Abstract
Plasmodium falciparum resistance to chloroquine, the former gold standard antimalarial drug, is mediated primarily by mutant forms of the ‘Chloroquine Resistance Transporter’ (PfCRT). These mutations impart upon PfCRT the ability to efflux chloroquine from the intracellular digestive vacuole, the site of drug action. Recent studies reveal that PfCRT variants can also affect parasite fitness, protect immature gametocytes against chloroquine action, and alter P. falciparum susceptibility to current first-line therapies. These results highlight the need to be vigilant in screening for the appearance of novel pfcrt alleles that could contribute to new multi-drug resistance phenotypes.
Keywords: Plasmodium falciparum, malaria, drug resistance, PfCRT, heme detoxification, haplotypes, selective sweep, transmission
The rise and fall of chloroquine
The discovery some 65 years ago of the exceptional antimalarial properties of chloroquine (CQ: Glossary) rapidly paved the way for its massive use worldwide. Treatment of Plasmodium falciparum malaria with CQ proved so successful that one of the greatest scourges of mankind was proposed to be “well on its way towards oblivion” (Preface to Paul Russell's “Man's Mastery of Malaria, 1955”, as cited in [1]). After a decade of use, however, CQ resistance (CQR) emerged in a handful of origins, including in Southeast Asia, South America, and the Western Pacific region (Figure 1), from there spreading progressively throughout malaria-endemic areas including Africa where surges in malaria mortality were reported [2, 3]. This has led in recent years to the global adoption of artemisinin-based combination therapies (ACTs) [4, 5]. None of the current first-line antimalarials, however, match the favorable efficacy, safety and affordability properties once held by CQ. Today, malaria still causes an estimated 216 million clinical episodes and 655 000 deaths per year, ~90% of which occur in Africa [6].
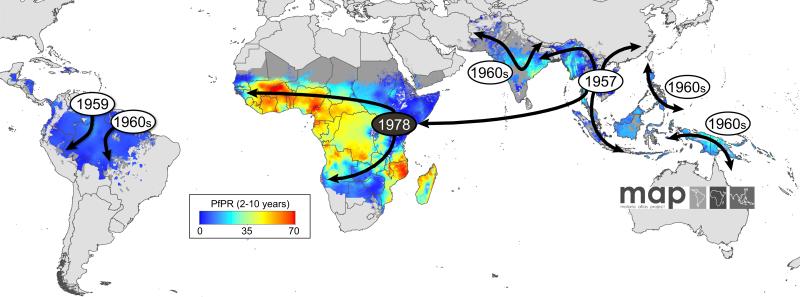
Figure 1.
The appearance and global spread of chloroquine resistance (CQR) in P. falciparum. Resistance is thought to have arisen in at least 6 independent origins (grey circles) and moved progressively as a CQ-driven selective sweep, including from Asia to Africa where it established itself on the East coast in the late 1970s (black circle) [3]. The geographic spread of CQR is overlaid onto a current map of P. falciparum endemicity modeled for 2010 [96]. This map was derived from P. falciparum parasite rate (PfPR) surveys, age standardized to the two to 10 year age range, using model-based geostatistics.
While CQ itself is officially no longer recommended for the treatment of P. falciparum malaria (it retains efficacy against Plasmodium vivax in most geographic regions [7]), studies into its mode of action and the molecular basis of CQR can provide important insights into this ’druggable‘ realm of parasite biology and aid future drug design. Furthermore, the exciting observations that CQ-sensitive parasites can re-emerge following the strict withdrawal of CQ [8-10], and that a double-dosing regimen can restore clinical efficacy of CQ even against CQ-resistant parasites [11], raise hopes that CQ (or derivatives thereof) may later rejoin our antimalarial armamentarium.
In this review we will discuss recent progress in our understanding of the mechanism of action of CQ and of CQR, with a focus on the contribution of the primary CQR determinant, pfcrt (Plasmodium falciparum chloroquine resistance transporter). We also highlight the potential that novel forms of PfCRT may continue to evolve to mediate parasite resistance to current first-line drugs being used to treat CQ-resistant malaria.
Mechanism of action of chloroquine
Parasitization of red blood cells (RBCs) by the pathogenic Plasmodium asexual blood stages results in the ingestion of large amounts of host cell hemoglobin into a lysosome-like organelle, the digestive vacuole (DV). Therein, hemoglobin is proteolytically cleaved, releasing globin moieties that are further degraded into small peptides that provide sources of amino acids for protein synthesis [12]. Hemoglobin degradation also releases ~20 mM toxic Fe2+-heme, which rapidly oxidizes to form Fe3+-heme (also known as ferriprotoporphyrin IX or FPIX). This in turn liberates an excess of electrons that can trigger the production of bio-reactive oxygen species including hydroxyl radicals (•OH) and hydrogen peroxide (H2O2). Fe3+-heme is largely insoluble and can disrupt membrane function. To avert this, parasites form iron-carboxylate coordinated FPIX dimers, also known as β-hematin, which bio-mineralize to form chemically inert crystals known as hemozoin (visible by microscopy as malaria pigment) (reviewed in [13, 14]).
CQ, a weak base, can freely diffuse across membranes in its neutral form, and concentrates inside the acidic DV, according to the Henderson-Hasselbach equilibrium, as membrane-impermeant CQH 2+2. Indeed, most (~85%) of the >1000-fold accumulated CQ in erythrocytes infected with CQ-sensitive parasites is found within the DV [15]. CQ has long been considered to be toxic as a result of it binding to FPIX, thereby helping to retain the drug at very high concentrations (up to low millimolar) and preventing heme detoxification, in essence poisoning the parasite with its own waste products [14]. As discussed below, an alternative model is that CQ may act at least in part by binding to PfCRT and inhibiting an endogenous function.
