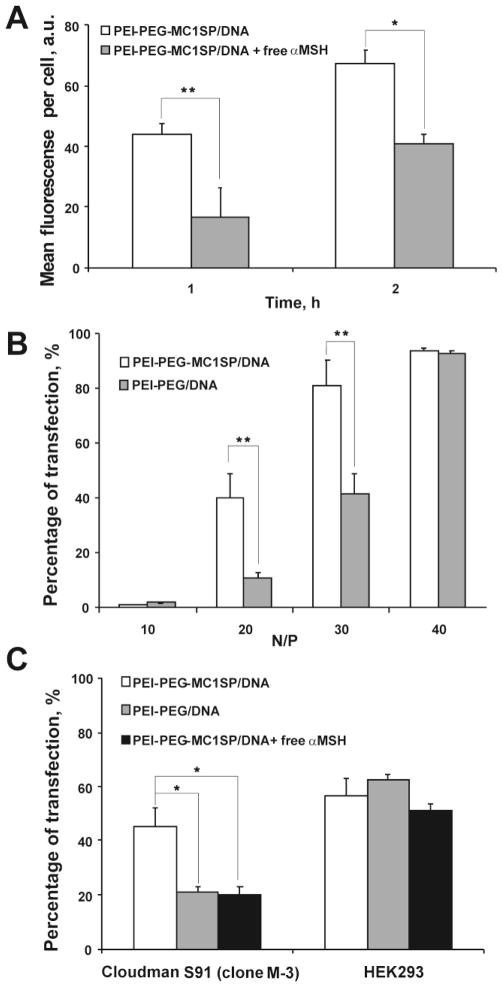
Our method for synthesis of peptide-containing PEI-based block-copolymers [16] yielded PEI-PEG-MC1SP product having similar ability to interact with MC1Rs as free MC1SP-peptide, which was demonstrated by induction of receptor-mediated melanogenesis using B16-F1 mouse melanoma cells (Supplementary Fig. 1). Mixing plasmid DNA with PEI-PEG-MC1SP or PEI-PEG (at N/P = 30) resulted in formation of polyplex nanoparticles with average hydrodynamic diameters of 47.1 ± 0.7 and 53.6 ± 1.0 nm, respectively (± S.E.M. for 9–10 runs). The ζ-potentials for the targeted and non-targeted polyplexes at N/P = 30 were 22.7 ± 1.0 and 19.4 ± 0.5 mV (± S.E.M. for 19–25 runs), respectively. For more detail information, see Supplementary Results and Supplementary Fig. 2. The uptake of targeted polyplexes was inhibited by free α-MSH (Fig. 1A). Competitive inhibition of intracellular uptake by the free ligand indicates high contribution of receptor-mediated endocytosis to entry of polyplexes into the cells.
Fig. 1.
Impact of free αMSH on cellular uptake of targeted polyplexes and TEs of M-3 and HEK293 cells with targeted and non-targeted polyplexes. Targeted polyplexes (PEI-PEG-MC1SP/DNA) with QD605 labeled plasmid DNA were added to M-3 cells in presence or absence of free αMSH (1μM) (A). M-3 cells were transfected with targeted (PEI-PEG-MC1SP/DNA) and non-targeted (PEI-PEG/DNA) polyplexes with N/P ratios of 10, 20, 30 and 40 (B). M-3 and HEK293 cells were transfected with i) targeted (PEI-PEG-MC1SP/DNA) polyplexes, ii) non-targeted (PEI-PEG/DNA) polyplexes, and iii) targeted polyplexes with 1 μM of αMSH (PEI-PEG-MC1SP/DNA + free αMSH); N/P ratio was 20 (C). All values are shown as mean ± S.E.M. (n = 6–9). *p < 0.05, **p < 0.03 (Mann-Whitney U test for A and B, one-way ANOVA followed by a post hoc Dunnett’s t-test for C).
For N/P 10, 20, 30, and 40, we evaluated transfection efficacies (TEs) with the EGFP reporter gene as the percentage of EGFP expressing cells (Fig. 1B) and cytotoxicities (Supplementary Fig. 3) of both targeted and non-targeted polyplexes. The targeted polyplexes carried the MC1SP peptide, with its C-part being a selective ligand [27] to MC1Rs overexpressed on many melanoma cells including Cloudman S91 murine melanoma cells (ca. 5,000 sites per cell) [18]. These polyplexes demonstrated significantly higher TE than non-targeted ones at N/P of 20 and 30. Addition of excess αMSH decreased the TE of targeted polyplexes to the level of the non-targeted ones on M-3 melanoma cells. We did not observe these effects (enhancement of TE with targeted polyplexes in comparison with non-targeted ones or competitive action of αMSH) on HEK293 cells lacking MC1Rs (Fig. 1C) [38].
The difference between the PEI-PEG-MC1SP and PEI-PEG polyplexes cannot be attributed to the influence of the polyplexes on promoter. We did not find statistically significant difference between these polyplexes with DNA not encoding fluorescent protein when they were added to cells permanently expressing green fluorescent protein under the CMV promoter (Supplementary Fig. 4).
The MC1SP peptide includes also a positively charged stretch, the SV40 large T-antigen nuclear localization sequence (NLS), at its N-terminus located after an –SGSG– spacer (see Materials and Methods). This stretch was included to check whether the NLS in the polymeric part of polyplex nanoparticles can enhance their transfection efficacy. We compared transfectabilities of the targeted polyplexes described above with polyplexes where MC1SP was replaced with its analog, CGYGPKTKRKVSGSGSSIISHFRWGKPV (MC1SP, NLS−), with 7Thr (in bold) instead of 7Lys. This replacement completely abolishes nuclear translocation of macromolecules with such a motif [39]. Experiments did not show any statistically significant difference between these polyplexes: firefly luciferase activity after transfection with MC1SP-containing polyplexes carrying the luciferase gene was (4.9 ± 1.9) × 108 relative light units (RLU) per mg protein (mean ± S.E.M., n = 5) and that for (MC1SP, NLS−)-containing ones was (6.3 ± 2.8) × 108 RLU per mg protein (n = 5); N/P = 30, 40-hr incubation with M-3 cells in both cases. This result shows little contribution of the packed targeted polyplex nanoparticle transport through nuclear pore complex for entry into the nuclei for the fast dividing cells.
