Abstract
With a rapid increase in the number of high-resolution protein-ligand structures, the known protein-ligand structures can be used to gain insight into ligand-binding modes in a target protein. Based on the fact that the structurally similar binding sites share information about their ligands, we have developed a local structure alignment tool, G-LoSA (Graph-based Local Structure Alignment). In G-LoSA, the known protein-ligand binding-site structure library is searched to detect binding-site structures with similar geometry and physicochemical properties to a query binding-site structure regardless of sequence continuity and protein fold. Then, the ligands in the identified complexes are used as templates (i.e., template ligands) to predict/design a ligand for the target protein. The performance of G-LoSA is validated against 76 benchmark targets from the Astex diverse set. Using the currently available protein-ligand structure library, G-LoSA is able to identify a single template ligand (from a non-homologous protein complex) that is highly similar to the target ligand in more than half of the benchmark targets. In addition, our benchmark analyses show that an assembly of structural fragments from multiple template ligands with partial similarity to the target ligand can be used to design novel ligand structures specific to the target protein. This study clearly indicates that a template-based ligand modeling has potential for de novo ligand design and can be a complementary approach to the receptor structure based methods.
Keywords: ligand binding site, G-LoSA, template-based ligand design, computer-aided drug design
INTRODUCTION
A fundamental task in bioinformatics is to acquire biologically meaningful information from available data sets using computational methods optimized for a given problem. A common approach is to analyze similarities between two biological entities. For example, in structural biology, such similarities can be recognized at different levels such as sequence, secondary structure, or three-dimensional (3D) structure1,2 This knowledge-based approach is one of the most powerful methods to elucidate the structure-function relationship of biological molecules.
Template-based structure modeling has become a key technique to obtain 3D structural information in the absence of experimental structures of interest. In protein structure prediction, based on sequence and/or fold similarity, single or multiple protein structures are used as templates3 The utility of the templates ranges from the de novo methods using a set of short structural fragments to the homology modeling methods using the entire structures4 Recently, template-based modeling has also been applied to predict 3D RNA structures5 A key element for successful template-based structure prediction is to efficiently identify correct templates from a known structure library.
With a rapid increase in the number of high-resolution protein-ligand structures in the Protein Data Bank (PDB, http://www.rcsb.org)6, it is possible to gain insight into the structures of ligands that are likely bound to a target protein from the known structures that have a similar ligand-binding environment to the target protein7 Therefore, it is important to develop an efficient computational method to detect structural conservation between ligand-binding sites from distinct proteins for identification of proper template protein-ligand complexes. Based on the fact that structurally similar ligand-binding sites share information about their ligands, one can use the ligands in the template complexes (i.e., template ligands) to predict/design a ligand for a target protein.
The ligand-binding site is a local structure. Dissimilarity in the global structures between two proteins need not imply different functions because their functions can be localized at conserved ligand-binding regions8–10 In addition, ligand-binding sites often share conserved surface regions with similar physicochemical properties and shapes, even if they do not show common sequence patterns11–13 However, aligning local structures, such as ligand-binding sites, is challenging because residues in the binding sites are mostly discontinuous (in terms of the residue number) and there is little secondary structure for fold recognition.
There are a handful of methods available for local structure alignment. SitesBase14 uses geometric hashing to identify geometrically equivalent atom pairs with the same element types. In a method by Minai et al.,15 feature vectors are utilized to discard atom pairs with dissimilar local physicochemical environments. In this method, a pair of surface atom triads is transformed with rotation and translation matrices, and this process is repeated to find the largest alignment score between the local structures. Cavbase16 and ProBiS17 adopt maximal common subgraph search methods. The local structures are represented as graphs of vertices and edges corresponding to functional groups and distances between the functional groups, respectively. Common subgraphs of maximal alignment in a pair of the local structures are then detected to superpose the structures. PocketAlign18 uses shape descriptors, which encode local structures using Cα atoms, to perform residue-wise pairing. The residue pairs are combined into a mapping to obtain an optimal alignment. A method by Feldman and Labute uses Cα atom coordinates to measure local structure similarity19 In this method, an optimal alignment is obtained by an exhaustive search of residue-wise pairs with similar physicochemical properties. The studies using these tools have mainly focused on validation of their local structure alignment methods and applications to binding-site detection and function prediction.
Here, we present a computational method for local structure alignment and its applications focusing on identification of template ligands. First, the algorithm adopted in our local structure alignment tool, G-LoSA (Graph-based Local Structure Alignment), is described. The G-LoSA algorithm uses the Cα atom coordinates and the maximal common subgraph search, similar to Cavbase16 and ProBiS,17 for structure alignment. The benchmark results using 76 targets from the Astex diverse set are then presented to show the performance of G-LoSA in detecting template ligands. Furthermore, G-LoSA is evaluated by performance comparison with TM-align20 and ProBiS. In addition, the representative examples of the template ligand search are described. The overall results indicate that G-LoSA is able to identify template ligands that share common structures and poses to the target ligand using the currently available protein-ligand structure library, illustrating its potential practical use for de novo drug design based on template ligands.
METHODS
G-LoSA algorithm
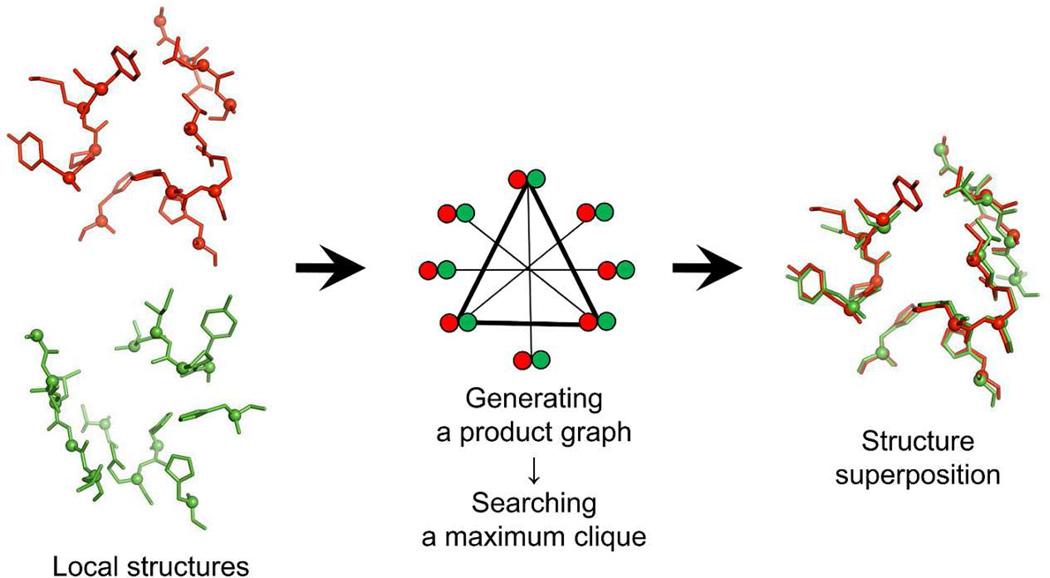
Figure 1 schematically shows the G-LoSA local structure alignment method that utilizes the nondirected graph. For two given local structures, all combinations of Cα-Cα pairs (Pij) are generated, where i and j are the first and the second local structures. If the first and the second local structures consist of M and N residues, the number of the generated pairs becomes M × N. The amino acid similarity between two residues in each pair is measured based on the BLOSUM62 matrix21 In order to perform a local structure superposition centered by conserved residues and to reduce the total number of pairs, a pair is removed from Pij if its BLOSUM62 score < 0. In the BLOSUM matrix, a positive score means more likely substitutions.
Figure 1.
A schematic representation of G-LoSA algorithm. Two local structures colored red and green are shown in stick representation with Cα atom spheres. A product graph is constructed based on the geometric and physicochemical similarity between the local structures. A subgraph representing the largest set of aligned residue pairs (maximum clique) is identified. The two local structures are then superposed using a translational and rotational matrix calculated from the identified residue pair set.
A product graph is then generated using Pij. A product graph consists of a set of vertices and a set of edges connecting the vertices. A vertex in the product graph corresponds to a residue pair p from Pij. Two pairs, p1(i1, j1) and p2(i2, j2), are selected from Pij, and then both distances d(i1, i2) and d(j1, j2) are calculated using the atomic coordinates. If | d(i1, i2) − d(j1, j2) | < 1 Å, p1(i1, j1) and p2(i2, j2) are assigned to product graph vertices and connected by an edge. These procedures are repeated to build a product graph for all the possible non-identical residue pairs in Pij.
The generated product graph is searched for a maximum clique. A clique is a subset of vertices in which each vertex is connected to all other vertices in the subset. A maximum clique is a clique with the largest number of vertices. In our case, solving a maximum clique problem for a product graph is to identify the largest set of structurally aligned residue pairs whose physicochemical properties are similar. To search a maximum clique in a product graph, we have utilized an algorithm developed by Konc and Janežič,22,23 where an approximate coloring algorithm is used to rapidly find a maximum clique in an undirected graph. The least-squared superposition of the aligned residue sets, which are determined by the maximum clique, is carried out using the Kabsch algorithm24 The G-LoSA structure alignment algorithm only requires atomic coordinates and residue names, and thus its performance does not depend on the sequence continuity and the fold similarity.