Acquiring CQ resistance via mutations in pfcrt
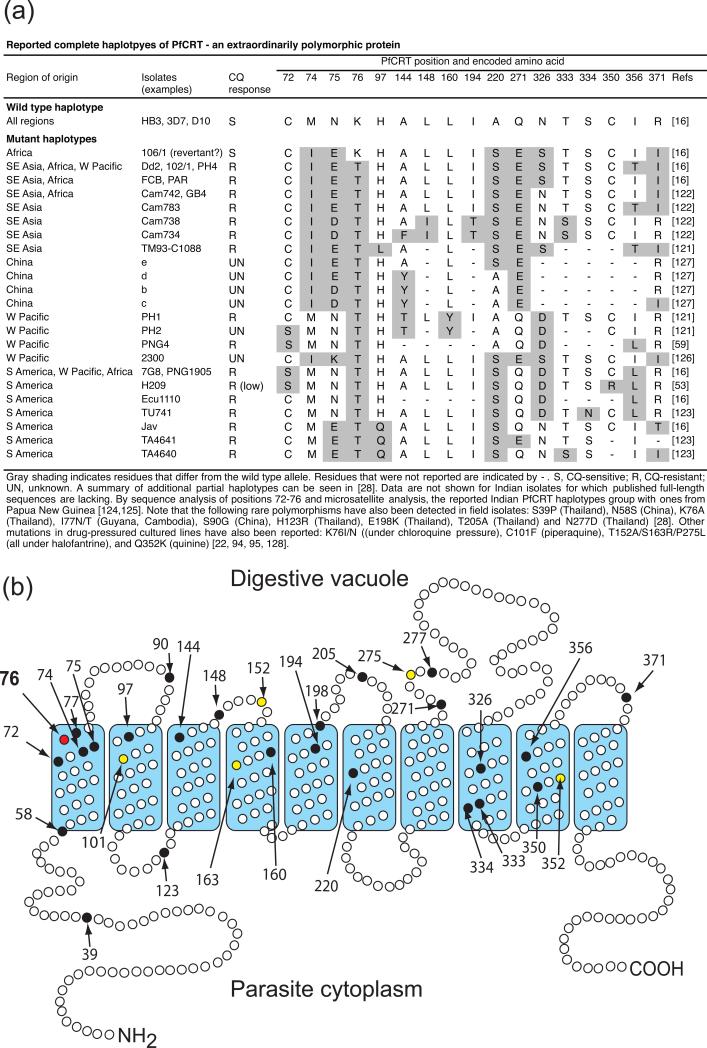
The identification of pfcrt (MAL7P1.27) as the primary determinant of CQR [16] was made possible by the detailed analysis of a genetic cross between the CQ-sensitive parent HB3 (Honduras) and the CQ-resistant parent Dd2 (Indochina), which mapped the CQR phenotype to a 48 kb chromosomal segment harboring this highly interrupted gene [17, 18]. Allelic exchange experiments provided conclusive evidence that Dd2 pfcrt (that carries 8 point mutations compared to the wild type allele present in HB3) could impart to a CQ-sensitive progeny clone all the features of CQR, namely elevated IC50 values, chemosensitization by verapamil, and reduced CQ accumulation [19]. Quantitative trait loci (QTL) analyses have confirmed that, in the CQ-resistant progeny, the CQR phenotype associates to a very high degree with inheritance of the pfcrt Dd2 allele [20, 21]. pfcrt encodes a 424 amino acid protein with 10 predicted transmembrane helices that localizes to the DV membrane [16, 22]. This localization appears to be dependent on phosphorylation of residue 416 [23]. A recent report of inducible expression of PfCRT-GFP fusions revealed expression beginning in ring stages in pre-DV compartments that then mature into the DV during the trophozoite stage of peak PfCRT expression and maximal hemoglobin digestion [24]. Phylogenetic analyses predict PfCRT to be a member of the drug/metabolite transporter superfamily of electrochemical potential-driven transporters [25, 26]. Geographic variants of pfcrt alleles harbor four to 10 non-synonymous mutations (Figure 2a) [27, 28]. To date no less than 30 variant residues have been identified, rendering PfCRT an extraordinarily polymorphic protein (Figure 2a,b). Of note, in all resistant parasites lysine 76 (K76) is replaced with an uncharged amino acid, either a threonine (76T) in the case of virtually all field isolates (with one reported exception of a 76A variant [29]) or an asparagine or isoleucine (76N/I) in lab-adapted lines pressured with CQ (starting with the strain 106/1 [16, 22]). Allelic exchange studies with Asian and South American P. falciparum parasites have shown that the K76T mutation is essential for in vitro CQR [30]. Field studies have repeatedly shown that K76T also provides a highly sensitive, albeit only moderately specific, marker for CQ treatment failure [31] (see below). A primary role for mutant PfCRT in driving CQR worldwide is consistent with population and genome-wide association studies that demonstrate the worldwide dissemination of mutant alleles from a handful of successful origins of resistance in a CQ-driven selective sweep [32, 33].
Figure 2.
PfCRT polymorphisms and predicted topology. (a) List of major PfCRT haplotypes identified in P. falciparum isolates from malaria-endemic regions. Additional rare mutations observed from patient isolates, as well as ones identified in drug-pressured culture-adapted parasites, are listed in the footnote. (b) Predicted topology showing the location of variant residues. Black and yellow shading indicate residues that vary in field isolates and drug-pressured laboratory lines, respectively. The K76T mutation, central to CQR and used as a molecular marker in endemic areas, is highlighted in red.
Studies of CQ uptake kinetics have shown that CQ-resistant P. falciparum parasites accumulate substantially less CQ than CQ-sensitive strains [34]. This difference in CQ accumulation is also observed with isolated DVs [35] and is associated with mutant PfCRT [14, 36, 37]. Due to its transporter-like structure it has been suggested that, having lost the positively charged lysine 76 in its second helical segment, mutant PfCRT has acquired the ability to transport protonated CQ. Indeed, a CQ-associated H+ leak from the DV has been observed in parasites with mutant PfCRT, consistent with the protein mediating the efflux of CQ in its protonated form and/or in symport with H+ [38, 39].
Evidence for CQ transport by mutant PfCRT (from the Dd2 strain) was first obtained in heterologous expression systems in Pichia pastoris and Dictyostelium discoideum [40, 41] and more recently has been demonstrated directly by heterologous expression of a codon-optimized, trafficking motif-depleted PfCRT on the surface of Xenopus laevis oocytes [42]. In oocytes, CQ transport was observed only with mutant PfCRT. Further evidence of CQ transport has also been obtained in Saccharomyces cerevisiae, although in this study it was reported with both mutant and wild type PfCRT [43].
Interestingly, in oocytes mutant PfCRT-mediated CQ transport was lost upon removal of the critical K76T mutation, but, conversely, introduction of just K76T into wild type PfCRT was insufficient to confer drug transport [42], illustrating an important role for additional mutations in this transporter. CQ transport was inhibited by the related quinoline-based antimalarial drugs quinine and amodiaquine, the CQR-reversal agent verapamil, and certain charged peptides [42]. Further studies are keenly awaited to shed light on whether peptides, emanating from degraded hemoglobin, and possibly the modified tri-peptide and reducing agent glutathione (GSH) can be one source of natural substrates for this transporter. Interestingly, a set of plastid-localized plant homologues of PfCRT, called CRT-like transporters (CLTs) have recently been reported to transport thiols including GSH, thereby influencing GSH levels and redox potential in the cytosol [44].
Based on these observations, a widely accepted model of mutant pfcrt-mediated CQR is that mutant PfCRT-mediated CQ efflux reduces access of CQ to its heme target in the DV. Transport and modeling studies tend to favor a model whereby mutant PfCRT acts as a saturable carrier [28, 34, 37, 45, 46], although other studies have argued for the mutant protein acting as a voltage-gated channel [36, 47]. Recent data from the Roepe laboratory, however, suggest additional layers of complexity for these models. One line of evidence is that as yet unidentified mutant PfCRT-mediated CQR mechanisms appear to be equally active in all stages of the asexual blood cell cycle, even schizonts that are thought to no longer digest hemoglobin [48]. Furthermore, recent experiments call into question the extent to which reduced CQ accumulation explains reduced killing of CQ-resistant strains [48, 49]. For example, CQ-resistant Dd2 parasites exposed to 750 nM CQ accumulated the same amount of CQ as CQ-sensitive HB3 parasites in the presence of 250 nM CQ. Those concentrations nevertheless killed 50% of the HB3 but none of the Dd2 parasites. Provided the same intravacuolar CQ concentration inhibited heme detoxification in the same manner and to the same extent in both parasite strains, this suggests that either the primary mechanism of CQ action in at least some CQ-resistant parasites is not inhibition of heme detoxification, or that CQ-resistant parasites cope better with the downstream consequences of the build-up of toxic heme and CQ-heme complexes [50]. For example, if heme and CQ-heme complexes kill parasites through the oxidation of biomolecules, one might envisage that CQ-resistant strains could possess better antioxidant defense mechanisms [51]. Differences in these defense mechanisms between strains could then account for the different levels of CQR observed in different genetic backgrounds. Another possibility relates to the report of CQ binding to PfCRT, discussed below.
Is mutant pfcrt sufficient to confer in vitro CQR?
Allelic exchange experiments earlier established that the replacement of wild type pfcrt with widely found variants (Dd2 and 7G8) sufficed to confer CQR to CQ-sensitive GC03 parasites (a progeny of the Dd2×HB3 cross) [19]. Further experiments have since shown that the degree of CQR imparted by mutant pfcrt appears to depend on the parasite genetic background. Indeed, in the D10 (Papua New Guinea) and 3D7 strains (isolated in The Netherlands but possibly of African origin [52]) the replacement of wild type pfcrt with the mutant 7G8 (Brazil) allele failed to introduce high-level CQR. Instead, these recombinant strains displayed a more subtle phenotype, termed ’CQ tolerance‘, which could be observed as a dose-response shift primarily at the IC90 level, decreased parasite susceptibility to the CQ metabolite monodesethyl-CQ more than to the parent drug, and the ability to recrudesce in the presence of CQ concentrations that were lethal to CQ-sensitive parasites [53]. This state of tolerance presumably would permit parasites to remain viable during a course of CQ treatment and recrudesce once CQ levels had become sub-therapeutic. Importantly, a CQ tolerance phenotype was recently observed in two culture-adapted isolates from French Guiana - the first example of lab-adapted field strains that would be customarily classified as CQ-sensitive despite harboring mutant pfcrt [53].