3.2. Effects of inhibitors of endocytosis on transfection efficacy and entry of polyplexes into cells
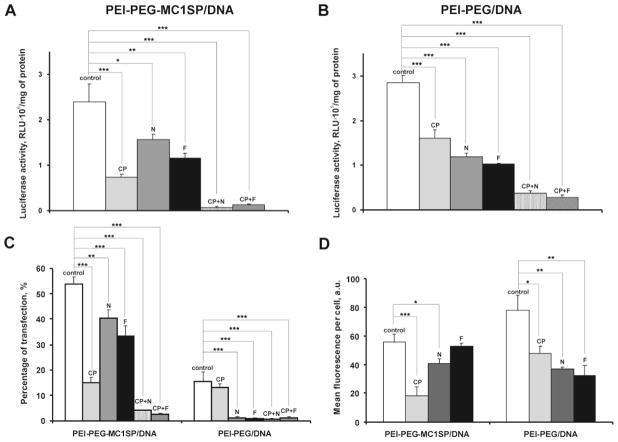
To determine the contribution of endocytosis pathways of targeted and non-targeted polyplexes to their TEs, we used chlorpromazine as an inhibitor of clathrin-dependent endocytosis and nystatin and filipin III as inhibitors of lipid raft-dependent endocytosis. The inhibitors were used in nontoxic concentrations (Supplementary Materials and Methods, Supplementary Fig. 5 and [40,41]).
Usually, incubation of cells with endocytosis inhibitors and polyplexes ends with washing of the cells either with fresh medium or with a simple replacement of the medium [11,42]. Our preliminary experiments (Supplementary Fig. 6) demonstrated that these operations are not sufficient to wash out polyplexes. Residual unwashed polyplexes in the absence of inhibitors as a result of replacement of medium could influence the experimental results, so we sought better washing conditions. We found that addition of heparin resulted in practically complete washing out of transfectable polyplexes, so the experiments were carried out under these conditions.
Transfection efficacy was evaluated using both determination of percentage of transfected cells with EGFP reporter gene and assay of firefly luciferase activity, and the two approaches gave similar results. Transfection of M-3 cells with targeted polyplexes was inhibited more by chlorpromazine than by nystatin or filipin III, whereas the opposite situation was observed with non-targeted polyplexes: their effect was mainly inhibited by nystatin or filipin III. The combination of inhibitors of both endocytosis pathways resulted in an almost complete suppression of transfection (Fig. 2A–C).
Fig. 2.
Transfection efficacies and cellular uptake of targeted and non-targeted polyplexes in the presence of inhibitors of endocytosis. M-3 cells were pretreated with chlorpromazine (CP), nystatin (N), filipin III (F), or their mixtures and targeted (PEI-PEG-MC1SP/DNA) or non-targeted (PEI-PEG/DNA) polyplexes with the plasmid encoding luciferase (A, B) or EGFP (C) reporter genes or plasmid DNA labeled with QD605 (D) for 4.5 h. Reporter gene expression was determined 48 h after transfection. Uptake of polyplexes was assessed by flow cytometry. Results are means ± S.E.M. (n = 4–6). *p < 0.05, **p < 0.01, ***p < 0.0001 (one-way ANOVA followed by a post hoc Dunnett’s t-test).
Cell entry of polyplexes, the DNA of which was labeled with QD605 quantum dots, was measured using flow cytometry. The data showed (Fig. 2D) that cell penetration of the targeted polyplexes was inhibited mainly by the inhibitor of the clathrin-dependent pathway, while suppression of entry of non-targeted polyplexes needed inhibitors of both types.
3.3. Assessment of tumor transgene expression after intratumoral administration of targeted and non-targeted polyplexes
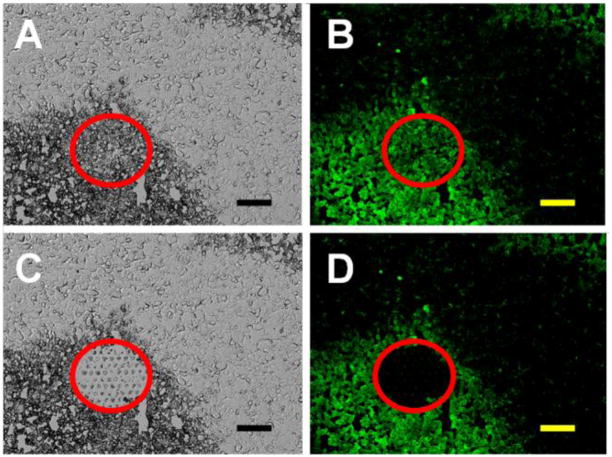
The difference in TE of targeted and non-targeted polyplexes was also demonstrated in vivo. The polyplexes carrying firefly luciferase reporter gene (in pCMV-Luc plasmid) were administered intratumorally into M-3 cell (transformed with the CopGFP gene) subcutaneous experimental tumors of DBA/2 mice. Total RNA was isolated from cancer cell areas removed from frozen sections of the tumors using laser dissection and catapulting (Fig. 3). Laser microdissection is a method for separating and extracting cells of interest out of tissue samples. For the separation we marked tumor tissue regions (M-3 melanoma cells stably transformed with the CopGFP gene were revealed by their pigmentation and green fluorescence) on the monitor and the laser cut them out and separated them from the residual tissue. The separated specimens were extracted with a software-controlled laser pulse and were catapulted into a microfuge tube.
Fig. 3.
Isolation of M-3 cells from frozen sections of M-3 melanoma tumors using laser dissection and catapulting. A typical image of a tumoral microscopic section before (A, B) and after (C, D) laser microdissection and catapulting using a PALM laser capture microscope. Images A and C represent transmitted light, and images B and D represent fluorescence. Red circles limit the dissection area of the cancerous CopGFP-expressing M-3 cells in the tumor. The bars indicate 50 μm.