Preparation of binding site-ligand structure library
We downloaded the PDB files of X-ray crystallographic structures and solution NMR structures containing at least one protein and one ligand. The X-ray structures with resolution of >3 Å were eliminated from the library. DNA and RNA molecules were also discarded, and ligand molecules in the PDB files were identified in the heteroatom section. Heteroatoms having an identical chain ID and sequence number were grouped into one heteroatom group. If a distance of any atom pair from different heteroatom groups was 1–2 Å, the two heteroatom groups were merged into one group and identified as multipart ligands. Metal ions, water molecules, and small molecular weight additives were removed by setting the minimum number of heavy atoms in a heteroatom group to 4. Duplicated proteins and ligands in a PDB file were removed except the first one. To only consider non-covalently bound ligands, if any atom in a heteroatom group was located within 2 Å from any protein atom, the heteroatom group was identified as a covalently-linked ligand and removed from the library. If any atom of a residue in a protein is within 4 Å of its cognate ligand, the residue is defined as the binding-site (BS) residue. A set of the BS residues is referred to as a BS-structure. Out of 50,487 downloaded structures, there were 38,755 structure files remained and 60,456 ligand/BS-structure pairs in our library (as of September, 2011).
Benchmark set
Benchmark proteins were taken from the Astex diverse set25 This set consists of diverse high-resolution protein-ligand complexes and represents interesting drug targets for the pharmaceutical and agrochemical industries. In the Astex diverse set, we excluded complex structures in which the ligand BS is shared by more than one protein chain. The final benchmark set consists of 76 complexes (Table S1 in Supporting Information). The BS-structure of each benchmark target was defined by a cutoff of 4 Å from the cognate ligand and used as a query (target) BS-structure to search template ligands from our ligand/BS-structure library using G-LoSA.
Template ligand search for benchmark targets
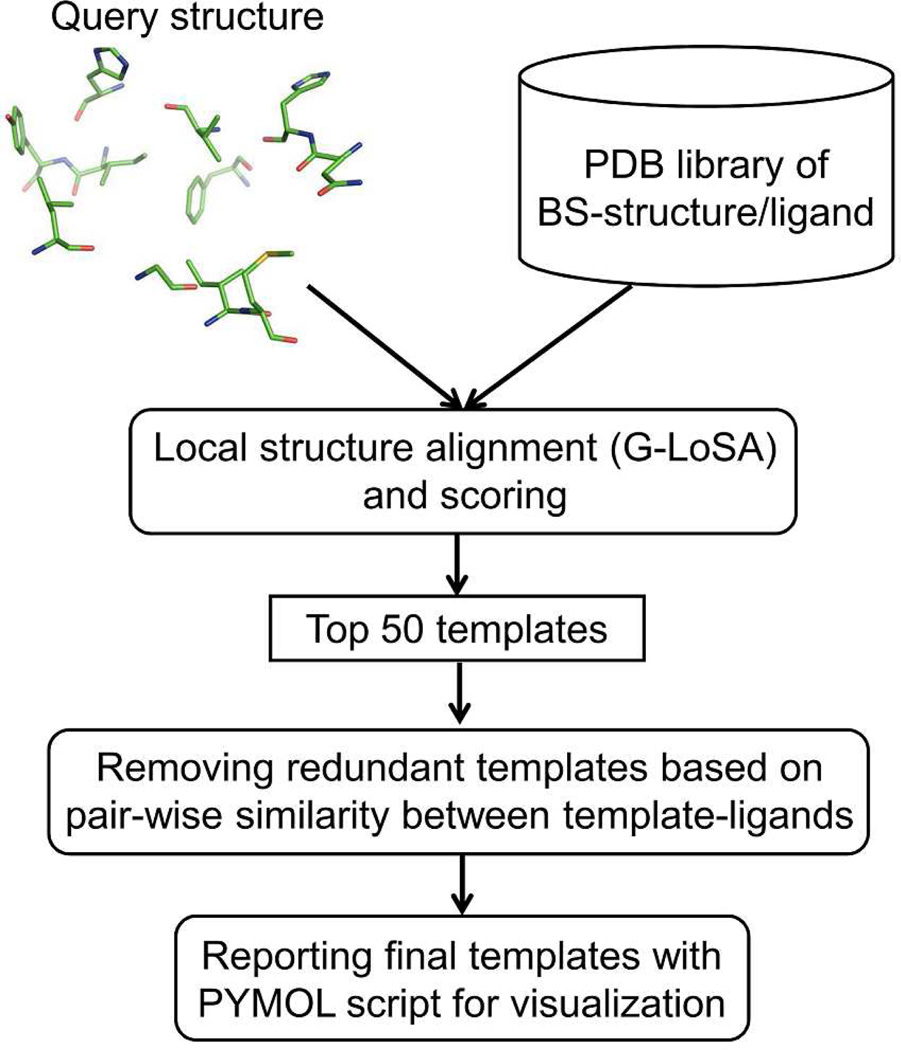
The overall scheme to search a template ligand is shown in Figure 2. For a given target BS-structure, a set of template ligands are identified in terms of a structural similarity score. The library BS-structure is first superposed onto the target structure by G-LoSA, and the similarity score (S) for the aligned local structures is calculated by
| (1) |
where N is the number of aligned library BS-structure residues. If the Cα distance between the target residue and the nearest library residue is within 2.5 Å, the library residue is assigned as the aligned residue. RMSD is the root-mean-square deviation of the aligned residues pairs and calculated using the coordinates of Cα atoms and side-chain centroids. To avoid numerical sensitivity of the score S with small RMSD, the RMSD value is set to 0.5 if RMSD < 0.5.
Figure 2.
The overall procedure to identify template ligands.
To put strict conditions on the template ligand search in this study, we excluded all homologous library proteins whose sequence identity is > 30% to the target protein. For each target, the top 50 template BS-structure/ligand pairs ranked by the score S were first collected. To obtain structurally diverse template ligands, we calculated a Tanimoto coefficient (TPO) representing the positional overlap between template ligands
| (2) |
where N1 and N2 are the number of non-hydrogen atoms in two template ligands. NO is the number of overlapped atoms. If the distance between an atom of the first template ligand and its nearest atom of the second template ligand is ≤ 1.2 Å, the atom of the first template is defined as an overlapped atom. A TPO cutoff value of 0.7 was applied to remove redundant template ligands except the template ligand with the highest S.
To quantify the structural and conformational similarity between an identified template ligand and a native (target) ligand, we calculated an “overlap ratio (RO)” defined by
| (3) |
where NOI and N are the number of overlapped identical atoms and a total number of atoms in the target ligand, respectively. If the distance between an atom of the target ligand and its nearest atom of the template ligand is ≤ 1.2 Å and their atom types are identical, the ligand atom is defined as the overlapped identical atom. A larger RO means that the larger structural fragments in the target ligand are regenerated by the template ligand.
Control programs
To assess the performance of G-LoSA, we carried out the performance comparison with TM-align20 and ProBiS17 TM-align20 is a computation method to identify the optimal structural alignment between protein pairs using the TM-score rotation matrix. The source code of the program was taken from the TM-align web site (http://zhanglab.ccmb.med.umich.edu/TM-align). All parameters were set to the default values.
ProBiS17 is a local structure alignment method to detect structurally similar sites on protein surface. An open source of parallel-ProBiS26 (version 2.3.8) was taken from the ProBiS web site (http://probis.cmm.ki.si)27 All BS-structures for the benchmark set and the structure library were converted into the surface format (.srf) files for the template ligand search using ProBiS. Z_SCORE and SURF_VECTOR_ANGLE were set to −10.0 and 4.0, respectively, to turn off filtering of local structure alignment results. Other parameters were set to the default values, and “alignment_score” was used to rank-order template BS-structures.