Strikingly, despite their significant differences in CQ susceptibility, as measured in standard [3H]-hypoxanthine incorporation assays, CQ-resistant and CQ-tolerant recombinant parasite lines revealed similar levels of reduced CQ accumulation and CQ:H+ efflux [50]. This was consistent with other evidence that parasite response to CQ cannot be fully accounted for by the level of CQ accumulation [49]. Notably, evidence that CQ can bind PfCRT was recently obtained using a fluorescently tagged analog [54]. Binding was localized to a loop between putative transmembrane helices 9 and 10 and has been speculated to interfere with a natural function of PfCRT (Box 1) [46, 55]. Conceivably, a strain-dependent impact of CQ binding on native PfCRT function might combine with variant-specific PfCRT-mediated drug efflux to determine the degree of CQR exerted by mutant forms of this transporter.
Mutant pfcrt as a driving force for CQ treatment failure
Most relevant to malaria therapy is of course the question of whether or not mutant pfcrt-mediated CQR is sufficient to cause CQ treatment failure. Thus, since the discovery of pfcrt, a large number of clinical studies have been carried out to determine the usefulness of mutant pfcrt (mainly the K76T mutation) as a predictive marker for CQ treatment failure.
Compared to the relatively straightforward in vitro measurements of drug susceptibility, clinical studies are complicated by the multiple factors in addition to parasite susceptibility that influence treatment outcome. These factors - which can be partially controlled for - include patient compliance, drug absorption and metabolism, nutritional status, acquired anti-plasmodial immunity (related to transmission intensity, and age as a surrogate in sub-Saharan Africa), and parasite biomass at the start of the treatment (reviewed in [56]). Despite these confounders numerous clinical studies have clearly shown that the presence of mutant pfcrt is highly correlated with CQ treatment failure. A recent meta-analysis of studies from Asia and Africa calculated that the presence of PfCRT K76T on the day of treatment initiation increases the risk of CQ treatment failure by 2.1-fold on day 14 post-treatment and by 7.2-fold on day 28 [31]. Our review of the literature confirms mutant pfcrt to be a highly sensitive marker for CQ treatment failure, i.e., it is detected in almost all patients that fail treatment. This correlation holds throughout the malaria-endemic world with the unique exception of Madagascar (Box 2). Nevertheless, the presence of mutant pfcrt cannot by itself predict treatment outcome. In many studies, particularly those carried out in areas with high transmission intensity, some patients infected with parasites carrying mutant pfcrt respond adequately to CQ treatment, most likely due to pre-existing immunity or potentially also because infection is caused by CQ-tolerant rather than CQ-resistant parasites [53, 57]. Mutant pfcrt thus only has moderate specificity as a marker for CQ treatment failure.
Of note, mutant pfcrt appears to only suffice to mediate resistance to the standard dosing regimen (25 mg/kg, delivered as 10 mg/kg on days 1 and 2 and 5 mg/kg on day 3). Studies from Guinea-Bissau have shown that doubling or tripling this dose can overcome PfCRT-mediated CQR [11, 58]. This dosing regimen is thought to account for the relatively low prevalence of mutant pfcrt (~20-30%) in that country compared to its neighbors Senegal and the Gambia (~80%), potentially because of the reduced selective advantage of mutant pfcrt when faced with these elevated drug doses and a reduced fitness of the mutant allele.
The search for secondary CQR determinants
Collectively, the current evidence argues that while pfcrt is the primary determinant of CQR, secondary determinants must also contribute. These may either directly augment the degree of CQR or generate a physiological environment in which mutant PfCRT can fully realize its resistance-conferring potential. Over the years several approaches have been tried to identify these elusive secondary determinants, including several genome-wide association studies as well as QTL analyses of two genetic crosses [20, 21, 59-62]. However, as yet only one such gene has been convincingly demonstrated, namely pfmdr1 (P. falciparum multidrug resistance-1). Mutations in this gene are thought to play a modulatory role in CQR in at least some parasite strains (Box 3).
In the Dd2×HB3 genetic cross that yielded pfcrt, the chromosome 7 peak around this locus accounted for >95% of the variation in CQ response, and no statistically significant additional peaks were initially identified [63]. Nevertheless, additional parasite factors influencing CQ susceptibility must exist, as illustrated by the 2.7-fold spread in CQ IC50 values amongst the resistant progeny [63]. QTL analysis of these values among the progeny harboring the Dd2 pfcrt allele identified two secondary loci, one on chromosome 5 mapping to the region of pfmdr1 and a novel locus on chromosome 7, which together accounted for ~70% of the IC50 variance among the resistant progeny [20]. Of note, this cross is not well-suited to assessing the role of pfmdr1 in parasite CQ response as Dd2 is unusual in having both the N86Y mutation and an amplified pfmdr1 copy number that have been associated with increased and decreased CQ IC50 values respectively [64]. Neither the Dd2 nor the HB3 forms of PfMDR1 displayed detectable CQ transport in an oocyte expression system [65]. Nonetheless, the 7G8 PfMDR1 haplotype (S1034C/ N1042D/ D1246Y) was recently reported to augment CQR in progeny that carried the pfcrt GB4 allele (Figure 2a) in the 7G8xGB4 cross [21, 66]. One conclusion would be that pfmdr1 mutations modulate CQR only in certain genetic backgrounds and that the linkage disequilibrium seen between pfcrt and pfmdr1 [67] reflects beneficial associations between certain pairs of haplotypes in terms of their functional impacts on intracellular physiology. Deciphering the natural substrates of these two transporters will be vital to understanding their epistatic interactions.
The influence of mutant PfCRT on parasite fitness and transmission
Failed attempts to disrupt the pfcrt gene have pointed to an essential role for PfCRT in the (haploid) asexual blood stages [68]. While the endogenous function of PfCRT has not yet been identified, it is likely that some of the mutations present in mutant PfCRT could interfere with its physiological function, and thereby reduce overall parasite fitness. Indeed, several field studies indicate a fitness cost imparted by certain mutant pfcrt alleles. For example, a study from Eastern Sudan reported significantly reduced parasite densities in infections with parasites carrying pfcrt K76T compared to wild type parasites [69]. Another study, from the Gambia, used parasite survival during the dry season as a measure of relative fitness [70]. During the dry season parasites persist mainly as asymptomatic low-density infections and drug pressure is largely absent. In each dry season, the prevalence of the pfcrt K76T mutation decreased, presumably due to intra-host competition with wild type parasites. However, the situation was reversed during the rainy season, when transmission began, drug use increased and the prevalence of parasites carrying pfcrt K76T increased significantly [70]. Most strikingly, field studies including from Malawi, Kenya and China have reported a long-term decline in the frequency of parasites with mutant pfcrt after CQ use had been discontinued ([71] and references therein). This decline was most notable in Malawi, where within ten years of CQ withdrawal wild type parasites have re-expanded and parasites possessing pfcrt K76T have become undetectable by standard methods [8, 71], although minority parasite populations possessing pfcrt K76T are still circulating [72]. Household surveys from Africa reveal that pfcrt K76T rates are decreasing in areas of low CQ use, however, this drug continues to be widely used at high rates in many African countries, thus keeping mutant pfcrt (typically with the 72-76 haplotype CVIET) at fairly high levels [73-75]. Of note, other forms of mutant pfcrt (harboring the 72-76 haplotype SVMNT) remain highly prevalent in South America in the absence of CQ pressure. Reasons for this could include a lesser fitness cost of the predominant 7G8 pfcrt allele (as proposed by [76]), region-specific differences in drug use (see below), or lesser opportunities for competition because of a lower rate of polyclonal infections and a relative lack of competing wild type parasites.