The RNA was purified and assayed using real-time RT-PCR. Detailed information about preparation of tumor specimens and analysis of transgene mRNA expression using RT-PCR are given in Supplementary Materials and Methods. Using the comparative ΔΔCt method and internal b2M for normalization [43], targeted polyplexes demonstrated significantly higher expression of firefly luciferase in cancer cells after in vivo transfection than non-targeted ones (Table. 1).
Table 1.
Transfection efficacies of M-3 cells with targeted and non-targeted polyplexes in vivo
| Polyplex | Mouse, no. | Reporter gene, CT | Reference gene, CT | ΔCT* | MeanΔΔCT |
|---|---|---|---|---|---|
| PEI-PEG-MC1SP/DNA | 1 | 24.70 ± 0.33 | 32.82 ± 0.32 | −8.12 | −8.31 |
| 2 | 19.34 ± 0.56 | 29.31 ± 0.53 | −9.97 | ||
| 3 | 17.03 ± 0.14 | 23.88 ± 0.56 | −6.85 | ||
| PEI-PEG/DNA | 4 | 20.32 ± 0.06 | 21.35 ± 0.03 | −1.04 | −1.66 |
| 5 | 24.98 ± 0.19 | 28.17 ± 0.14 | −3.19 | ||
| 6 | 20.04 ± 0.29 | 20.79 ± 0.15 | −0.75 |
ΔCT are differences of threshold cycle numbers (CT) for reporter luciferase gene and reference beta-2-microglobulin gene. Cancer cells were isolated using a PALM laser capture microscope from M-3 melanoma tumors. Results are means ± S.E.M.
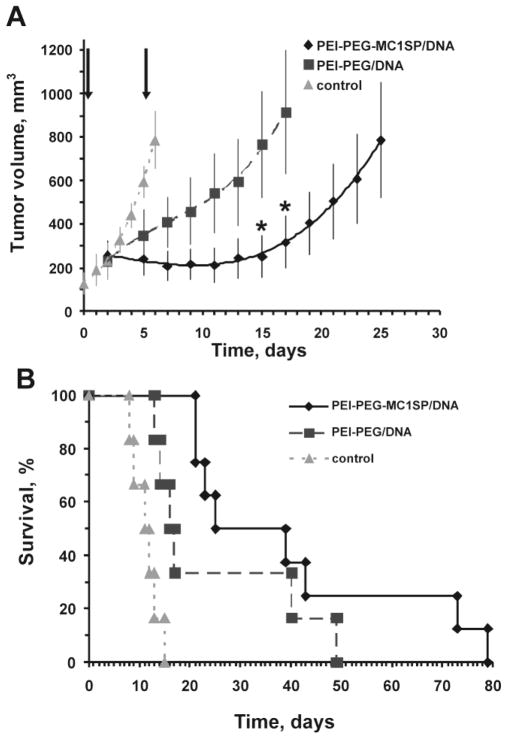
Based on optimal composition of polyplexes that provided high TE (Fig. 1B), we investigated in vivo delivery of the HSVtk therapeutic gene under the CMV promoter by the polyplexes after their intratumoral injection into experimental M-3 melanoma tumors subcutaneously produced in DBA/2 mice. The Herpes simplex virus thymidine kinase gene (HSVtk) is widely used for suicide therapy. This enzyme is responsible for converting the rather nontoxic drug ganciclovir into its triphoshorylated form, which interferes with DNA synthesis leading to death of transfected cells [44]. Two intratumoral injections of polyplexes together with intraperitoneal ganciclovir administration resulted in significant inhibition of tumor growth; moreover, the targeted polyplexes induced tumor regression during the treatment (Fig. 4A). Treatment with the targeted polyplexes resulted in a 3.6-fold enhancement of the lifespan of the mice, which is 1.6-fold higher than the effect of control non-targeted polyplexes (Fig. 4B).
Fig. 4.
Tumor growth (A) and survival of tumor-bearing mice (B) after intratumoral injection of targeted and non-targeted polyplexes with HSVtk gene under CMV promoter and ganciclovir. The injections of targeted (PEI-PEG-MC1SP/DNA) (n = 8) or non-targeted (PEI-PEG/DNA) (n = 6) polyplexes, indicated with arrows, were made intratumorally to DBA/2 mice with subcutaneous M-3 melanoma tumors. These were followed by administrations of ganciclovir (25 mg/kg, twice per day for 3.5 days). The control group (n = 6) was intraperitoneally injected with ganciclovir only. Results are means ± S.E.M. p < 0.05 for relative tumor growth with administration of targeted polyplexes compared to non-targeted ones. *p < 0.05 (Mann-Whitney U test).
Our data demonstrate both in vitro and in vivo higher TE of targeted polyplexes in comparison with control non-targeted ones. The data, especially those obtained in vitro, also serve as an indicator that the causes of this difference can be sought in features of cell entry and trafficking of the polyplexes.
3.4. Intracellular transport and unpacking kinetics of polyplexes
Using FRET technology as described earlier [16], polyplexes with QD605-labeled DNA and Alexa Fluor647-labeled PEI-based block-copolymer were colocalized with nuclei or cytoplasm using in-house computer-aided particle localization (CAPL) software (Supplementary Fig. 7), and kinetic curves of polyplex accumulation in these compartments (five experiments with targeted polyplexes and three experiments with non-targeted polyplexes) were obtained. Objects exhibiting FRET-mediated AlexaFluor647 signals were considered as condensed DNA within polyplexes (“packed DNA”), and objects exhibiting only Qdot signals were considered as released, repacked, or free DNA (“unpacked DNA”). Transport rates and unpacking kinetics of polyplexes were assessed as described in the Supplementary Materials and Methods.