RESULTS
Identification of template ligands by local structure alignment
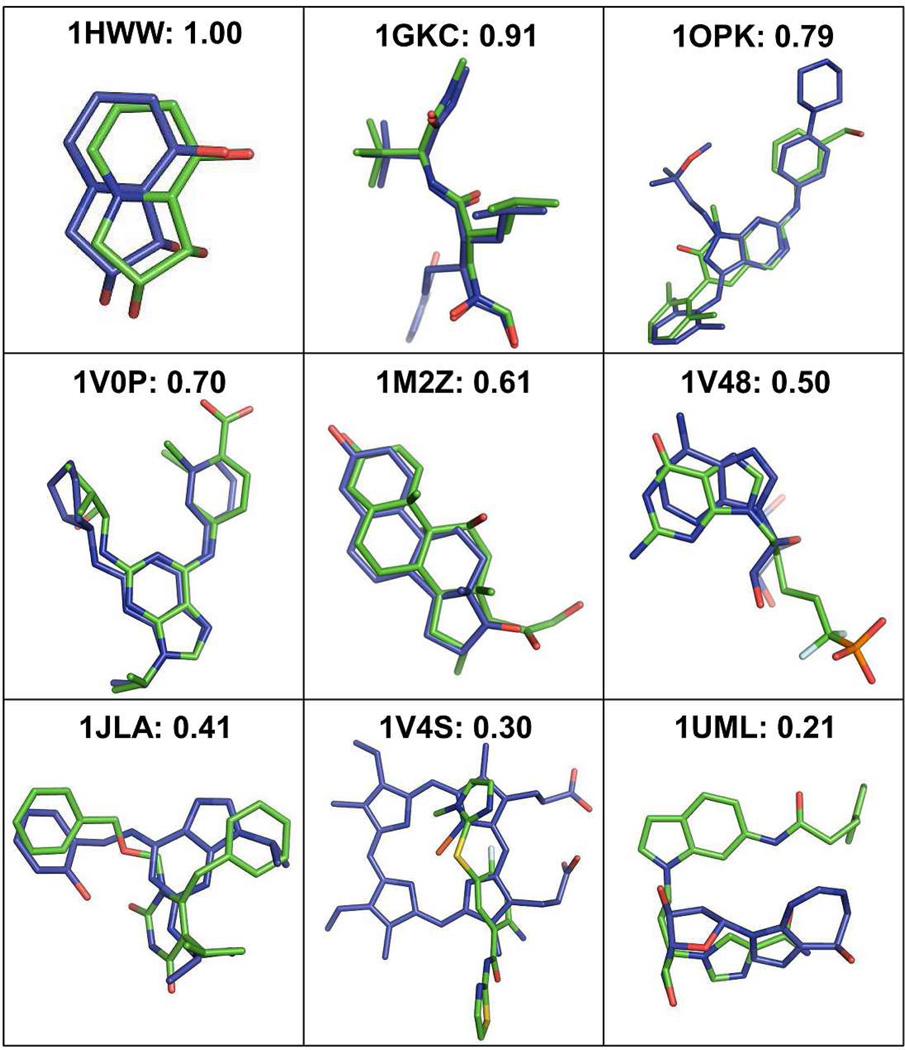
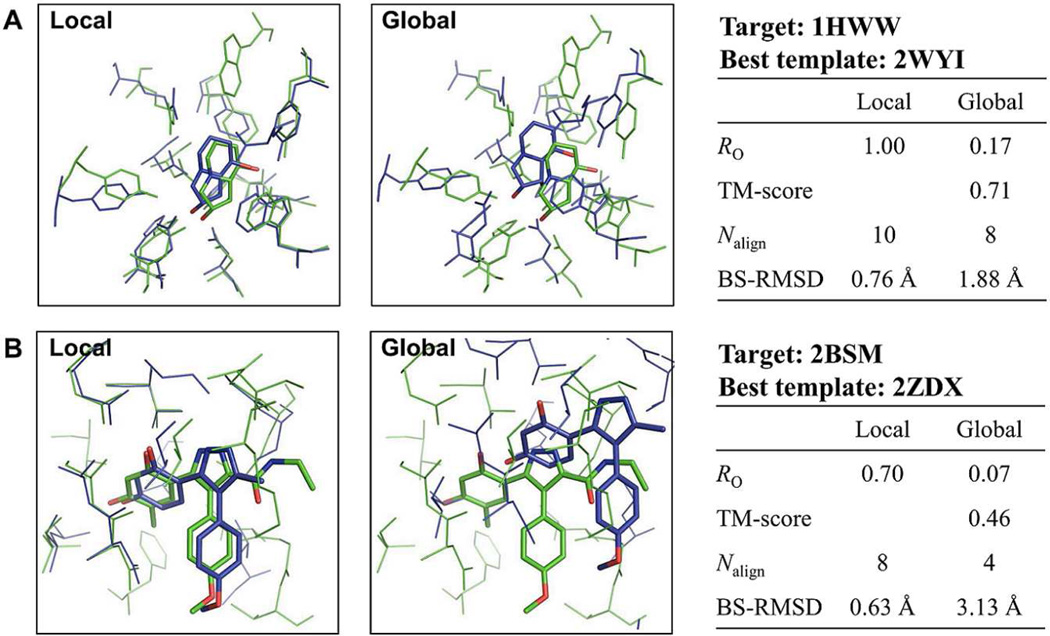
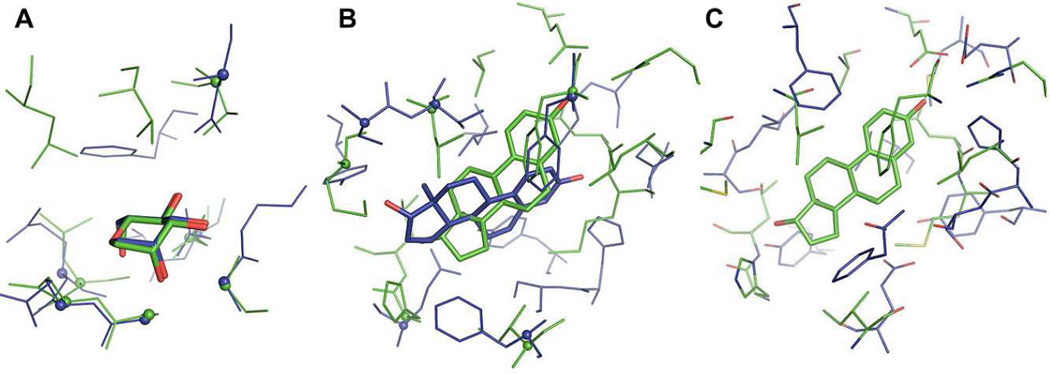
The template ligand search using G-LoSA has been implemented for the 76 benchmark targets (Table S1) from the Astex diverse set25 Figure 3 shows representative examples to illustrate the relationship between the overlap ratio RO in Eq. 3 and target-template ligand structural similarity. The best template ligand, which has the best RO among top 10 templates (ranked by the similarity score S in Eq. 1), was used for the structural comparison. The PDB IDs of the best templates for each benchmark target are also listed in Table S1. In the case of target PDB:1HWW where RO is 1.0, the template ligand (PDB:2WYI) is identical to the cognate ligand in terms of binding pose as well as the overall structure and chemical composition. However, RO = 1.0 does not always mean the exact match between the target and template ligands because a larger template ligand may have a value of RO = 1.0. As RO decreases, the target-template ligand structural similarity decreases. The figures intuitively show that template ligands with RO ≥ 0.5 share geometrically well-overlapped conserved structures to the corresponding target ligands. In some cases, however, the scaffold similarity and/or partial structure matches are still noticeable even for the template ligands with RO < 0.5.
Figure 3.
Representative examples to illustrate the relationship between the overlap ratio value and the structural similarity between target ligands (green) and template ligands (blue). The template ligand of the best overlap ratio among top 10 templates was used for the structural comparison.
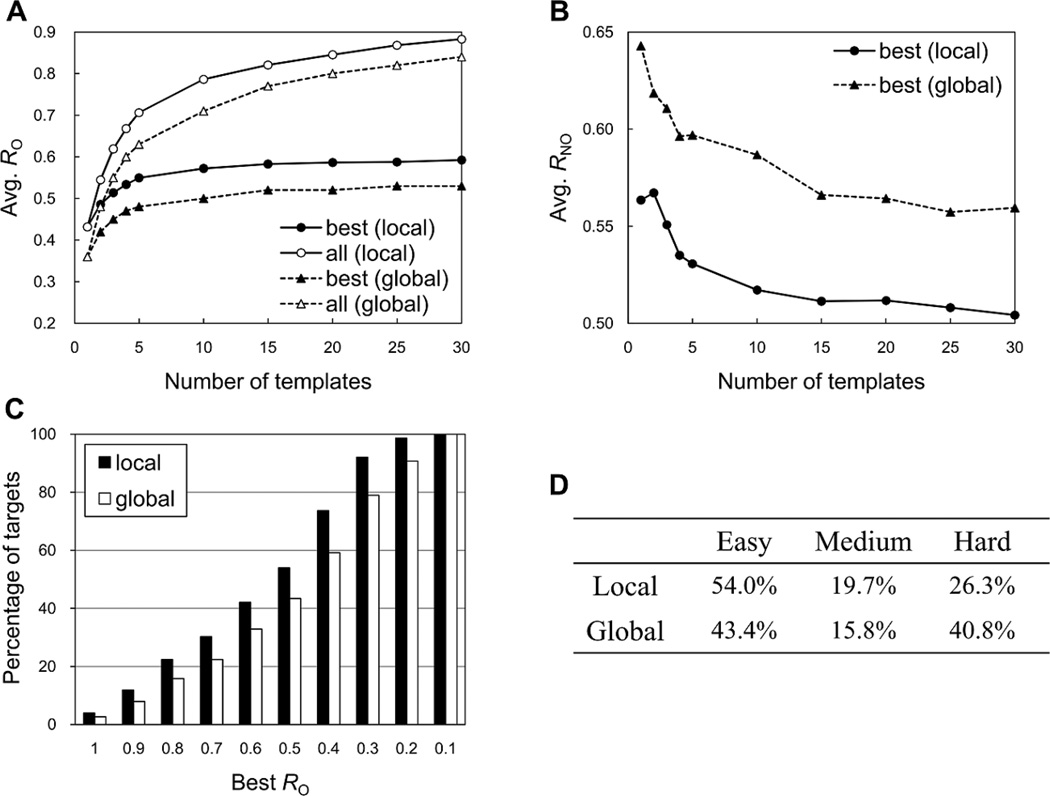
Figure 4A shows the average RO between the best template ligand and the target ligand over the benchmark targets as a function of the number of the top templates. The best template was chosen among given top N templates in terms of RO. The average RO increases as more multiple template ligands are used for the best template selection, though the effect is not significant after five templates. On the other hand, as shown in Figure 4B, the average non-overlap ratio (RNO) of the best template ligands decreases in terms of the number of the top templates. RNO is calculated by NNO/N, where NNO is the number of the template ligand atoms whose distance from the nearest atom of the target ligand is > 1.2 Å or whose atom types are not identical, and N is the total number of the target ligand atoms. When the RO values were measured by a collection of all the top template ligands instead of a single best template ligand, the average structural coverage to the target ligands significantly increased (Figure 4A). For example, the average RO by the best template ligand of top 10 templates was 0.57, but the value increased to 0.79 with the addition of the multiple templates. This indicates that if a set of substructures (or fragments) can be effectively extracted from a set of identified template ligands and assembled into a single molecule, the assembled molecule could be a ligand that is very similar to the target ligand (also see Representative Example 2 and 3 below).