Interestingly, parasites carrying mutant pfcrt may compensate for a fitness cost in the asexual blood stages by increasing investment in the transmission stages (gametocytes). Indeed, the Sudanese study discussed above found a significantly higher rate of gametocyte production in parasites with pfcrt K76T compared to wild type parasites [69]. Parasites carrying pfcrt K76T were also shown to be more infectious to mosquitoes following CQ treatment [77], suggesting that mutant pfcrt protects immature gametocytes from CQ action. We have recently confirmed this protective effect in transgenic rodent malaria parasites expressing either wild type or mutant pfcrt [78], and suggest that this may have played an important role in facilitating the worldwide spread of mutant pfcrt.
Reversing PfCRT-mediated drug resistance – a strategy for novel therapeutics
The central role of mutant PfCRT in verapamil-reversible CQR has recently led to renewed efforts to leverage this resistance reversibility as a ’druggable‘ feature for new drugs. One exciting new chemical series developed by Riscoe and colleagues, known as acridones, contains a dual functionality that acts both to target heme and chemosensitize CQ- and amodiaquine-resistant parasites, apparently by interfering with heme detoxification as well as targeting mutant PfCRT [79]. Egan and colleagues have also recently reported a similar dual functionality with a distinct quinoline-dibemethin series [80]. The HIV protease inhibitor saquinavir was also recently reported to have antimalarial activity by virtue of inhibiting PfCRT and acting as a CQR reversal agent [81]. Chemical modifications to CQ are also being engineered to develop potent analogs that overcome mutant PfCRT-mediated drug efflux [82, 83]. These chemical approaches illustrate the practical benefit gained from defining the molecular basis of CQR.
PfCRT and its impact on other antimalarials
The recent adoption of ACTs, spurred by the spread of resistance to CQ and the relatively short-lived replacement drug sulfadoxine-pyrimethamine, will create new selective pressures on the P. falciparum genome that need to be anticipated [84]. A consideration of the potential role of pfcrt is particularly relevant in light of a recent study that genetically mapped the determinants of parasite susceptibility to more than 2800 compounds and that reported that 96% of the differential susceptibilities could be mapped to pfcrt, pfmdr1, or the antifolate target dhfr (dihydrofolate reductase) [85]. Furthermore, most of the current ACT partner drugs (amodiaquine, piperaquine, and pyronaridine) share with CQ a quinoline scaffold [86]. Studies with artemether-lumefantrine (CoArtem®), the most widely-used ACT [5, 86], have shown that lumefantrine (an arylaminoalcohol related to mefloquine) selects for wild type pfcrt; as was confirmed in vitro with pfcrt-modified lines [87]. By contrast, the amodiaquine partner in the second most widely-used ACT, amodiaquine-artesunate (Coarsucam™), appears to select for mutant forms of pfcrt in field isolates (along with selection for mutant pfmdr1) [88]. In Africa, mutant pfcrt is predominantly CVIET at positions 72-76, and its cross-resistance to amodiaquine is consistent with data from pfcrt-modified isogenic lines [19, 30]. Of particular concern is the recent discovery in several African countries of the SVMNT haplotype present in the 7G8 strain that originated in South America and the Western Pacific region [89-91]. The 7G8 allele is known to mediate fairly high-level resistance to amodiaquine and its primary metabolite (monodesethyl-amodiaquine), as ascertained from the progeny of the 7G8×GB4 genetic cross as well as transgenic lines [19, 53, 66, 92]. The warning has thus correctly been raised that the increasing use of artesunate-amodiaquine in Africa will select for mutant pfcrt (especially the SVMNT type) [76]. In this light, one can argue that the use of both artemether-lumefantrine and artesunate-amodiaquine as multiple first-line therapies in the marketplace [93] presents the most beneficial combination in terms of balancing selection for and against mutant pfcrt.
For mefloquine an increase in susceptibility mediated by mutant pfcrt has been documented [19]. For the other ACT partner drugs, further studies are clearly required. In the case of piperaquine, mutant pfcrt Dd2 parasites selected for resistance were found to have a novel PfCRT mutation, C101F, in addition to a change in pfmdr1 copy number and amplification of a novel locus on chromosome 5 [94]; studies are ongoing to study the impact of this PfCRT mutation on drug response. Of note, studies with genetically modified lines have shown that mutant pfcrt can increase parasite susceptibility to artemisinins [19, 53]. Overall, given the complexity of polymorphisms in PfCRT (Figure 2) and its wide ranging effect on many antimalarials including CQ, amodiaquine, lumefantrine, mefloquine, and quinine [95], there is a major concern that additional mutations will continue to arise that can contribute to parasite resistance to the new first-line therapies as well as candidate replacement compounds in the preclinical and clinical development pipeline.
Concluding remarks and future perspectives
The loss of CQ had a devastating impact on malaria mortality and morbidity rates, and only in the past few years has the situation begun to improve substantially with the worldwide adoption of ACTs. A vigorous examination of CQR, combining field studies with laboratory research that takes advantage of powerful genomic and genetic tools, has clearly implicated mutant pfcrt as the principal determinant, and identified pfmdr1 as a modulatory factor. Much more remains to be learned about precisely how CQ leads to parasite death, how mutant PfCRT mediates resistance, and what the natural functions of PfCRT and its DV neighbor PfMDR1 are. The evidence that mutant PfCRT can accommodate mutations at multiple residues, and that it affects parasite susceptibility to a wide range of antimalarial drugs, creates a compelling need to further investigate the potential for novel mutations to arise that can contribute to resistance to the new first-line therapies. Leveraging this understanding to define new antimalarial agents will also benefit from ongoing biochemical and chemical studies to further clarify the CQR mechanism and identify novel compounds that bind PfCRT and/or interfere with its drug efflux properties. Metabolomic and structural studies will also be important to define function and resolve this at an atomic level as a way to develop new therapeutic strategies. It took the research community decades to discover how P. falciparum had acquired CQR. The hope is that we can apply the vastly improved capacities in biology and chemistry to our advantage in the ongoing efforts to reduce the burden of malaria worldwide.
Box 1: PfCRT and parasite physiology.
Much remains to be discovered about how PfCRT mutations affect parasite physiology. An early hypothesis that PfCRT-mediated CQR might be a consequence of an altered pH inside the DV (pHDV) was supported by reports that the DVs of CQ-resistant parasites are more acidic than those of CQ-sensitive parasites [16, 97-99]. Other groups, however, found no significant difference in pHDV between CQ-sensitive and CQ-resistant parasites [100-102]. While pHDV could conceivably play a role in some strains, mutant PfCRT is generally considered to cause CQR more directly via its ability to efflux CQ from the DV [37].
A number of heterologous expression studies have raised the possibility that mutations in PfCRT might affect the transport of H+ ions across the DV membrane [40, 41, 103, 104]. This is supported by the demonstration that the rate of leakage of H+ from the DV (determined by monitoring the rate of DV alkalinization on inhibition of the V-type H+-ATPase with concanamycin A) is greater in pfcrt-modified parasites expressing mutant PfCRT than in an isogenic line expressing wild type PfCRT [39]. An increased leak of H+ from the DV has the potential to short-circuit the H+ pump; the H+-ATPase would have to operate faster to allow the parasite to maintain pHDV at the same level as in wild type parasites, otherwise if it were unable to pump H+ at a sufficient rate then pHDV might be expected to increase.
In a recent study using drug-selected lines generated in the laboratory, it was shown that the acquisition of mutations in pfcrt was accompanied by changes in the expression levels of many genes, including a V-type H+ pyrophosphatase (PfVP2) [105]. Physiological evidence supports the presence of a H+-pumping PPase on the DV membrane [106], and it is possible that the level of PfVP2 might be increased to counter a mutant PfCRT-mediated increase in the endogenous leak of H+ from the DV.
Mutations in PfCRT have also been reported to increase the volume of the DV [99]. The molecular basis for this is not understood; one possibility is that mutant PfCRT has a reduced capacity to transport one or more of its (unknown) natural substrates from the DV, resulting in osmotic swelling of the organelle [99].
Box 2. Is mutant pfcrt necessary for CQR?
As discussed, the possession of mutant pfcrt does not necessarily result in in vitro CQR and in vivo CQ treatment failure. Is the opposite true as well, i.e., can P. falciparum parasites achieve in vitro and/or in vivo CQR in the absence of pfcrt mutations?