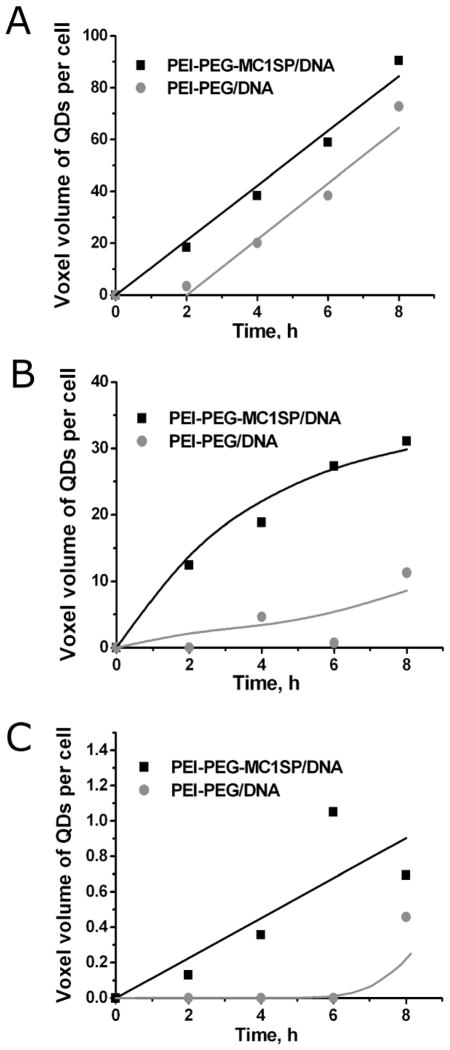
A 2-h delay in accumulation was estimated for non-targeted polyplexes (PEI-PEG/DNA) (Fig. 5A) as opposed to no delay for targeted ones (PEI-PEG-MC1SP/DNA).
Fig. 5.
Kinetics of accumulation of polyplexes. Overall accumulation (A) and accumulation of packed (B) targeted (PEI-PEG-MC1SP/DNA) and non-targeted polyplexes (PEI-PEG/DNA) inside cells and overall accumulation of polyplexes in nuclei (C). Effective concentration of internalized polyplexes is expressed as a voxel volume of quantum-dot signal (sum of pixel areas on each slice of 3D-image) per cell [46]. Solid curves are model curves for A and B (see below) and trend lines for C.
Similar results were obtained for packed polyplexes, i.e. no entry delay was observed for the targeted polyplexes, and a 6-h delay was found for the non-targeted polyplexes (Fig. 5B). This delay may be due to the fact that beginning from the first time points, most of the non-targeted polyplexes localized at the cell surface produced no detectable FRET signal: the result that can be interpreted as a loss of the polymeric part of the polyplexes. Early dissociation of polyplexes leads to DNA being in an unpacked state, which might be followed by DNA packaging with a natural polycation. The unpacked DNA state may be a cause for its degradation by nucleases in acidifying compartments [45], thus lowering transfection.
There is also a 6–8 h time lag before the entry of the non-targeted polyplexes into the nuclei, whereas the targeted polyplexes start accumulating in the nuclei from the first hours of incubation (Fig. 5C).
4. Discussion
As we mentioned in the introduction, the action of endocytosis inhibitors can be cell-type dependent, so this is important when polyplex effects are compared on different cell lines. In this work, effects of targeted and non-targeted polyplexes were compared using one cell line, M-3 cells. It is noteworthy that when investigating endocytosis of disulfide-based poly(amido amine) polyplexes by ARPE-19 retinal cells, Vercauteren et al. [2] used three different approaches: (1) endocytosis inhibitors; (2) RNAi to block the corresponding endocytotic events; (3) fluorescence colocalization microscopy to confirm association of the polyplexes with endocytotic proteins. They obtained complementary (rather than contradictory) data that still allows considering endocytosis inhibitors as efficient tools for experiments carried out on the same cell line. The other reason for using inhibitor analysis in our work was the necessity to obtain at least semiquantitative data about impacts of different endocytosis pathways in polyplex uptake and transfection efficacy, which is impossible or very time-consuming if one uses fluorescence colocalization microscopy or RNAi approaches.
As expected, inclusion of an α-melanocyte stimulating hormone analog into the polymeric part of the polyplexes, though it comprised not more than 4% of the total PEI-PEG molecular weight, imparted receptor specificity to the polyplexes: (1) the peptide ligand on targeted block-copolmers is able to cause receptor-mediated melanogenesis, so the ligand is accessible to melanocortin receptors (Supplementary Fig. 1); (2) enhanced transfection with targeted polyplexes was observed on melanocortin-receptor bearing M-3 cells but not on HEK293 cells lacking these receptors (Fig. 1C); (3) the enhanced transfection with targeted polyplexes was decreased to the level of transfection with non-targeted polyplexes by addition of a free ligand to melanocortin receptors on M-3 cells (Fig. 1C); (4) this enhanced transfection with targeted polyplexes was inhibited by chlorpromazine (Fig. 2), an inhibitor of clathrin-dependent endocytosis, which is characteristic of receptor-mediated endocytosis of melanocortin receptors [47,48]. Increase in transfection with targeted polyplexes in comparison with non-targeted ones cannot be due to the influence of the polymeric part of the polyplexes on the CMV promoter: we found only a slight activation of the promoter in cells permanently transformed with CopGFP under the CMV promoter by PEI-PEG-MC1SP polyplexes (see Supplementary Fig. 4), which was remarkably less than the 2–3-fold enhancement of transfection observed with the targeted polyplexes if compared with the non-targeted ones. A possible explanation of this slight CMV promoter activation could be the NFkB-mediated effect of PEI on the promoter that was found by Merkel et al. [49] in certain cell lines. It is worth emphasizing that the contribution of nonspecific pathways comprises from 30–40% to 90% of the total transfection effect of targeted polyplexes (e.g. for polyplexes with such receptor ligands as EGF [50,51], transferrin [52,53], folate [54], and pentacyclic RDG-mimetic [55]), so understanding of these pathways and minimization of their contribution are the major requirements for amendment of polyplexes.