Figure 4.
Performance in identifying template ligands by the local (G-LoSA) and global (TM-align) structure alignments for the benchmark targets. (A) The number of top templates vs. the average overlap ratio (RO) over the benchmark targets, calculated for the best (closed circle and triangle) and multiple templates (open circle and triangle). The best template was chosen among given top N templates in terms of RO. The average RO for the multiple templates was measured by using a collection of the coordinates of all the top N template ligands. (B) The number of top templates vs. the average non-overlap ratio (RNO) for the best templates over the benchmark targets. RNO is the fraction of the best template which is not overlapped with the target ligand. (C) Percentage of targets vs. the best RO. The best RO was obtained from top 10 template ligands. (D) Percentages of easy, medium, and hard targets, calculated using the best RO of top 10 template ligands.
Based on the relationship between the best RO and the structural similarity of the top 10 template ligands in Figure 3, the template-based ligand prediction can be approximately classified into three classes: easy (best RO ≥ 0.5), medium (0.5 > best RO ≥ 0.4), and hard targets (best RO < 0.4). With this definition and the currently available protein-ligand structure library, the G-LoSA results show that there are 54.0% easy, 19.7% medium, and 26.3% hard targets in our benchmark set (Figures 4C and 4D).
Comparison with global structure alignment
It is known that conserved substrate binding sites are observed in certain protein folds despite little evidence of a common ancestor for the proteins, suggesting that analogous or very distantly homologous proteins having similar folds can have common binding sites28 Brylinski and Skolnick have developed FINDSITE, a method for ligand BS prediction on the basis of the significant BS similarity across groups of weakly homologous template structures detected by threading methods29 Lee and Zhang have developed BSP-SLIM, a method for protein-ligand blind docking30 in which putative ligand binding sites were successfully derived from template holo protein structures identified from the PDB library based on global structure alignment by TM-align20 These studies indicate that the structure alignment based on global topology similarity between different proteins can also be a powerful approach to identify template ligands.
Diverse tools for the global structural alignment such as DALI,31 CE,32 SAL33 and TM-align20 are available. Because of its advantage in accuracy,20 TM-align was chosen as a global structure alignment tool to compare its performance with that of G-LoSA. To identify template ligands based on TM-align, we carried out the identical procedure used for the G-LoSA search except that the initial structure alignment between the whole structures of target and library proteins was implemented by TM-align. Figure 4C shows that the TM-align search can successfully detect template ligands that are highly similar to the target ligands for 43.4% of the benchmark targets (easy targets in Figure 4D). Nonetheless, the G-LoSA performance is better in every property evaluated (Figure 4). For example, the total percentage of the easy and medium targets classified by RO dropped by 14.5%, indicating that there are ~15% less chances to yield the template ligand with the best RO ≥ 0.4 in the global alignment-based method.
The following two representative examples are used to elucidate the better performance of G-LoSA. Figure 5A shows the superposed BS-structures between Drosophila Golgi α-mannosidase II (PDB:1HWW) and bacterial α-mannosidase (PDB:2WYI) by G-LoSA and TM-align. Both proteins are in complex with an identical ligand SWA. The sequence identity between the two proteins is low (0.18), but their global topology is similar (TM-score = 0.71). When the two proteins were locally aligned, the measured RO between the template and target ligands was 1.0. In contrast, the global structural alignment by TM-align yielded an RO of 0.17. In the structures aligned by TM-align, not only the number of aligned residues decreased, but also the BS-RMSD significantly increased, resulting in a low similarity score between the BS-structures. Consequently, the low quality structural alignment within the binding site leads to the poor performance in identifying good template ligands.
Figure 5.
Comparison of structure superposition results by the local (G-LoSA) and global (TM-align) alignment methods for benchmark target 1HWW and 2BSM (green). The corresponding best templates (blue) were chosen from top 10 templates by G-LoSA.
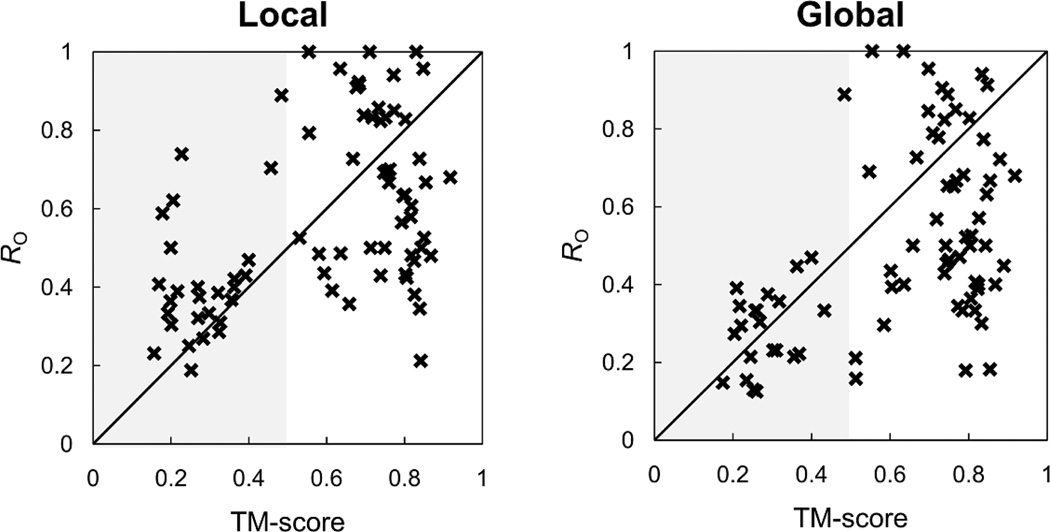
Such inaccuracy of the global structural alignment method in superposing BS-structures becomes more severe when global topologies between target and template proteins are largely different. Figure 5B shows the superposed BS-structures between the N-terminal domain of molecular chaperone Hsp90 with ligand BSM (PDB:2BSM) and human pyruvate dehydrogenase kinase 4 with ligand P4A (PDB:2ZDX) by G-LoSA and TM-align. The TM-score is 0.46. Compared to the previous example (Figure 5A), the smaller number of aligned residues with increased BS-RMSD is observed in the global alignment result. Such poor performance of the global alignment method can be explained by the RO of the best template ligand (among the top 10) with respect to the TM-score between the target and template proteins for the benchmark targets (Figure 6). For both G-LoSA and TM-align, highly similar template ligands tend to be detected from proteins with similar global topology (TM-score ≥ 0.5). However, compared to the global structure alignment, the ability of the local structure alignment in identifying good templates is less sensitive to global fold similarity. Among the easy and medium targets identified by G-LoSA, 12 benchmark targets showed TM-score < 0.5, whereas there are only 3 such benchmark proteins among the easy and medium targets identified by TM-align. The results indicate that the quality of the template ligand selection is limited by the global fold similarity when the global alignment is used, and the local structure alignment by G-LoSA is more effective in detecting structural similarity within binding sites between proteins having distinct folds.
Figure 6.
Overlap ratio (RO) as a function of TM-score for local (G-LoSA) and global (TM-align) alignment methods. The RO and TM-score were calculated using the best template ligand of the top 10 template ligands and its cognate receptor, respectively.