Several studies, both from Africa and Southeast Asia, have reported the occasional patient isolate that lacked the critical pfcrt K76T mutation, but nevertheless demonstrated in vitro CQR (defined, somewhat arbitrarily, as an IC50 value > 80–100 nM) [107-111]. In these studies drug susceptibility measurements were done directly on polyclonal, non-culture adapted patient isolates, which are therefore unfortunately not available for more detailed investigations. However, in all of these studies, CQ-resistant parasites with wild type pfcrt alleles were isolated cases (1.0-10.5% of all CQ-resistant isolates) and thus clearly the exception to the rule.
By contrast, in vivo CQ-resistant P. falciparum parasites lacking mutations in pfcrt appear to be the norm in Madagascar. In Madagascar, clinical and parasitological treatment failure rates for CQ as high as 35-44% have been reported [112, 113], but the pfcrt K76T genotype remains exceedingly rare and in vitro CQR is infrequently observed. For example, a recent large-scale study identified the pfcrt K76T mutation in only three of 693 (0.4%) Malagasy patient isolates collected in 2006 and 2007, and only 3.2% of 372 tested isolates from 2006-2008 were resistant to CQ in vitro [113]. Using a more sensitive heteroduplex tracking assay, a slightly higher prevalence of parasites carrying pfcrt K76T was reported (2 in 17 patients following CQ treatment; i.e., 11.7%); however, even in these CQ-treated patients parasites with mutant pfcrt remained a minority population, and their contribution to CQ treatment failure remains unclear [114]. Of note, a recent study has now revealed that in Madagascar, in vivo CQ treatment failure, but not in vitro CQR, appears to associate with pfmdr1 N86Y (Box 3) [115].
It is worth noting that in Madagascar almost all CQ treatment failures are late clinical or parasitological failures, while early treatment failures are rare [112]. Moreover, while CQ treatment failure rates are high, they have remained more or less stable since having first been described in 1975, and have not reached complete saturation as in other malaria-endemic countries [113]. Madagascar thus clearly presents a unique situation in which to investigate a pfcrt-unrelated basis of CQ treatment failure.
Box 3. pfmdr1, a secondary determinant of CQR.
Like pfcrt, pfmdr1 encodes a transporter that localizes to the DV membrane (for recent reviews on pfmdr1 see [37, 64, 116]). PfMDR1 (also known as P-glycoprotein homolog 1; Pgh1) is an ortholog of mammalian P-glycoproteins that mediate verapamil-reversible tumor multidrug resistance. Based on the observation that CQR is similarly verapamil-reversible, pfmdr1 became the main CQR candidate gene, until a genetic cross showed that CQR did not segregate with the pfmdr1 locus on chromosome 5 [117].
Nevertheless, PfMDR1 can indeed transport several antimalarial drugs including CQ, with substrate specificity varying between different haplotypes, as recently demonstrated in a Xenopus oocyte expression system [65]. Furthermore, there is convincing evidence for a role of PfMDR1 in parasite response to various antimalarials, such as mefloquine (resistance to which is correlated with pfmdr1 amplification [118]). However, its contribution to CQR is less clear. A single point mutation, pfmdr1 N86Y, that is dominant in Africa and Asia, has been associated with a 1.8-fold increased risk of CQ treatment failure on day 28 in a recent meta-analysis [31]. This compares to a 7.2-fold increased risk with pfcrt K76T. Moreover, while almost all patients that fail CQ treatment carry the pfcrt K76T mutation, pfmdr1 N86Y is usually found in a much lower percentage of post-treatment samples.
South American pfmdr1 haplotypes carry different mutations (Y184F, S1034C, N1042D and D1246Y); however due to the ubiquitous nature of CQR in South America the contribution of these mutations to CQ treatment failure cannot be studied clinically. Two allelic exchange studies have assessed the role of these mutations in CQR. Reversal of S1034C, N1042D and D1246Y in the CQ-resistant 7G8 strain reduced its CQ IC50 value, but the introduction of the same three mutations did not alter CQ susceptibility in another CQ-resistant, and two CQ-sensitive strains [119, 120]. This suggests that the contribution of these mutations to CQR is strongly dependent on the genetic background. Interestingly, a recent in-depth analysis of two genetic crosses revealed that the South American 7G8 pfcrt and pfmdr1 alleles interact to confer greater resistance to monodesethyl-amodiaquine than to CQ [66]. In contrast, pfmdr1 had less influence on the level of CQR when combined with African pfcrt alleles [66]. South American pfcrt and pfmdr1 haplotypes may therefore have evolved primarily in response to amodiaquine drug pressure.
Thus, while pfmdr1 mutations by themselves appear to be insufficient to confer CQR to a CQ-sensitive strain, they can modulate the degree of mutant pfcrt-mediated CQR in a strain-dependent manner. This interplay is also evidenced by the linkage disequilibrium between pfcrt and pfmdr1 observed in numerous field studies [67]. How mutant pfmdr1 contributes to CQR is unclear. It appears that certain mutations result in the loss of a CQ transport function [65] and might therefore result in a lowered accumulation of CQ in the DV. pfmdr1 mutations may also compensate for physiological perturbations induced by expression of mutant pfcrt. Nevertheless, even with identical pfcrt and pfmdr1 mutations parasites can show a range of CQ IC50 values [66], suggesting that additional, as yet unidentified, factors influence parasite susceptibility to CQ.
Acknowledgements
We thank Drs. Thomas Wellems (NIH, USA) and Simon Hay (University of Oxford, UK) for kindly providing the figure files used to generate Figure 1. The endemicity map was generated as part of the Malaria Atlas Project http://www.map.ox.ac.uk), headed by Drs. Bob Snow and Simon Hay. We also thank Ettie Lipner for help with assembling PfCRT haplotype data. Partial funding for this work was provided by the National Institutes of Health (R01 AI50234, to D.A.F.). A.E., J.C. and A.L. gratefully acknowledge funding from a Human Frontiers Science Programme long-term fellowship, a French National University Council fellowship and an Australian National Health and Medical Research Council Overseas Biomedical Fellowship (585519) respectively.
Glossary
- Artemisinin-based combination therapy (ACT)
The current first-line antimalarials, which combine a potent, short-acting artemisinin derivative with a longer-acting partner drug. Examples include artemether-lumefantrine (CoArtem®) and amodiaquine-artesunate (Coarsucam™)
- Chloroquine (CQ)
A 4-aminoquinoline antimalarial drug, formerly extremely effective as first-line therapy
- Chloroquine resistance (CQR)
The ability of parasites to proliferate in the presence of CQ at concentrations that are inhibitory to sensitive strains
- Digestive vacuole (DV)
A lysosome-like acidic organelle that is the site of hemoglobin proteolysis and CQ action
- Gametocytes
sexual blood stages that arrest early in their cell cycle prior to nuclear division and that circulate in the blood as mature forms that are infectious for the Anopheles mosquito vector. P. falciparum gametocytes require ~10-12 days to attain infectivity
- 50%/90% Inhibitory concentration (IC50/IC90)
The drug concentration that inhibits parasite growth by 50% or 90% respectively. In the laboratory this is usually measured by the reduction in the incorporation of [3H]-hypoxanthine or the intercalation of SYBR Green I into parasite nucleic acids
- Isogenic lines
parasites genetically engineered at a specific locus. In the case of pfcrt, this has allowed specific alleles to be investigated in the same genetic background for their effect on drug response
- Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT)
A transporter on the P. falciparum digestive vacuole that when mutated constitutes the primary determinant of CQR
- Plasmodium falciparum parasite rate (PfPR)
A commonly reported index of malaria transmission intensity that describes the proportion of the population found to carry asexual blood-stage parasites
- Quantitative trait loci analysis (QTL analysis)
A method of comparing the inheritance of genetic and phenotypic traits such that log of difference probability scores can be assigned across the genome to define genetic regions that track with the inheritance of a particular phenotype
- Rings, trophozoites, schizonts
Successive stages of the Plasmodium replicative asexual cycle in the host red blood cell. Each cycle consists of invasion, replication, and egress and requires ~48 hr, resulting in the production of ~8-36 infectious merozoites per red blood cell
- Verapamil
A calcium channel blocker that can reverse CQR (at supra-therapeutic doses)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hay SI, et al. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 2004;4:327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trape JF, et al. Combating malaria in Africa. Trends Parasitol. 2002;18:224–230. doi: 10.1016/s1471-4922(02)02249-3. [DOI] [PubMed] [Google Scholar]
- 3.Wellems TE, et al. The impact of malaria parasitism: from corpuscles to communities. J. Clin. Invest. 2009;119:2496–2505. doi: 10.1172/JCI38307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nosten F, White NJ. Artemisinin-based combination treatment of falciparum malaria. Am. J. Trop. Med. Hyg. 2007;77:181–192. [PubMed] [Google Scholar]
- 5.Wells TN, et al. New medicines to improve control and contribute to the eradication of malaria. Nat. Rev. Drug Discov. 2009;8:879–891. doi: 10.1038/nrd2972. [DOI] [PubMed] [Google Scholar]
- 6.WHO Global Malaria Programme World malaria report 2011. 2011.