Our results conform to those of de Bruin et al. [56] who also observed a time delay in internalization of non-targeted PEI-based polyplexes in Huh7 cells and prolonged attachment of non-targeted polyplexes to the cell membrane as compared with targeted, EGF-containing ones. Their analysis [56] of the internalization process by single-particle tracking and quenching experiments revealed that targeted (EGF) and non-targeted PEI-based polyplexes differed in the duration of attachment to the cell membrane prior to internalization. They suggested that the prolonged attachment of non-targeted polyplexes to the cell membrane implied a more complex or time-consuming internalization mechanism, which may involve clustering of heparan sulfate proteoglycans at the cell surface followed by endocytosis. Kopatz et al. [57] showed that penetration of non-targeted polyplexes into HeLa cells proceeds via the following steps: (1) electrostatic binding of the cationic polyplex particle to syndecan heparan-sulfate proteoglycans; (2) polyplex-induced syndecan clustering into cholesterol-rich rafts; (3) actin-mediated engulfment of the polyplex particle into the cell. Syndecans can also interact with Frizzled receptors [58], which undergo clathrin-mediated endocytosis [59]. It is noteworthy that penetration of PEI polyplexes into HEK293 cells via syndecan-1 occurred within minutes after addition of the polyplexes, whereas endocytosis via syndecan-2 took hours. Moreover, coexpression of syndecan-2 together with syndecan-1 resulted in a negative effect on syndecan-1-mediated PEI transfection [60]. On the other hand, extracellular proteoglycans cause aggregation of polyplexes and release of DNA from them [61,62]. So, cell surface proteoglycans can play a double role: they contribute (1) to internalization of mainly non-targeted polyplexes via long and complicated pathways; (2) to unpacking of the polyplexes on the cell surface. Both processes make the non-targeted polyplexes more susceptible to degradation. A balance between these contrary effects of surface proteoglycans seems to be one of determinants of the intracellular fate of non-targeted polyplexes. We assume that similar processes take place in our case because melanoma cells also overexpress at least three types of heparan sulfate proteoglycans, i.e. syndecan-1 and syndecan-4 [63] and especially syndecan-2 [64], the latter being a general protumorigenic receptor in melanoma cells and possessing Frizzled receptors [65].
The main difference between targeted and non-targeted polyplexes were observed when their N/P ratios were between 20 and 30, i.e. when there was a significant excess of free PEI [66,67], which helps to overcome cell-surface proteoglycan binding that might otherwise lead to a block in transfection [67]. Or else, the difference was observed under the conditions favorable for the non-targeted polyplexes.
During past decade, several models providing quantitative information (such as rate constants of entry/exit from subcellular compartments) from experimental data have been developed [68–72]. As one can see from the above discussion, the situation on the cell surface, such as state of polyplexes (packed or unpacked, their surface concentrations) should be taken into account to obtain a complete picture of the processes of polyplex transport within the cell. Though this idea had been expressed [73] in the beginning of the quantitative investigations of polyplex transport, it did not attract attention until our recent publication [16] where quantitative consideration of intracellular transport kinetics of polyplexes was combined with quantitative description of their unpacking based on the works of Leong et al. [13,14]. Here we used a similar approach to estimate polyplex accumulation and unpacking rates.
As both direct measurements and calculations made on the basis of developed kinetics models (Supplementary Materials and Methods, Supplementary Fig. 8, and Supplementary Table 1) demonstrated, the non-targeted polyplexes differ from the targeted ones in: (1) later entry of packed polyplexes into the murine melanoma cells and their nuclei; (2) different relations between entry velocities of packed and unpacked polyplexes. These differences can be possibly attributed to different velocities [59] of different interactions of polyplexes with components of the cell surface. Whereas targeted polyplexes relatively quickly interact with the corresponding receptors on the cell surface and undergo subsequent clathrin-mediated endocytosis, the non-targeted polyplexes need to interact with surface proteoglycans including syndecan-2 (which is overexpressed on melanoma cells [63], see also above), being partially unpacked meanwhile (see above) in order to be finally endocytosed.
As the comparison between transfectabilities of MC1SP- and (MC1SP, NLS−)-containing polyplexes showed, entry of the packed targeted polyplex nanoparticles into the nuclei of dividing melanoma cells is hardly accomplished through nuclear pore complex. So, one needs to investigate other pathways of nuclear entry of the polyplex DNA.
5. Conclusion
The presence of the ligand to MC1R provides more efficient transfection by polyplexes under both in vivo and in vitro conditions. According to our experimental data, the difference in TE between targeted and non-targeted polyplexes could be explained by difference in rates of intracellular accumulation of packed polyplex nanoparticles. The data revealed a significant contribution of the clathrin-dependent endocytosis pathway for internalization of the targeted polyplexes. It provides fast accumulation of polyplex nanoparticles inside target cells and avoids the interaction of the polyplexes with surface proteoglycans that can lead to polyplex unpacking and subsequent degradation. We believe that knowledge of the detailed intracellular transport of polyplexes will help to construct more efficient polyplex nanoparticles.
Supplementary Material
Acknowledgments
The experiments were performed using equipment of the Institute of Gene Biology of the Russian Academy of Sciences facilities supported by the Ministry of Science and Education of the Russian Federation (grant No. 16.552.11.7067). We are grateful to Prof. S. Lukyanov (Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow) for M-3 CopGFP cells. We are thankful to V. Malysheva (Institute of Gene Biology, Russian Academy of Sciences, Moscow) for help with transfection experiments, and E. Karandashov (Institute of Gene Biology, Russian Academy of Sciences, Moscow) for help with the flow cytometry experiments. This work was supported by Russian Federation State contracts nos. 16.512.12.2002, 16.512.12.2004, and 11411.1008700.13.084, the Russian Foundation for Basic Research grant no. 10-04-01037-a, as well as by NIH grant P50 NS20023.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mikhail O Durymanov, Email: mdurymanov@gmail.com.
Elena A Beletkaia, Email: elenebelle@gmail.com.
Alexey V Ulasov, Email: bornuwa@gmail.com.
Yuri V Khramtsov, Email: ykhram2000@mail.ru.
Georgiy A Trusov, Email: georgetrusov@yandex.ru.
Nikita S Rodichenko, Email: bornikita@gmail.com.
Tatiana A Slastnikova, Email: slacya@gmail.com.
Tatiana V Vinogradova, Email: tv@ibch.ru.
Natalia Y Uspenskaya, Email: natusp@gmail.com.
Eugene P Kopantsev, Email: kopantzev@ibch.ru.
Andrey A Rosenkranz, Email: aar@igb.ac.ru.
Eugene D Sverdlov, Email: sverd@ibch.ru.
Alexander S Sobolev, Email: sobolev@igb.ac.ru.