Comparison with TM-align-BS
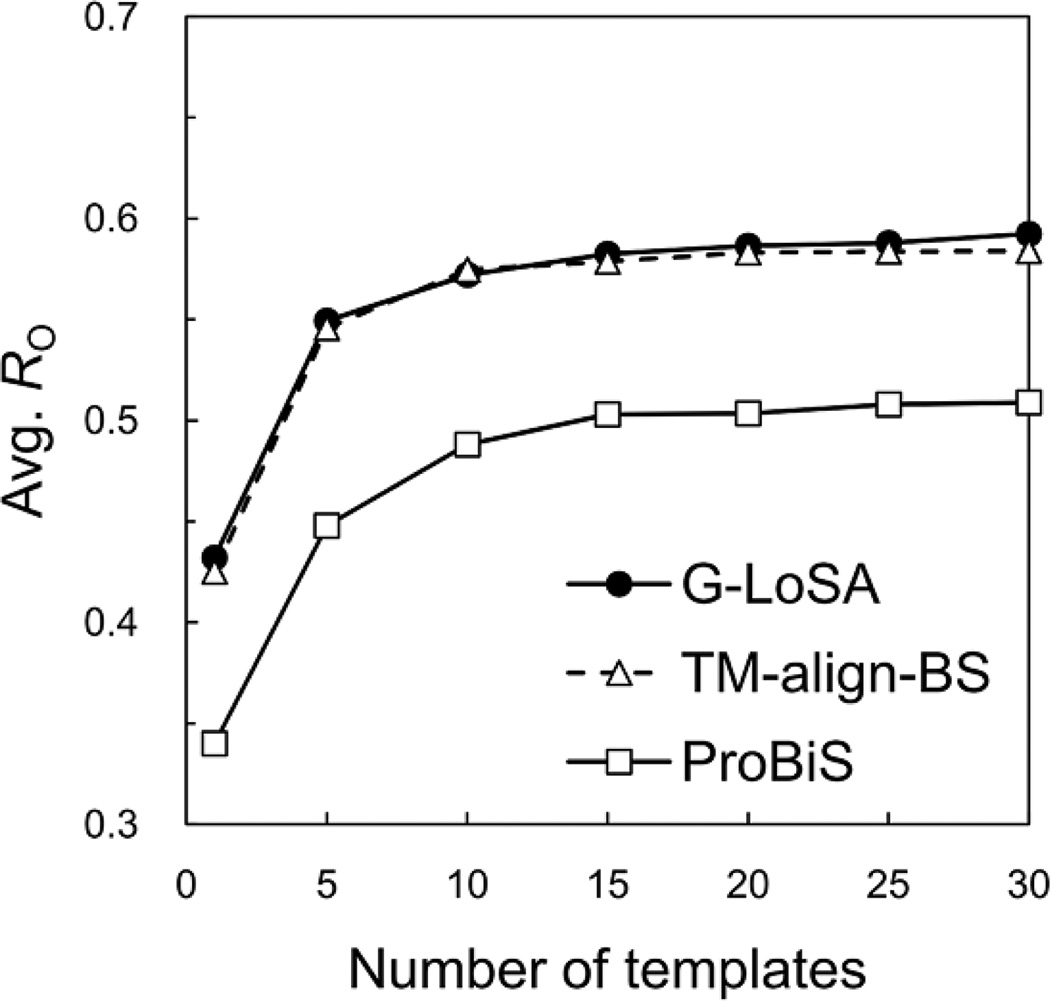
In the previous section, we used TM-align to align the whole structures of target and library proteins, and the (local) BS structural similarities were calculated on the basis of the superposed pairs. Here, G-LoSA is compared with the local structural alignment by TM-align (TM-align-BS), i.e., only the BS-structures (for both targets and library proteins) were used in TM-align. Unexpectedly, the average RO by TM-align-BS are comparable to those by G-LoSA (Figure 7). The TM-align algorithm was originally developed to detect the similarity of the global structure topology between proteins. The alignments by TM-align are obtained from the secondary structure alignment using dynamics programming, gapless threading using TM-score matrix, dynamics programming allowing gap using a combination of secondary structure score and the distance score matrix in the gapless threading alignment, and dynamics programming using TM-score similarity matrix20 Therefore, when the local structures rather than the whole structures are used in TM-align, the quality of the structure alignments is expected to be badly affected by mismatch of sequence continuity and poorly defined secondary structures.
Figure 7.
Performance comparison of G-LoSA with other local structure alignment methods (TM-align-BS and ProBiS). The number of top templates vs. the average RO over the benchmark targets, calculated for the best templates. The best template was chosen among given top N templates in terms of RO.
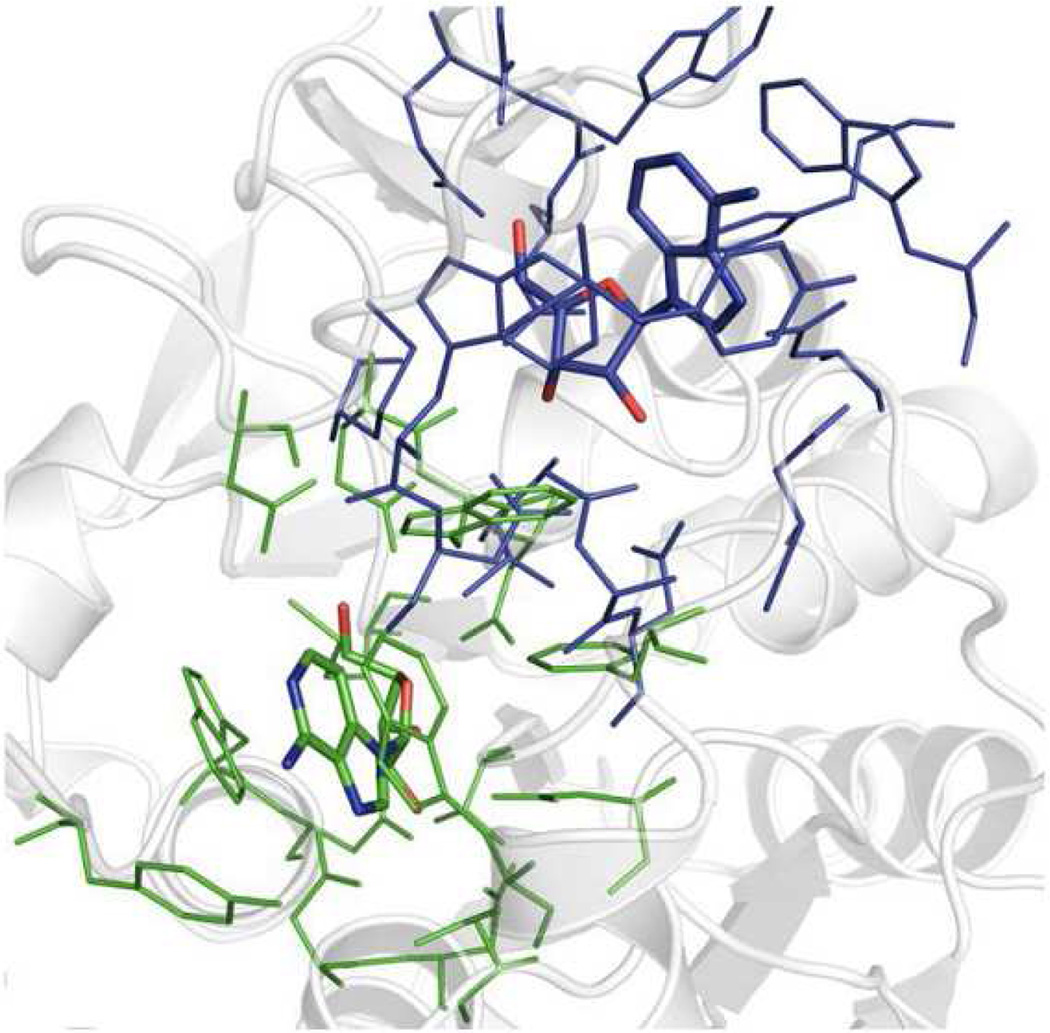
To further evaluate the alignment quality by G-LoSA and TM-align-BS, we carried out two different self-alignment experiments for each benchmark BS-structure: the BS-structure vs. the BS-structure itself, and the BS-structure vs. the whole structure of the corresponding protein. The success rate of these self-alignments was evaluated by RO between the cognate ligands in the superposed pair. If RO = 1, the self-alignment is defined as a success. When the BS-structures were self-aligned onto the BS-structures themselves, both G-LoSA and TM-align yielded 100% success rate over the benchmark targets. However, when the BS-structures were self-aligned onto the whole structures of the corresponding proteins, the success rate of TM-align was only 42%, whereas G-LoSA still showed 100%. As expected, the structure alignment quality by TM-align is determined not only by the geometric similarity between two structures, but also by the sequence and secondary structure continuity. An example of the unsuccessful alignment by TM-align, where RO = 0, is illustrated in Figure 8.
Figure 8.
Representative example (PDB:1HP0) of unsuccessful structure alignment by TM-align-BS between a BS-structure and a whole structure of its corresponding protein.
The local structure alignment between BS-structures consisting of a comparable number of residues can minimize the problem with the alignments by TM-align. In addition, the sequence continuity can be largely conserved between the target-template BS-structures because highly similar local structures likely have similar global folds (Figure 6), resulting in the reliable performance of TM-align-BS. Our results demonstrate that TM-align can be utilized for local structure alignment to identify template ligands, but it is only applicable to pairs of local structures with conserved sequence continuity and geometry. In contrast, G-LoSA uses only geometry to superpose two structures and can be applied to more diverse local structure alignment purposes even with distinct structures in their sizes. A potential application of G-LoSA could be an alignment between the whole structure of a target protein and BS-structures in the structure library to predict the ligand binding site of the target protein. The work in this direction is in progress.