- 7.Price RN, et al. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr. Opin. Infect. Dis. 2009;22:430–435. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 8.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, et al. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am. J. Trop. Med. Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- 10.Mwai L, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar. J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ursing J, et al. Similar efficacy and tolerability of double-dose chloroquine and artemether-lumefantrine for treatment of Plasmodium falciparum infection in Guinea-Bissau: a randomized trial. J. Infect. Dis. 2011;203:109–116. doi: 10.1093/infdis/jiq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg DE. Hemoglobin degradation. Curr. Top. Microbiol. Immunol. 2005;295:275–291. doi: 10.1007/3-540-29088-5_11. [DOI] [PubMed] [Google Scholar]
- 13.Egan TJ. Recent advances in understanding the mechanism of hemozoin (malaria pigment) formation. J. Inorg. Biochem. 2008;102:1288–1299. doi: 10.1016/j.jinorgbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Roepe PD. Molecular and physiologic basis of quinoline drug resistance in Plasmodium falciparum malaria. Future Microbiol. 2009;4:441–455. doi: 10.2217/fmb.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bray PG, et al. PfCRT and the trans-vacuolar proton electrochemical gradient: regulating the access of chloroquine to ferriprotoporphyrin IX. Mol. Microbiol. 2006;62:238–251. doi: 10.1111/j.1365-2958.2006.05368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wellems TE, et al. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc. Natl. Acad. Sci. USA. 1991;88:3382–3386. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su X, et al. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- 19.Sidhu AB, et al. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel JJ, et al. Chloroquine susceptibility and reversibility in a Plasmodium falciparum genetic cross. Mol. Microbiol. 2010;78:770–787. doi: 10.1111/j.1365-2958.2010.07366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez CP, et al. Genetic linkage analyses redefine the roles of PfCRT and PfMDR1 in drug accumulation and susceptibility in Plasmodium falciparum. Mol. Microbiol. 2011;82:865–878. doi: 10.1111/j.1365-2958.2011.07855.x. [DOI] [PubMed] [Google Scholar]
- 22.Cooper RA, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol. Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn Y, et al. Trafficking of the phosphoprotein PfCRT to the digestive vacuolar membrane in Plasmodium falciparum. Traffic. 2010;11:236–249. doi: 10.1111/j.1600-0854.2009.01018.x. [DOI] [PubMed] [Google Scholar]
- 24.Ehlgen F, et al. Investigation of the Plasmodium falciparum food vacuole through inducible expression of the chloroquine resistance transporter (PfCRT). PLoS One. 2012;7:e38781. doi: 10.1371/journal.pone.0038781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin RE, Kirk K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol. Biol. Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- 26.Tran CV, Saier MH., Jr. The principal chloroquine resistance protein of Plasmodium falciparum is a member of the drug/metabolite transporter superfamily. Microbiol. 2004;150:1–3. doi: 10.1099/mic.0.26818-0. [DOI] [PubMed] [Google Scholar]
- 27.Ecker A, et al. Molecular markers of Plasmodium resistance to antimalarials. In: Staines HM, Krishna S, editors. Treatment and Prevention of Malaria – Antimalarial Drug Chemistry, Action and Use. Springer; 2012. pp. 249–280. [Google Scholar]
- 28.Summers RL, et al. Know your enemy: understanding the role of PfCRT in drug resistance could lead to new antimalarial tactics. Cell. Mol. Life Sci. 2012;69:1967–1995. doi: 10.1007/s00018-011-0906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaijaroenkul W, et al. Sequence and gene expression of chloroquine resistance transporter (pfcrt) in the association of in vitro drugs resistance of Plasmodium falciparum. Malar. J. 2011;10:42. doi: 10.1186/1475-2875-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakshmanan V, et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. Embo J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picot S, et al. A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar. J. 2009;8:89. doi: 10.1186/1475-2875-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mu J, et al. Recent progress in functional genomic research in Plasmodium falciparum. Curr. Genomics. 2010;11:279–286. doi: 10.2174/138920210791233081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volkman SK, et al. Harnessing genomics and genome biology to understand malaria biology. Nature reviews. Genetics. 2012;13:315–328. doi: 10.1038/nrg3187. [DOI] [PubMed] [Google Scholar]
- 34.Chinappi M, et al. On the mechanism of chloroquine resistance in Plasmodium falciparum. PLoS One. 2010;5:e14064. doi: 10.1371/journal.pone.0014064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saliba KJ, et al. Role for the Plasmodium falciparum digestive vacuole in chloroquine resistance. Biochem. Pharmacol. 1998;56:313–320. doi: 10.1016/s0006-2952(98)00140-3. [DOI] [PubMed] [Google Scholar]
- 36.Bray PG, et al. Defining the role of PfCRT in Plasmodium falciparum chloroquine resistance. Mol. Microbiol. 2005;56:323–333. doi: 10.1111/j.1365-2958.2005.04556.x. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez CP, et al. Transporters as mediators of drug resistance in Plasmodium falciparum. Int. J. Parasitol. 2010;40:1109–1118. doi: 10.1016/j.ijpara.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Lehane AM, et al. A verapamil-sensitive chloroquine-associated H+ leak from the digestive vacuole in chloroquine-resistant malaria parasites. J. Cell Sci. 2008;121:1624–1632. doi: 10.1242/jcs.016758. [DOI] [PubMed] [Google Scholar]
- 39.Lehane AM, Kirk K. Chloroquine resistance-conferring mutations in pfcrt give rise to a chloroquine-associated H+ leak from the malaria parasite's digestive vacuole. Antimicrob. Agents Chemother. 2008;52:4374–4380. doi: 10.1128/AAC.00666-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H, et al. The antimalarial drug resistance protein Plasmodium falciparum chloroquine resistance transporter binds chloroquine. Biochemistry. 2004;43:8290–8296. doi: 10.1021/bi049137i. [DOI] [PubMed] [Google Scholar]
- 41.Naude B, et al. Dictyostelium discoideum expresses a malaria chloroquine resistance mechanism upon transfection with mutant, but not wild-type, Plasmodium falciparum transporter PfCRT. J. Biol. Chem. 2005;280:25596–25603. doi: 10.1074/jbc.M503227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin RE, et al. Chloroquine transport via the malaria parasite's chloroquine resistance transporter. Science. 2009;325:1680–1682. doi: 10.1126/science.1175667. [DOI] [PubMed] [Google Scholar]
- 43.Baro NK, et al. Analysis of chloroquine resistance transporter (CRT) isoforms and orthologues in S. cerevisiae yeast. Biochemistry. 2011;50:6701–6710. doi: 10.1021/bi200922g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maughan SC, et al. Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc. Natl. Acad. Sci. USA. 2010;107:2331–2336. doi: 10.1073/pnas.0913689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summers RL, Martin RE. Functional characteristics of the malaria parasite's “chloroquine resistance transporter”: implications for chemotherapy. Virulence. 2010;1:304–308. doi: 10.4161/viru.1.4.12012. [DOI] [PubMed] [Google Scholar]