References
- 1.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15:1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 2.Vercauteren D, Piest M, van der Aa LJ, Al Soraj M, Jones AT, Engbersen JFJ, De Smedt SC, Braeckmans K. Flotillin-dependent endocytosis and a phagocytosis-like mechanism for cellular internalization of disulfide-based poly(amido amine)/DNA polyplexes. Biomaterials. 2011;32:3072–3084. doi: 10.1016/j.biomaterials.2010.12.045. [DOI] [PubMed] [Google Scholar]
- 3.Lai WF. In vivo nucleic acid delivery with PEI and its derivatives: current status and perspectives. Expert Rev Med Devices. 2011;8:173–185. doi: 10.1586/erd.10.83. [DOI] [PubMed] [Google Scholar]
- 4.Wagner E, Culmsee C, Boeckle S. Targeting of Polyplexes: Toward Synthetic Virus Vector Systems. Adv Genet. 2005;53PA:333–354. doi: 10.1016/S0065-2660(05)53013-X. [DOI] [PubMed] [Google Scholar]
- 5.Mok H, Park TG. Functional polymers for targeted delivery of nucleic acid drugs. Macromol Biosci. 2009;9:731–743. doi: 10.1002/mabi.200900044. [DOI] [PubMed] [Google Scholar]
- 6.Sadeqzadeh E, Rahbarizadeh F, Ahmadvand D, Rasaee MJ, Parhamifar L, Moghimi SM. Combined MUC1-specific nanobody-tagged PEG-polyethylenimine polyplex targeting and transcriptional targeting of tBid transgene for directed killing of MUC1 over-expressing tumour cells. J Control Release. 2011;156:85–91. doi: 10.1016/j.jconrel.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery—Past, present, and future. Prog Polym Sci. 2007;32:799–837. [Google Scholar]
- 8.Elfinger M, Geiger J, Hasenpusch G, Uzgun S, Sieverling N, Aneja MK, Maucksch C, Rudolph C. Targeting of the beta(2)-adrenoceptor increases nonviral gene delivery to pulmonary epithelial cells in vitro and lungs in vivo. J Control Release. 2009;135:234–241. doi: 10.1016/j.jconrel.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Khalil IA, Futaki S, Niwa M, Baba Y, Kaji N, Kamiya H, Harashima H. Mechanism of improved gene transfer by the N-terminal stearylation of octaarginine: enhanced cellular association by hydrophobic core formation. Gene Ther. 2004;11:636–644. doi: 10.1038/sj.gt.3302128. [DOI] [PubMed] [Google Scholar]
- 10.Farrell LL, Pepin J, Kucharski C, Lin X, Xu Z, Uludag H. A comparison of the effectiveness of cationic polymers poly-L-lysine (PLL) and polyethylenimine (PEI) for non-viral delivery of plasmid DNA to bone marrow stromal cells (BMSC) Eur J Pharm Biopharm. 2007;65:388–397. doi: 10.1016/j.ejpb.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 11.Dhaliwal A, Maldonado M, Han Z, Segura T. Differential uptake of DNA-poly(ethylenimine) polyplexes in cells cultured on collagen and fibronectin surfaces. Acta Biomater. 2010;6:3436–3447. doi: 10.1016/j.actbio.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pichon C, Billiet L, Midoux P. Chemical vectors for gene delivery: uptake and intracellular trafficking. Curr Opin Biotechnol. 2010;21:640–645. doi: 10.1016/j.copbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Ho YP, Chen HH, Leong KW, Wang TH. Evaluating the intracellular stability and unpacking of DNA nanocomplexes by quantum dots-FRET. J Control Release. 2006;116:83–89. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum dot-FRET. Mol Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thibault M, Nimesh S, Lavertu M, Buschmann MD. Intracellular trafficking and decondensation kinetics of chitosan-pDNA polyplexes. Mol Ther. 2010;18:1787–1795. doi: 10.1038/mt.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulasov AV, Khramtsov YV, Trusov GA, Rosenkranz AA, Sverdlov ED, Sobolev AS. Properties of PEI-based polyplex nanoparticles that correlate with their transfection efficacy. Mol Ther. 2011;19:103–112. doi: 10.1038/mt.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trusov GA, Ulasov AV, Beletkaia EA, Khramtsov YV, Durymanov MO, Rosenkranz AA, Sverdlov ED, Sobolev AS. Investigation of transport and unpacking mechanisms of polyplexes for transfection efficacy on different cell lines. Dokl Biochem Biophys. 2011;437:77–79. doi: 10.1134/S1607672911020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Borrón JC, Sánchez-Laorden BL, Jiménez-Cervantes C. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res. 2005;18:393–410. doi: 10.1111/j.1600-0749.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 19.Chatzinasiou F, Lill CM, Kypreou K, Stefanaki I, Nicolaou V, Spyrou G, Evangelou E, Roehr JT, Kodela E, Katsambas A, Tsao H, Ioannidis JP, Bertram L, Stratigos AJ. Comprehensive field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma. J Natl Cancer Inst. 2011;103:1227–1235. doi: 10.1093/jnci/djr219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegrist W, Solca F, Stutz S, Giuffre L, Carrel S, Girard J, Eberle AN. Characterization of receptors for alpha-melanocyte-stimulating hormone on human melanoma cells. Cancer Res. 1989;49:6352–6358. [PubMed] [Google Scholar]
- 21.Jiang J, Sharma SD, Fink JL, Hadley ME, Hruby VJ. Melanotropic peptide receptors: membrane markers of human melanoma cells. Exp Dermatol. 1996;5:325–333. doi: 10.1111/j.1600-0625.1996.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 22.Salazar-Onfray F, López M, Lundqvist A, Aguirre A, Escobar A, Serrano A, Korenblit C, Petersson M, Chhajlani V, Larsson O, Kiessling R. Tissue distribution and differential expression of melanocortin 1 receptor, a malignant melanoma marker. Br J Cancer. 2002;87:414–422. doi: 10.1038/sj.bjc.6600441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López MN, Pereda C, Ramírez M, Mendoza-Naranjo A, Serrano A, Ferreira A, Poblete R, Kalergis AM, Kiessling R, Salazar-Onfray F. Melanocortin 1 receptor is expressed by uveal malignant melanoma and can be considered a new target for diagnosis and immunotherapy. Invest Ophthalmol Vis Sci. 2007;48:1219–1227. doi: 10.1167/iovs.06-0090. [DOI] [PubMed] [Google Scholar]