Comparison with ProBiS
G-LoSA is also compared with ProBiS17 that uses a maximal common subgraph search method for detecting a structurally similar region, which is also adopted in G-LoSA. A major difference is that while G-LoSA uses only Cα atoms for structure alignment, ProBiS uses the coordinates of specific groups of atoms (so-called functional groups) belonging to a residue. The functional groups are classified into five physicochemical properties, and structural similarity is determined by the geometry of the functional groups with identical physicochemical properties. Therefore, the ProBiS algorithm is more sophisticated than G-LoSA in that the receptor’s surface atoms rather than its Cα atoms mainly determine a ligand-binding site.
Interestingly, Figure 7 indicates that ProBiS does not outperform G-LoSA for the template ligand search and the simple Cα-based G-LoSA yields reliable performance (also see the next section). Unfortunately, it is not clear why G-LoSA significantly outperforms ProBiS for the benchmark targets. Perhaps, the ProBiS algorithm is optimized for detection of structurally similar protein binding sites rather than detection of template ligands, resulting in missing some good templates. Therefore, we would not emphasize the better performance of G-LoSA over ProBiS in the template ligand search.
Rationale of Cα superposition in G-LoSA for template ligand search
The results in the previous “Comparison with global structure alignment” section have shown that the local alignment by G-LoSA is more effective in identifying template ligands from proteins with distinct folds than the global alignment by TM-align. Here, we further examine the structure alignment performance of G-LoSA against evolutionary non-related proteins.
Figure 9A is the BS-structures of Artocarpus Integer artocarpin (PDB:1VBP) and Pterocarpus Angolensis lectin (PDB:1Q8Q), aligned by G-LoSA. The two proteins bind with an identical ligand, α-D-mannose and the TM-score is 0.36. The figure shows that G-LoSA produced well-aligned ligands (RO = 1.0) as well as the BS-structures. In the superposed structures, the number of aligned Cα-atom pairs was six between the eight residues of the artocarpin BS-structure and the nine residues of the lectin BS-structures. This high conservation of Cα-geometry in their BS-structures enabled G-LoSA to yield high RO despite the distinct folds of the evolutionary non-related proteins.
Figure 9.
The structure alignment performance of G-LoSA against evolutionary non-related proteins. (A) BS-structures of Artocarpus Integer artocarpin (PDB:1VBP) and Pterocarpus Angolensis lectin (PDB:1Q8Q), aligned by G-LoSA. (B) BS-structures of human estrogen receptor α ligand-binding domain (PDB:1A52) and human 17-β-hydroxysteroid-dehydrogenase type 1 (PDB:1FDT), aligned by G-LoSA. (C) The same structures to Figure B, but the structures are superposed using the ligand coordinates. In Figure A and B, the aligned Cα-atoms (inter-Cα distance ≤ 2.5 Å) are represented as spheres.
Figure 9B shows the G-LoSA-aligned structures of human estrogen receptor α ligand-binding domain (PDB:1A52) and human 17-β-hydroxysteroid-dehydrogenase type 1 (PDB:1FDT). Both have the same ligand, estradiol. In this example, their global topology similarity is lower (TM-score = 0.26) than the previous example. Only five pairs of aligned Cα-atoms were found between the BS-structures consisting of 12 residues in each protein, yielding RO = 0.3 and indicating that an accurate overlap between the ligands is hard to be achieved by G-LoSA for this case. To compare the side-chain geometries of the two BS-structures, we superposed the BS-structure/ligand pair only based on the ligand coordinates (Figure 9C). In the superposed structures, highly conserved side-chain geometry between the BS-structures was not observed either. When we used ProBiS to align the BS structures, it did not produce any outputs because significant structural similarity was not detected in the structure pair. These examples suggest that the bad alignment by G-LoSA does not result from its Cα-based approach. In addition, the current local structure alignment methods using atom coordinates may have a problem in aligning the BS-structures with completely different geometries, even though all the surface atoms are explicitly considered for the structure alignment. Nevertheless, there still might be cases in which only side-chains in completely non-related proteins are conserved geometrically, but not Cα atoms. In these cases, G-LoSA would fail to detect the structural similarity.
Evaluation of the computation speed of G-LoSA
To measure computation time by G-LoSA, pairwise alignments using the benchmark BS-structures were executed on a single 2.33 GHz processor. For the structure alignment, each BS-structure was superposed onto the BS-structure itself and the computation times were measured. The structure alignments performed in this way are the cases where the longest time would take in superposing two local structures with the corresponding number of residues. The average computation time over the 76 benchmark structures was 0.18 sec. We expect a better performance through code optimization.
Representative examples of template-based drug design approach
In this section, we present three representative examples from the template ligand search for the benchmark targets in order to illustrate the potential application of G-LoSA to template-based rational drug design. In this study, the BS-structures of the benchmark targets were prepared a priori (based on cognate ligand). Therefore, it should be noted that an additional computational step or experimental evidence is needed to define ligand-binding sites for the practical applications, which is further elaborated in CONCLUDING DISCUSSION.
EXAMPLE 1. Rational design based on a template ligand
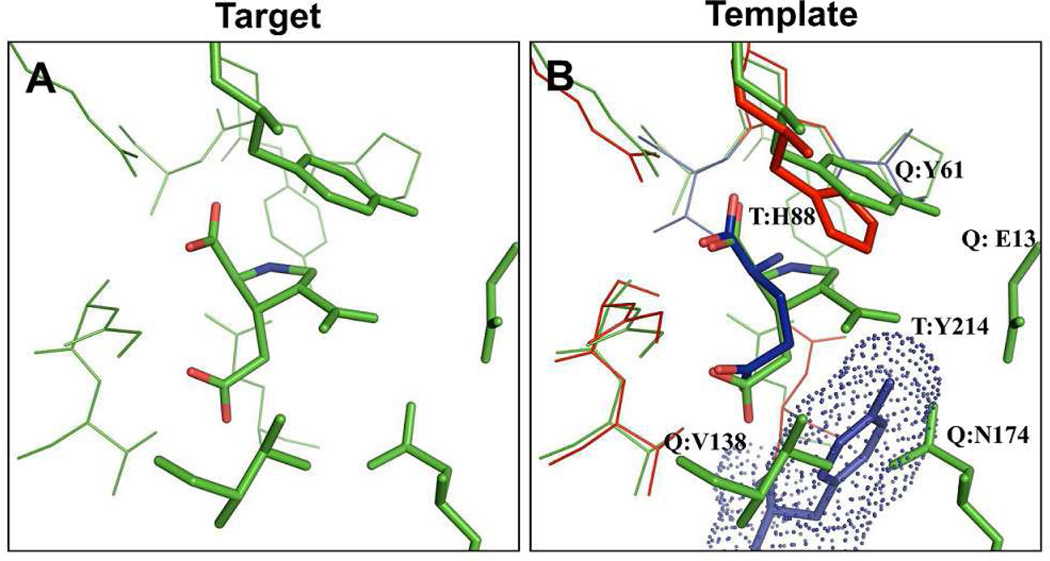
A set of template BS-structure/ligand was identified for the BS-structure of rat glutamate receptor 6 in complex with kainite, a drug isolated from red algae Digenea simplex (PDB:1TT1, Figure 10A). The top-ranked template by the similarity score S was obtained from rat glutamate NMDA receptor subtype 2D in complex with L-glutamate (PDB:3OEN, TM-score: 0.85, Figure 10B). A major structural difference was observed when the template BS structure was compared to the target BS structure (Figure 10B). The residues E13, V138, and N174 consist of the binding site of the target BS-structure. However, in the template BS-structure, a single residue (Y214) at a more inward position replaces these three residues. This finding implies that bulkier structures can be introduced at the template ligand to fill the space, and in particular, a hydrophobic group that can interact with V138 may be an option to design a ligand specific to the target protein. In addition, the target BS-structure Y61, which is substituted by H88 in the template BS-structure, strongly suggests that a scaffold enabling ring-ring interactions with Y61 may also be a design approach to modify the template ligand structure. A ligand designed by such template-based lead optimization strategies should be comparable to the target ligand, kainite (Figure 10A). The overlap ratio RO between the target and template ligands was 0.67.
Figure 10.
Representative example of lead optimization using a single template ligand. (A) Target PDB:1TT1. (B) Template PDB:3OEN. Critical residues determining ligand specificity are shown in stick representation. In (B), T and Q represent “template” and “query” (target), respectively. Template residue Y214 is displayed in dot-and-stick representation. If a template residue has similar physicochemical property to its aligned target residue, the template residue is colored red.