- 46.Papakrivos J, et al. Functional characterization of the Plasmodium falciparum chloroquine-resistance transporter (PfCRT) in transformed Dictyostelium discoideum vesicles. PLoS One. 2012;7:e39569. doi: 10.1371/journal.pone.0039569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paguio MF, et al. Chloroquine transport in Plasmodium falciparum. 2. Analysis of PfCRT-mediated drug transport using proteoliposomes and a fluorescent chloroquine probe. Biochemistry. 2009;48:9482–9491. doi: 10.1021/bi901035j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gligorijevic B, et al. Stage independent chloroquine resistance and chloroquine toxicity revealed via spinning disk confocal microscopy. Mol. Biochem. Parasitol. 2008;159:7–23. doi: 10.1016/j.molbiopara.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabrera M, et al. Reduced digestive vacuolar accumulation of chloroquine is not linked to resistance to chloroquine toxicity. Biochemistry. 2009;48:11152–11154. doi: 10.1021/bi901765v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehane AM, et al. Differential drug efflux or accumulation does not explain variation in the chloroquine response of Plasmodium falciparum strains expressing the same isoform of mutant PfCRT. Antimicrob. Agents Chemother. 2011;55:2310–2318. doi: 10.1128/AAC.01167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehane AM, et al. Degrees of chloroquine resistance in Plasmodium - is the redox system involved? Int. J. Parasitol. Drugs Drug Resist. 2012;2:47–57. doi: 10.1016/j.ijpddr.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mu J, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS biology. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valderramos SG, et al. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog. 2010;6:e10000887. doi: 10.1371/journal.ppat.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lekostaj JK, et al. Photoaffinity labeling of the Plasmodium falciparum chloroquine resistance transporter with a novel perfluorophenylazido chloroquine. Biochemistry. 2008;47:10394–10406. doi: 10.1021/bi8010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roepe PD. PfCRT-mediated drug transport in malarial parasites. Biochemistry. 2011;50:163–171. doi: 10.1021/bi101638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ekland EH, Fidock DA. In vitro evaluations of antimalarial drugs and their relevance to clinical outcomes. Int. J. Parasitol. 2008;38:743–747. doi: 10.1016/j.ijpara.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Djimde AA, et al. Clearance of drug-resistant parasites as a model for protective immunity in Plasmodium falciparum malaria. Am. J. Trop. Med. Hyg. 2003;69:558–563. [PubMed] [Google Scholar]
- 58.Ursing J, et al. Carriers, channels and chloroquine efficacy in Guinea-Bissau. Trends Parasitol. 2008;24:49–51. doi: 10.1016/j.pt.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Wootton JC, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 60.Kidgell C, et al. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2006;2:e57. doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 62.Mu J, et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat. Genet. 2010;42:268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferdig MT, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 64.Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanchez CP, et al. Polymorphisms within PfMDR1 alter the substrate specificity for anti-malarial drugs in Plasmodium falciparum. Mol. Microbiol. 2008;70:786–798. doi: 10.1111/j.1365-2958.2008.06413.x. [DOI] [PubMed] [Google Scholar]
- 66.Sa JM, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc. Natl. Acad. Sci. USA. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 68.Waller KL, et al. Chloroquine resistance modulated in vitro by expression levels of the Plasmodium falciparum chloroquine resistance transporter. J. Biol. Chem. 2003;278:33593–33601. doi: 10.1074/jbc.M302215200. [DOI] [PubMed] [Google Scholar]
- 69.Osman ME, et al. Field-based evidence for linkage of mutations associated with chloroquine (pfcrt/pfmdr1) and sulfadoxine-pyrimethamine (pfdhfr/pfdhps) resistance and for the fitness cost of multiple mutations in P. falciparum. Infect. Genet. Evol. 2007;7:52–59. doi: 10.1016/j.meegid.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 70.Ord R, et al. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum imply reduced fitness of chloroquine-resistant parasites. J. Infect. Dis. 2007;196:1613–1619. doi: 10.1086/522154. [DOI] [PubMed] [Google Scholar]
- 71.Laufer MK, et al. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J. Infect. Dis. 2010;202:801–808. doi: 10.1086/655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Juliano JJ, et al. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg. Infect. Dis. 2007;13:872–877. doi: 10.3201/eid1306.061182. [DOI] [PubMed] [Google Scholar]
- 73.Djimde AA, et al. A molecular map of chloroquine resistance in Mali. FEMS Immunol. Med. Microbiol. 2010;58:113–118. doi: 10.1111/j.1574-695X.2009.00641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frosch AE, et al. Patterns of chloroquine use and resistance in sub-Saharan Africa: a systematic review of household survey and molecular data. Malar. J. 2011;10:116. doi: 10.1186/1475-2875-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ly O, et al. Evolution of the pfcrt T76 and pfmdr1 Y86 markers and chloroquine susceptibility 8 years after cessation of chloroquine use in Pikine, Senegal. Parasitol Res. 2012 doi: 10.1007/s00436-012-2994-7. in press. [DOI] [PubMed] [Google Scholar]
- 76.Sa JM, Twu O. Protecting the malaria drug arsenal: halting the rise and spread of amodiaquine resistance by monitoring the PfCRT SVMNT type. Malar. J. 2010;9:374. doi: 10.1186/1475-2875-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hallett RL, et al. Combination therapy counteracts the enhanced transmission of drug-resistant malaria parasites to mosquitoes. Antimicrob. Agents Chemother. 2004;48:3940–3943. doi: 10.1128/AAC.48.10.3940-3943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ecker A, et al. Evidence that mutant PfCRT facilitates the transmission to mosquitoes of chloroquine-treated Plasmodium gametocytes. J. Infect. Dis. 2011;203:228–236. doi: 10.1093/infdis/jiq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kelly JX, et al. Discovery of dual function acridones as a new antimalarial chemotype. Nature. 2009;459:270–273. doi: 10.1038/nature07937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zishiri VK, et al. Quinoline antimalarials containing a dibemethin group are active against chloroquinone-resistant Plasmodium falciparum and inhibit chloroquine transport via the P. falciparum chloroquine-resistance transporter (PfCRT). J. Med. Chem. 2011;54:6956–6968. doi: 10.1021/jm2009698. [DOI] [PubMed] [Google Scholar]
- 81.Martin RE, et al. Saquinavir inhibits the malaria parasite's chloroquine resistance transporter. Antimicrob. Agents Chemother. 2012;56:2283–2289. doi: 10.1128/AAC.00166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hocart SJ, et al. 4-aminoquinolines active against chloroquine-resistant Plasmodium falciparum: basis of antiparasite activity and quantitative structure-activity relationship analyses. Antimicrob. Agents Chemother. 2011;55:2233–2244. doi: 10.1128/AAC.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pou S, et al. Sontochin as a guide to the development of drugs against chloroquine-resistant malaria. Antimicrob. Agents Chemother. 2012;56:3475–3480. doi: 10.1128/AAC.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crabb BS, et al. Perspectives: The missing pieces. Nature. 2012;484:S22–23. doi: 10.1038/484S22a. [DOI] [PubMed] [Google Scholar]