- 24.Xia Y, Muceniece R, Wikberg JE. Immunological localisation of melanocortin 1 receptor on the cell surface of WM266-4 human melanoma cells. Cancer Lett. 1996;98:157–162. [PubMed] [Google Scholar]
- 25.Bohm M, Metze D, Schulte U, Becher E, Luger TA, Brzoska T. Detection of melanocortin-1 receptor antigenicity on human skin cells in culture and in situ. Exp Dermatol. 1999;8:453–461. doi: 10.1111/j.1600-0625.1999.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 26.Mountjoy KG. The human melanocyte stimulating hormone receptor has evolved to become “super-sensitive” to melanocortin peptides. Mol Cell Endocrinol. 1994;102:7–11. doi: 10.1016/0303-7207(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 27.Szardenings M, Muceniece R, Mutule I, Mutulis F, Wikberg JE. New highly specific agonistic peptides for human melanocortin MC(1) receptor. Peptides. 2000;21:239–243. doi: 10.1016/s0196-9781(99)00207-7. [DOI] [PubMed] [Google Scholar]
- 28.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 29.Perrine TD, Landis WR. Analysis of polyethylenimine by spectrophotometry of its copper chelate. J Polym Sci A1. 1967;5:1993–2003. doi: 10.1002/pol.1967.150050816. [DOI] [PubMed] [Google Scholar]
- 30.Kunath K, von Harpe A, Petersen H, Fischer D, Voigt K, Kissel T, Bickel U. The structure of PEG-modified poly(ethylene imines) influences biodistribution and pharmacokinetics of their complexes with NF-kappaB decoy in mice. Pharm Res. 2002;19:810–817. doi: 10.1023/a:1016152831963. [DOI] [PubMed] [Google Scholar]
- 31.Gong XW, Wei DZ, He ML, Xiong YC. Discarded free PEG-based assay for obtaining the modification extent of pegylated proteins. Talanta. 2007;71:381–384. doi: 10.1016/j.talanta.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 32.De Silva JA, Strojny N. Spectrofluorometric determination of pharmaceuticals containing aromatic or aliphatic primary amino groups as their fluorescamine (fluram) derivatives. Anal Chem. 1975;47:714–718. doi: 10.1021/ac60354a044. [DOI] [PubMed] [Google Scholar]
- 33.van Gaal EV, Oosting RS, van Eijk R, Bakowska M, Feyen D, Kok RJ, Hennink WE, Crommelin DJ, Mastrobattista E. DNA nuclear targeting sequences for non-viral gene delivery. Pharm Res. 2011;28:1707–1722. doi: 10.1007/s11095-011-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dey D, Inayathullah M, Lee AS, Limiuex M, Zhang X, Wu Y, Nag D, De Almeida PE, Han L, Rajadas J, Wu JC. Efficient Gene Delivery of Primary Human Cells Using Peptide Linked Polyethylenimine Polymer Hybrid. Biomaterials. 2011;32:4647–4658. doi: 10.1016/j.biomaterials.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahor S, Collin E, Dash BC, Pandit A. Controlled release of plasmid DNA from hyaluronan nanoparticles. Curr Drug Deliv. 2011;8:354–362. doi: 10.2174/156720111795768031. [DOI] [PubMed] [Google Scholar]
- 36.Vermes I, Haanen C, Reutelingsperger C. Flow cytometry of apoptotic cell death. J Immunol Methods. 2000;243:167–190. doi: 10.1016/s0022-1759(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 37.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 38.Mountjoy KG, Kong PL, Taylor JA, Willard DH, Wilkison WO. Melanocortin receptor-mediated mobilization of intracellular free calcium in HEK293 cells. Physiol Genomics. 2001;5:11–19. doi: 10.1152/physiolgenomics.2001.5.1.11. [DOI] [PubMed] [Google Scholar]
- 39.Zanta MA, Belguise-Valladier P, Behr J-P. Gene delivery: A single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci U S A. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Smedt SC, Wagner E, Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol Ther. 2006;14:745–753. doi: 10.1016/j.ymthe.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Stuart AD, Brown TD. Entry of feline calicivirus is dependent on clathrin-mediated endocytosis and acidification in endosomes. J Virol. 2006;80:7500–7509. doi: 10.1128/JVI.02452-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ming X, Sato K, Juliano RL. Unconventional internalization mechanisms underlying functional delivery of antisense oligonucleotides via cationic lipoplexes and polyplexes. J Control Release. 2011;153:83–92. doi: 10.1016/j.jconrel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cummings J, Ward TH, La Casse E, Lefebvre C, St-Jean M, Durkin J, Ranson M, Dive C. Validation of pharmacodynamic assays to evaluate the clinical efficacy of an antisense compound (AEG 35156) targeted to the X-linked inhibitor of apoptosis protein XIAP. Br J Cancer. 2005;14:532–538. doi: 10.1038/sj.bjc.6602363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Delivery Revs. 2002;54:715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 45.Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O’Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 46.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9:443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Cai MY, Varga EV, Stankova M, Mayorov A, Perry JW, Yamamura HI, Trivedi D, Hruby VJ. Cell signaling and trafficking of human melanocortin receptors in real time using two-photon fluorescence and confocal laser microscopy: Differentiation of agonists and antagonists. Chem Biol Drug Des. 2006;68:183–193. doi: 10.1111/j.1747-0285.2006.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Laorden BL, Jimenez-Cervantes C, Garcia-Borron JC. Regulation of human melanocortin 1 receptor signaling and trafficking by Thr-308 and Ser-316 and its alteration in variant alleles associated with red hair and skin cancer. J Biol Chem. 2007;282:3241–3251. doi: 10.1074/jbc.M606865200. [DOI] [PubMed] [Google Scholar]