EXAMPLE 2. Fragment assembly using multiple templates
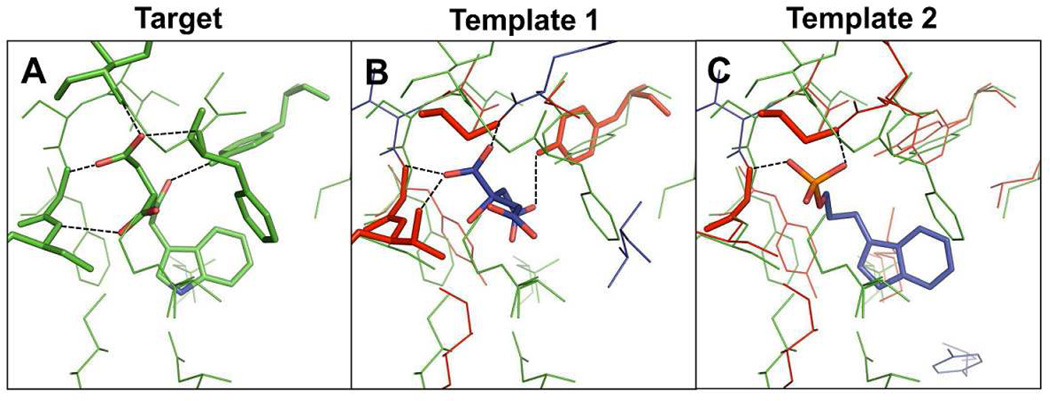
In this example, the BS-structure from S. typhimurium tryptophan synthase α-subunit complexed with indole-3-acetyl aspartic acid (PDB:1K3U, Figure 11A) was used as a query local structure. When a template search was performed against the structure library, the first-ranked template ligand by the score S was citric acid bound in T. thermophilus HB8 tryptophan synthase α-subunit (template 1, PDB:1UJP, TM-score: 0.83, Figure 11B). Nine out of 11 residues in the template BS-structure were aligned with the target residues and eight of the aligned residues had similar physicochemical properties, strongly indicating the identified template ligand may become a critical element for ligand design. However, the target BS-structure consists of 16 residues, and thus the seven residues were not aligned with the template BS residues, showing the target BS-structure were partially overlapped. An additional template ligand was selected from T. thermophilus HB8 tryptophan synthase α-subunit complexed with indole-3-propanol phosphate (template 2, PDB:1WXJ, TM-score: 0.83, Figure 11C). This template was ranked third by the S scores. The acetyl aspartic acid group of the native ligand in Figure 9A is very similar to template ligand 1 and the indole group can be derived from the template ligand 2. The RO of the template ligands 1 and 2 for the target ligand were 0.29 and 0.38, respectively, but the combined overlap ratio became 0.62. This example demonstrates that the fragment assembly of the multiple template ligands may be a promising approach to design a novel ligand.
Figure 11.
Representative example of ligand design by fragment assembly using multiple templates. (A) Target PDB:1K3U. (B) Template 1 PDB: 1UJP. (C) Template 1 PDB: 1WXJ. Residues forming hydrogen bonds with ligands are shown in stick representation. Proteins are colored based on the same scheme in Figure 10.
EXAMPLE 3. Fragment assembly using iterative template search
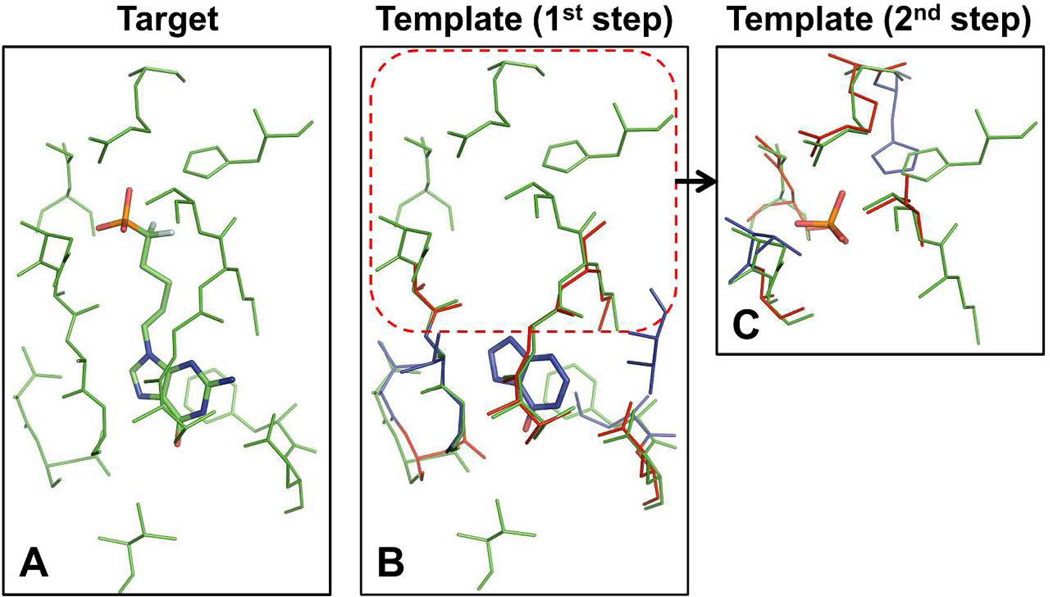
In this example, the target BS-structure was extracted from bovine purine-nucleoside phosphorylase (bPNP) (PDB:1V48, Figure 12A). The template binding site/ligand structures by the best similarity score S was obtained from P. aeruginosa 5´-methylthioinosine phosphorylase in complex with hypoxanthine (template 1, PDB:3OZB, TM-score: 0.74, Figure 12B). The target BS-structure was composed of 17 residues. Among them, 10 target residues were aligned well onto the template residues (RMSD = 0.46 Å), and seven target residues out of the aligned residues had similar physicochemical properties to the template residues. As seen in Figure 12B, no aligned residues were detected at the top region of the target BS-structure. The coordinates of the top region were extracted from the target BS-structure and then the second round of template search against the structure library was implemented. A best template in terms of the score S was obtained from human 5´-methylthioadenosine phosphorylase in complex with phosphate ion (template 2, PDB:1K27, TM-score: 0.77, Figure 12C).
Figure 12.
Representative example of ligand design by fragment assembly using iterative template search. (A) Target PDB:1V48. (B) Template 1 PDB:3OZB. (C) Template 2 PDB:1K27. Proteins are colored based on the same scheme in Figure 10.
The two identified template ligands clearly show structural similarity to the native ligand (Figure 12A). The target receptor bPNP catalyses the reversible phosphorolytic cleavage of the glycosidic bond of 6-oxopurine nucleosides,34 and the native ligand of the target protein is a ground-state analogue inhibitor of trimeric bPNPs. This ligand is the so-called multisubstrate analogue inhibitor, which consists of three structural parts linked together corresponding to a guanine base, a difluoropentyl moiety, and a phosphate35 Each template ligand identified from the first and second round template search is structurally identical to the guanine base and the phosphate of the target ligand, except that the template ligand from the first round search does not have the amine group at C-2. The target ligand has a structure where these two template ligands are connected by a linker. The RO of the template ligands 1 and 2 for the target ligand were 0.46 and 0.18, respectively, but the combined overlap ratio became 0.64. This example indicates that an iterative template ligand search using partial BS-structures may be a promising approach for de novo ligand design.
CONCLUDING DISCUSSION
Structure-based drug design approaches can be broadly divided into two classes: those using receptor coordinates and those using ligand coordinates. If the structural information of a target receptor with active compounds is available, the drug design process can be dramatically facilitated. However, there is no available ligand structural information in most cases of the known drug target structures. Thus, time-consuming costly experiments need to be implemented to identify a lead compound. To tackle this issue, we have developed a computational method to identify template ligands from a library of known protein-ligand complex structures using the BS-structure of the target receptor. The knowledge of 3D structures of template ligands can play a critical role in designing and optimizing compounds that may be highly active to the target protein.
Our study is based on a hypothesis that template ligands can be detected from conserved ligand recognition patterns (i.e., similarity in the BS-structures) between receptor structures. In particular, an efficient local structure alignment tool is necessary to measure BS structural similarity. We have presented G-LoSA, a local structure alignment tool, and its applications to search the protein-ligand PDB library and identify template ligands. The performance of G-LoSA is independent of sequence continuity, secondary structure, and global topology. The benchmark results for top 10 template ligands demonstrate that for more than half of benchmark targets, G-LoSA is able to detect a (single) template ligand (from a non-homologous protein complex) that is highly similar to the target ligand in terms of overall scaffold structure, chemical composition, and binding pose (overlap ratio RO ≥ 0.5). The RO is significantly improved by using multiple template ligands. Highly conserved substructures (or fragments) can be derived from multiple template ligands, even though the template ligand shows low RO (RO < 0.4), suggesting that a fragment assembly approach from the multiple templates can be used to design novel scaffold ligands specific to the target proteins.
The performance comparison with TM-align showed that the local alignment by G-LoSA is more efficient in identifying proper template ligands than the global alignment by TM-align, resulting from the ability of G-LoSA in accurately superposing the local structures regardless of the global protein topology. Unexpectedly, the local alignment by TM-align was comparable to G-LoSA in the template ligand search for the current benchmark targets. Nonetheless, G-LoSA can be utilized for alignments of structures pairs whose sequence and secondary-structure continuity are not conserved. The comparison with ProBiS using all surface functional groups indicates that the Cα-based G-LoSA is reliable for identifying template ligands. Furthermore, the computationally less-expensive G-LoSA is meritorious for large-scale protein-library search.