- 85.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eastman RT, Fidock DA. Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat. Rev. Microbiol. 2009;7:864–874. doi: 10.1038/nrmicro2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sisowath C, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J. Infect. Dis. 2009;199:750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Folarin OA, et al. In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop. 2011;120:224–230. doi: 10.1016/j.actatropica.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Danquah I, et al. Selection of pfmdr1 and pfcrt alleles in amodiaquine treatment failure in north-western Burkina Faso. Acta Trop. 2010;114:63–66. doi: 10.1016/j.actatropica.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 90.Frank M, et al. A thirteen-year analysis of Plasmodium falciparum populations reveals high conservation of the mutant pfcrt haplotype despite the withdrawal of chloroquine from national treatment guidelines in Gabon. Malar. J. 2011;10:304. doi: 10.1186/1475-2875-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gbotosho GO, et al. Different patterns of pfcrt and pfmdr1 polymorphisms in P. falciparum isolates from Nigeria and Brazil: the potential role of antimalarial drug selection pressure. Am. J. Trop. Med. Hyg. 2012;86:211–213. doi: 10.4269/ajtmh.2012.11-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sa JM, et al. Malaria drug resistance: new observations and developments. Essays Biochem. 2011;51:137–160. doi: 10.1042/bse0510137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boni MF, et al. Benefits of using multiple first-line therapies against malaria. Proc. Natl. Acad. Sci. USA. 2008;105:14216–14221. doi: 10.1073/pnas.0804628105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eastman RT, et al. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob. Agents Chemother. 2011;55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cooper RA, et al. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol. Microbiol. 2007;63:270–282. doi: 10.1111/j.1365-2958.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 96.Gething PW, et al. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar. J. 2010;10:378. doi: 10.1186/1475-2875-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dzekunov SM, et al. Digestive vacuolar pH of intact intraerythrocytic P. falciparum either sensitive or resistant to chloroquine. Mol. Biochem. Parasitol. 2000;110:107–124. doi: 10.1016/s0166-6851(00)00261-9. [DOI] [PubMed] [Google Scholar]
- 98.Bennett TN, et al. Drug resistance-associated pfCRT mutations confer decreased Plasmodium falciparum digestive vacuolar pH. Mol. Biochem. Parasitol. 2004;133:99–114. doi: 10.1016/j.molbiopara.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 99.Gligorijevic B, et al. Spinning disk confocal microscopy of live, intraerythrocytic malarial parasites. 2. Altered vacuolar volume regulation in drug resistant malaria. Biochemistry. 2006;45:12411–12423. doi: 10.1021/bi0610348. [DOI] [PubMed] [Google Scholar]
- 100.Hayward R, et al. The pH of the digestive vacuole of Plasmodium falciparum is not associated with chloroquine resistance. J. Cell Sci. 2006;119:1016–1025. doi: 10.1242/jcs.02795. [DOI] [PubMed] [Google Scholar]
- 101.Klonis N, et al. Evaluation of pH during cytostomal endocytosis and vacuolar catabolism of haemoglobin in Plasmodium falciparum. Biochem. J. 2007;407:343–354. doi: 10.1042/BJ20070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuhn Y, et al. Quantitative pH measurements in Plasmodium falciparum-infected erythrocytes using pHluorin. Cell Microbiol. 2007;9:1004–1013. doi: 10.1111/j.1462-5822.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 103.Nessler S, et al. Evidence for activation of endogenous transporters in Xenopus laevis oocytes expressing the Plasmodium falciparum chloroquine resistance transporter, PfCRT. J. Biol. Chem. 2004;279:39438–39446. doi: 10.1074/jbc.M404671200. [DOI] [PubMed] [Google Scholar]
- 104.Reeves DC, et al. Chloroquine-resistant isoforms of the Plasmodium falciparum chloroquine resistance transporter acidify lysosomal pH in HEK293 cells more than chloroquine-sensitive isoforms. Mol. Biochem. Parasitol. 2006;150:288–299. doi: 10.1016/j.molbiopara.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang H, et al. Genome-wide compensatory changes accompany drug- selected mutations in the Plasmodium falciparum crt gene. PLoS One. 2008;3:e2484. doi: 10.1371/journal.pone.0002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saliba KJ, et al. Acidification of the malaria parasite's digestive vacuole by a H+-ATPase and a H+-pyrophosphatase. J. Biol. Chem. 2003;278:5605–5612. doi: 10.1074/jbc.M208648200. [DOI] [PubMed] [Google Scholar]
- 107.Thomas SM, et al. In vitro chloroquine susceptibility and PCR analysis of pfcrt and pfmdr1 polymorphisms in Plasmodium falciparum isolates from Senegal. Am. J. Trop. Med. Hyg. 2002;66:474–480. doi: 10.4269/ajtmh.2002.66.474. [DOI] [PubMed] [Google Scholar]
- 108.Basco LK. Molecular epidemiology of malaria in Cameroon. XIII. Analysis of pfcrt mutations and in vitro chloroquine resistance. Am. J. Trop. Med. Hyg. 2002;67:388–391. doi: 10.4269/ajtmh.2002.67.388. [DOI] [PubMed] [Google Scholar]
- 109.Basco LK. Molecular epidemiology of malaria in Cameroon. XII. In vitro drug assays and molecular surveillance of chloroquine and proguanil resistance. Am. J. Trop. Med. Hyg. 2002;67:383–387. doi: 10.4269/ajtmh.2002.67.383. [DOI] [PubMed] [Google Scholar]
- 110.Mayxay M, et al. In vitro antimalarial drug susceptibility and pfcrt mutation among fresh Plasmodium falciparum isolates from the Lao PDR (Laos). Am. J. Trop. Med. Hyg. 2007;76:245–250. [PMC free article] [PubMed] [Google Scholar]
- 111.Sarr O, et al. In vivo and in vitro analysis of chloroquine resistance in Plasmodium falciparum isolates from Senegal. Parasitol Res. 2005;97:136–140. doi: 10.1007/s00436-005-1406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Randrianarivelojosia M, et al. Lessons learnt from the six decades of chloroquine use (1945-2005) to control malaria in Madagascar. Trans. R. Soc. Trop. Med. Hyg. 2009;103:3–10. doi: 10.1016/j.trstmh.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 113.Andriantsoanirina V, et al. Plasmodium falciparum drug resistance in Madagascar: facing the spread of unusual pfdhfr and pfmdr-1 haplotypes and the decrease of dihydroartemisinin susceptibility. Antimicrob. Agents Chemother. 2009;53:4588–4597. doi: 10.1128/AAC.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Juliano JJ, et al. Nonradioactive heteroduplex tracking assay for the detection of minority-variant chloroquine-resistant Plasmodium falciparum in Madagascar. Malar. J. 2009;8:47. doi: 10.1186/1475-2875-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Andriantsoanirina V, et al. Chloroquine clinical failures in P. falciparum malaria are associated with mutant Pfmdr-1, not Pfcrt in Madagascar. PLoS One. 2010;5:e13281. doi: 10.1371/journal.pone.0013281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koenderink JB, et al. The ABCs of multidrug resistance in malaria. Trends Parasitol. 2010;26:440–446. doi: 10.1016/j.pt.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 117.Wellems TE, et al. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 118.Price RN, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reed MB, et al. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 120.Sidhu AB, et al. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 121.Chen N, et al. pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob. Agents Chemother. 2003;47:3500–3505. doi: 10.1128/AAC.47.11.3500-3505.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Durrand V, et al. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol. Biochem. Parasitol. 2004;136:273–285. doi: 10.1016/j.molbiopara.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 123.Echeverry DF, et al. Short report: polymorphisms in the pfcrt and pfmdr1 genes of Plasmodium falciparum and in vitro susceptibility to amodiaquine and desethylamodiaquine. Am. J. Trop. Med. Hyg. 2007;77:1034–1038. [PubMed] [Google Scholar]
- 124.Lumb V, et al. Differential genetic hitchhiking around mutant pfcrt alleles in the Indian Plasmodium falciparum population. J. Antimicrob. Chemother. 2011;67:600–608. doi: 10.1093/jac/dkr532. [DOI] [PubMed] [Google Scholar]
- 125.Mixson-Hayden T, et al. Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum isolates in India. Antimicrob. Agents Chemother. 2010;54:997–1006. doi: 10.1128/AAC.00846-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nagesha HS, et al. New haplotypes of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene among chloroquine-resistant parasite isolates. Am. J. Trop. Med. Hyg. 2003;68:398–402. [PubMed] [Google Scholar]
- 127.Yang Z, et al. Molecular analysis of chloroquine resistance in Plasmodium falciparum in Yunnan Province, China. Trop. Med. Int. Health. 2007;12:1051–1060. doi: 10.1111/j.1365-3156.2007.01882.x. [DOI] [PubMed] [Google Scholar]
- 128.Johnson DJ, et al. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol. Cell. 2004;15:867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]