- 49.Merkel OM, Beyerle A, Beckmann BM, Zheng M, Hartmann RK, Stöger T, Kissel TH. Polymer-related off-target effects in non-viral siRNA delivery. Biomaterials. 2011;32:2388–2398. doi: 10.1016/j.biomaterials.2010.11.081. [DOI] [PubMed] [Google Scholar]
- 50.Zhang B, Mallapragada S. The mechanism of selective transfection mediated by pentablock copolymers; part I: investigation of cellular uptake. Acta Biomater. 2011;7:1570–1579. doi: 10.1016/j.actbio.2010.11.032. [DOI] [PubMed] [Google Scholar]
- 51.Kloeckner J, Boeckle S, Persson D, Roedl W, Ogris M, Berg K, Wagner E. DNA polyplexes based on degradable oligoethylenimine-derivatives: combination with EGF receptor targeting and endosomal release functions. J Control Release. 2006;116:115–122. doi: 10.1016/j.jconrel.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Kakimoto S, Moriyama T, Tanabe T, Shinkai S, Nagasaki T. Dual-ligand effect of transferrin and transforming growth factor alpha on polyethyleneimine-mediated gene delivery. J Control Release. 2007;120:242–249. doi: 10.1016/j.jconrel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Bellocq NC, Pun SH, Jensen GS, Davis ME. Transferrin-containing, cyclodextrin polymer-based particles for tumor-targeted gene delivery. Bioconjug Chem. 2003;14:1122–1132. doi: 10.1021/bc034125f. [DOI] [PubMed] [Google Scholar]
- 54.Luten J, van Steenbergen MJ, Lok MC, de Graaff AM, van Nostrum CF, Talsma H, Hennink WE. Degradable PEG-folate coated poly(DMAEA-co-BA)phosphazene-based polyplexes exhibit receptor-specific gene expression. Eur J Pharm Sci. 2008;33:241–251. doi: 10.1016/j.ejps.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Merkel OM, Germershaus O, Wada CK, Tarcha PJ, Merdan T, Kissel T. Integrin alphaVbeta3 targeted gene delivery using RGD peptidomimetic conjugates with copolymers of PEGylated poly(ethylene imine) Bioconjug Chem. 2009;20:1270–1280. doi: 10.1021/bc9001695. [DOI] [PubMed] [Google Scholar]
- 56.de Bruin K, Ruthardt N, von Gersdorff K, Bausinger R, Wagner E, Ogris M, Bräuchle C. Cellular dynamics of EGF receptor-targeted synthetic viruses. Mol Ther. 2007;15:1297–1305. doi: 10.1038/sj.mt.6300176. [DOI] [PubMed] [Google Scholar]
- 57.Kopatz I, Remy JS, Behr J-P. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 58.Multhaupt HA, Yoneda A, Whiteford JR, Oh ES, Lee W, Couchman JR. Syndecan signaling: when, where and why? J Physiol Pharmacol. 2009;60(Suppl 4):31–38. [PubMed] [Google Scholar]
- 59.Blitzer JT, Nusse R. A critical role for endocytosis in Wnt signaling. BMC cell biology. 2006;7:28. doi: 10.1186/1471-2121-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paris S, Burlacu A, Durocher Y. Opposing roles of syndecan-1 and syndecan-2 in polyethyleneimine-mediated gene delivery. J Biol Chem. 2008;283:7697–7704. doi: 10.1074/jbc.M705424200. [DOI] [PubMed] [Google Scholar]
- 61.Ruponen M, Ronkko S, Honkakoski P, Pelkonen J, Tammi M, Urtti A. Extracellular glycosaminoglycans modify cellular trafficking of lipoplexes and polyplexes. J Biol Chem. 2001;276:33875–33880. doi: 10.1074/jbc.M011553200. [DOI] [PubMed] [Google Scholar]
- 62.Burke RS, Pun SH. Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjug Chem. 2008;19:693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 63.O’Connell MP, Fiori JL, Kershner EK, Frank BP, Indig FE, Taub DD, Hoek KS, Weeraratna AT. Heparan sulfate proteoglycan modulation of Wnt5A signal transduction in metastatic melanoma cells. J Biol Chem. 2009;284:28704–28712. doi: 10.1074/jbc.M109.028498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JH, Park H, Chung H, Choi S, Kim Y, Yoo H, Kim TY, Hann HJ, Seong I, Kim J, Kang KG, Han IO, Oh ES. Syndecan-2 regulates the migratory potential of melanoma cells. J Biol Chem. 2009;284:27167–27175. doi: 10.1074/jbc.M109.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Connell MP, Weeraratna AT. Hear the Wnt Ror: how melanoma cells adjust to changes in Wnt. Pigment cell & melanoma research. 2009;22:724–739. doi: 10.1111/j.1755-148X.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 67.Hanzlikova M, Ruponen M, Galli E, Raasmaja A, Aseyev V, Tenhu H, Urtti A, Yliperttula M. Mechanisms of polyethylenimine-mediated DNA delivery: free carrier helps to overcome the barrier of cell-surface glycosaminoglycans. J Gene Med. 2011;13:402–409. doi: 10.1002/jgm.1587. [DOI] [PubMed] [Google Scholar]
- 68.Varga CM, Wickham TJ, Lauffenburger DA. Receptor-mediated targeting of gene delivery vectors: insights from molecular mechanisms for improved vehicle design. Biotechnol Bioeng. 2000;70:593–605. doi: 10.1002/1097-0290(20001220)70:6<593::aid-bit1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 69.Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4:438–446. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- 70.Banks GA, Roselli RJ, Chen R, Giorgio TD. A model for the analysis of nonviral gene therapy. Gene Ther. 2003;10:1766–1775. doi: 10.1038/sj.gt.3302076. [DOI] [PubMed] [Google Scholar]
- 71.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 72.Dinh AT, Pangarkar C, Theofanous T, Mitragotri S. Understanding intracellular transport processes pertinent to synthetic gene delivery via stochastic simulations and sensitivity analyses. Biophys J. 2007;92:831–846. doi: 10.1529/biophysj.106.095521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schaffer DV, Lauffenburger DA. Optimization of cell surface binding enhances efficiency and specificity of molecular conjugate gene delivery. J Biol Chem. 1998;273:28004–28009. doi: 10.1074/jbc.273.43.28004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.