Template ligands identified by G-LoSA can also be applicable to other computer-aided drug discovery purposes. Template ligands can be used to improve accuracy of molecular docking. To constrain the ligand’s conformation and orientation in the binding site upon molecular docking, CORES36 uses the substructures of a template ligand extracted from 3D structures of a holo-receptor that has a sequence homologous to the target protein. FINDSITELHM 37 superimposes the target ligand onto the consensus substructure identified from weakly related proteins to perform rapid ligand docking. These methods showed reliable performance in predicting ligand pose, demonstrating that template ligand-based docking may be a complementary approach to classical docking methods to improve docking accuracy and speed. Lee and Zhang have developed a template ligand-based blind docking (TLBD) method30 In the TLBD method, a set of template ligands are extracted from library holo-structures based on global structural alignment and then used to measure their shape and chemical feature similarity to target ligands. The TLBD method showed an enhanced ability in prioritizing active compounds compared to docking methods in a large-scale EGFR virtual screening experiment against a modeled protein structure. This result suggests that template ligand-based virtual screening can be a complementary method to receptor-based virtual screening. In the aforementioned methods, the successful identification of good template ligand is a crucial step to yield high performance. Our method should be useful in identifying good template ligands for such applications. In addition, as shown in the three representative examples for template-based ligand design (Figures 10−12), our method can also be applied to protein function prediction because we found common functions shared by proteins in complex with top-ranked template ligands that are evolutionarily remotely related to the target protein.
In this study, the residues consisting of a BS-structure were prepared by applying a 4 Å cutoff from its cognate ligand. In most real problems, however, the BS-residues are not well characterized and thus it may be an issue how the BS-residues are defined. In addition to utilizing available experimental evidences on the BS-residues, this problem could be solved by using various computational approaches for the detection of putative ligand binding sites. The computational approaches include geometry-based methods combined with evolutionary sequence conservation,38,39 geometry-based methods combining with druggability assessment,40 global topology similarity-based method,30 and the threading-based method29 Once putative ligand binding sites are determined, the BS-residues can be easily extracted using the inner shape of the binding pocket.
In the study by Feldman and Labute,19 they developed a local structure alignment tool to cluster a set of BS-structures from different protein families. Although their approach is similar to G-LoSA in that Cα-atom positions are used to detect conserved BS-structures, the processes for identifying aligned pairs from two local structures and the scoring system are significantly different from our approach. Moreover, an ultimate goal of our research is to identify template ligands based on the similarity between BS-structures for de novo ligand design. Since our focus on ligand structure and pose similarity is unique, we expect that our methodology will be useful to the medicinal chemist.
The average overlap ratio RO of the best template among the selected top 10 templates and the top-ranked template by the similarity score S were 0.57 and 0.43 over the benchmark targets (Figure 4A), indicating that the current scoring method has difficulty in perfectly prioritizing the good template ligands. Although the average RO increased as more multiple template ligands were used, the difference between five templates (0.55) and 10 templates (0.57) was not large. It suggests that using only five top templates is an efficient in finding the best template. However, the results also indicate that visual inspection of multiple template BS-structures and a more sophisticated scoring system are needed to effectively identify potential template ligands.
The results in this study open up the potential use of a template-based ligand design method to complement the approaches only using receptor-coordinates. Our method, where the G-LoSA local structure alignment is integrated with the PDB library search, can efficiently detect the template BS-structures similar to a query structure and thus provide reliable template ligands. We believe that the identified template ligands should be useful in designing and optimizing ligand structures specific to a target protein, and it could improve effectiveness of drug discovery research.
Supplementary Material
ACKNOWLEDGMENT
The authors are grateful to Janez Konc and Dusanka Janežič for helpful discussions and suggestions. This work was supported in part by the University of Kansas General Research Fund allocation #2301388-003, NIH U54-GM087519, NIH R01-GM092950, and TeraGrid/XSEDE resources (TG-MCB110072).
Footnotes
Supporting Information. Structure information of the benchmark set and the best template ligand identified by G-LoSA are available in Table S1. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Thornton JM, Todd AE, Milburn D, Borkakoti N, Orengo CA. From structure to function: approaches and limitations. Nat. Struct. Biol. 2000;7991(Suppl):994. doi: 10.1038/80784. [DOI] [PubMed] [Google Scholar]
- 2.Kolodny R, Petrey D, Honig B. Protein structure comparison: implications for the nature of 'fold space', and structure and function prediction. Curr. Opin. Struct. Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiser A. Template-based protein structure modeling. Methods Mol. Biol. 2010;673:73–94. doi: 10.1007/978-1-60761-842-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rother M, Rother K, Puton T, Bujnicki JM. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011;39:4007–4022. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bravo J, Aloy P. Target selection for complex structural genomics. Curr. Opin. Struct. Biol. 2006;16:385–392. doi: 10.1016/j.sbi.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Dodson G, Wlodawer A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 9.Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 10.Gherardini PF, Wass MN, Helmer-Citterich M, Sternberg MJ. Convergent evolution of enzyme active sites is not a rare phenomenon. J. Mol. Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Moodie SL, Mitchell JB, Thornton JM. Protein recognition of adenylate: an example of a fuzzy recognition template. J. Mol. Biol. 1996;263:486–500. doi: 10.1006/jmbi.1996.0591. [DOI] [PubMed] [Google Scholar]
- 12.Denessiouk KA, Rantanen VV, Johnson MS. Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins. Proteins. 2001;44:282–291. doi: 10.1002/prot.1093. [DOI] [PubMed] [Google Scholar]
- 13.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J. Mol. Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold ND, Jackson RM. SitesBase: a database for structure-based protein-ligand binding site comparisons. Nucleic Acids Res. 2006;34:D231–D234. doi: 10.1093/nar/gkj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minai R, Matsuo Y, Onuki H, Hirota H. Method for comparing the structures of protein ligand-binding sites and application for predicting protein-drug interactions. Proteins. 2008;72:367–381. doi: 10.1002/prot.21933. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 17.Konc J, Janežič D. ProBiS algorithm for detection of structurally similar protein binding sites by local structural alignment. Bioinformatics. 2010;26:1160–1168. doi: 10.1093/bioinformatics/btq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeturu K, Chandra N. PocketAlign a novel algorithm for aligning binding sites in protein structures. J. Chem. Inf. Model. 2011;51:1725–1736. doi: 10.1021/ci200132z. [DOI] [PubMed] [Google Scholar]
- 19.Feldman HJ, Labute P. Pocket similarity: are alpha carbons enough? J. Chem. Inf. Model. 2010;50:1466–1475. doi: 10.1021/ci100210c. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konc J, Janežič D. Branch and Bound Algorithm for Matching Protein Structures. Lect. Notes. Comput. Sci. 2007;4432:399–406. [Google Scholar]
- 23.Konc J, Janežič D. An improved branch and bound algorithm for the maximum clique problem. MATCH Commun. Math. Comput. Chem. 2007;58:569–590. [Google Scholar]
- 24.Kabsch W. Solution for the Best Rotation to Relate Two Sets of Vectors. Acta Crystallogr., Sect. A. 1976;32:922–923. [Google Scholar]
- 25.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 26.Konc J, Depolli M, Trobec R, Rozman K, Janežič D. Parallel-ProBiS: Fast parallel algorithm for local structural comparison of protein structures and binding sites. J. Comput. Chem. 2012 doi: 10.1002/jcc.23048. [DOI] [PubMed] [Google Scholar]
- 27.Konc J, Janežič D. ProBiS-2012: web server and web services for detection of structurally similar binding sites in proteins. Nucleic Acids Res. 2012;40:W214–W221. doi: 10.1093/nar/gks435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell RB, Sasieni PD, Sternberg MJ. Supersites within superfolds. Binding site similarity in the absence of homology. J. Mol. Biol. 1998;282:903–918. doi: 10.1006/jmbi.1998.2043. [DOI] [PubMed] [Google Scholar]
- 29.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc. Natl. Acad. Sci. USA. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HS, Zhang Y. BSP-SLIM: a blind low-resolution ligand-protein docking approach using predicted protein structures. Proteins. 2012;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 32.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 33.Kihara D, Skolnick J. The PDB is a covering set of small protein structures. J. Mol. Biol. 2003;334:793–802. doi: 10.1016/j.jmb.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 34.Bzowska A, Kulikowska E, Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Ther. 2000;88:349–425. doi: 10.1016/s0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 35.Luic M, Koellner G, Yokomatsu T, Shibuya S, Bzowska A. Calf spleen purine-nucleoside phosphorylase: crystal structure of the binary complex with a potent multisubstrate analogue inhibitor. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1417–1424. doi: 10.1107/S0907444904013861. [DOI] [PubMed] [Google Scholar]
- 36.Hare BJ, Walters WP, Caron PR, Bemis GW. CORES: an automated method for generating three-dimensional models of protein/ligand complexes. J. Med. Chem. 2004;47:4731–4740. doi: 10.1021/jm0499054. [DOI] [PubMed] [Google Scholar]
- 37.Brylinski M, Skolnick J. FINDSITELHM: a threading-based approach to ligand homology modeling. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000405. e1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang B, Schroeder M. LIGSITECSC: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct. Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000585. e1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halgren TA. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.