Abstract
The common genetic loci that independently influence the risk of type 1 diabetes have largely been determined. Their interactions with age-at-diagnosis of type 1 diabetes, sex, or the major susceptibility locus, HLA class II, remain mostly unexplored. A large collection of more than 14,866 type 1 diabetes samples (6,750 British diabetic individuals and 8,116 affected family samples of European descent) were genotyped at 38 confirmed type 1 diabetes-associated non-HLA regions and used to test for interaction of association with age-at-diagnosis, sex, and HLA class II genotypes using regression models. The alleles that confer susceptibility to type 1 diabetes at interleukin-2 (IL-2), IL2/4q27 (rs2069763) and renalase, FAD-dependent amine oxidase (RNLS)/10q23.31 (rs10509540), were associated with a lower age-at-diagnosis (P = 4.6 × 10−6 and 2.5 × 10−5, respectively). For both loci, individuals carrying the susceptible homozygous genotype were, on average, 7.2 months younger at diagnosis than those carrying the protective homozygous genotypes. In addition to protein tyrosine phosphatase nonreceptor type 22 (PTPN22), evidence of statistical interaction between HLA class II genotypes and rs3087243 at cytotoxic T-lymphocyte antigen 4 (CTLA4)/2q33.2 was obtained (P = 7.90 × 10−5). No evidence of differential risk by sex was obtained at any loci (P ≥ 0.01). Statistical interaction effects can be detected in type 1 diabetes although they provide a relatively small contribution to our understanding of the familial clustering of the disease.
Knowledge of the genetic architecture of type 1 diabetes has increased recently owing to large-scale genome-wide association (GWA) studies (1–3). Estimates of the contributions of the HLA region and numerous non-HLA loci across the genome now account for a sizeable proportion of familial clustering of the disorder (4–6). However, there remains substantial familial clustering that is not explained by the known loci (likely to be in excess of 40%) (4–6). Interactions between risk loci beyond that of a multiplicative model on the odds ratio (OR) scale (or additive on the log odds scale (7)) could account for some of the “missing heritability.” In addition, the existence of differential effects according to age-at-diagnosis and sex remains relatively unexplored.
The HLA region on chromosome 6p21 is the major source of familial clustering in type 1 diabetes (4). HLA-DRB1 and HLA-DQB1 are associated with ORs in excess of 10 for susceptible genotypes (or less than 0.1 for protective genotypes) (8). The risk genotype HLA-DRB1*03/HLA-DRB1*04-HLA-DQB1*0302 (referred to as DR3/DR4-DQ302) with greatest effect has been shown to have the highest frequency in the individuals with youngest onset (9). An age-at-diagnosis interaction has also been reported for HLA-DRB1*04 (10) and the HLA class I alleles HLA-A*24 and HLA-B*39 (11,12).
In contrast, reports of age-at-diagnosis interaction effects at non-HLA loci are contradictory, with positive reports largely confined to studies involving small sample sets (3,13–15). Similarly, reports of gene–gene interaction of type 1 diabetes–associated regions are also mainly conflicting (16–19), we presume due to inadequate sample sizes, with most positive reports likely to be false because the false-discovery rate would be high in these underpowered studies. The only convincing gene–gene interaction reported, is between a major non-HLA locus, protein tyrosine phosphatase nonreceptor type 22 (PTPN22) and DR3/DR4-DQ302 genotypes (20–23).
The incidence of childhood type 1 diabetes is similar in males and females, unlike other autoimmune diseases such as Graves disease, celiac disease, or multiple sclerosis. Despite similar frequencies of childhood type 1 diabetes by sex, there have been reports of genetic risk factors differing between males and females (22,24).
Given that most studies of gene–gene interaction, age-at-diagnosis effects, and sex effects on type 1 diabetes risk have not been addressed in sufficiently well-powered studies, the Type 1 Diabetes Genetics Consortium (T1DGC) has collected more than 16,000 type 1 diabetes–affected samples and tested them for interaction effects with sex and age-at-diagnosis at 38 non-HLA type 1 diabetes–associated regions (Supplementary Table 2). Gene–gene interaction was also tested between HLA class II and the 38 non-HLA loci. With this very large sample set, the study had at least 80% power to detect effects as small as an interaction OR = 1.12 for sex and 1.19 for interactions with age-at-diagnosis or HLA. These calculations assume a multiplicative (log additive) effects model, an OR = 1.15 for association with type 1 diabetes for the test locus and a minor allele frequency of 0.2 and α = 0.0004. In contrast, with 5,000 samples, which is twice as large as any other study testing for interaction effects in type 1 diabetes published to date, the study would only be powered at 80% to detect interaction effects larger than an OR = 1.3 with sex (with the same assumptions as above). For age-at-diagnosis interaction, an OR ≥ 1.37 could be detected; for HLA interaction, an OR ≥ 1.38 could be detected (Supplementary Figs. 1–6).
RESEARCH DESIGN AND METHODS
Subjects.
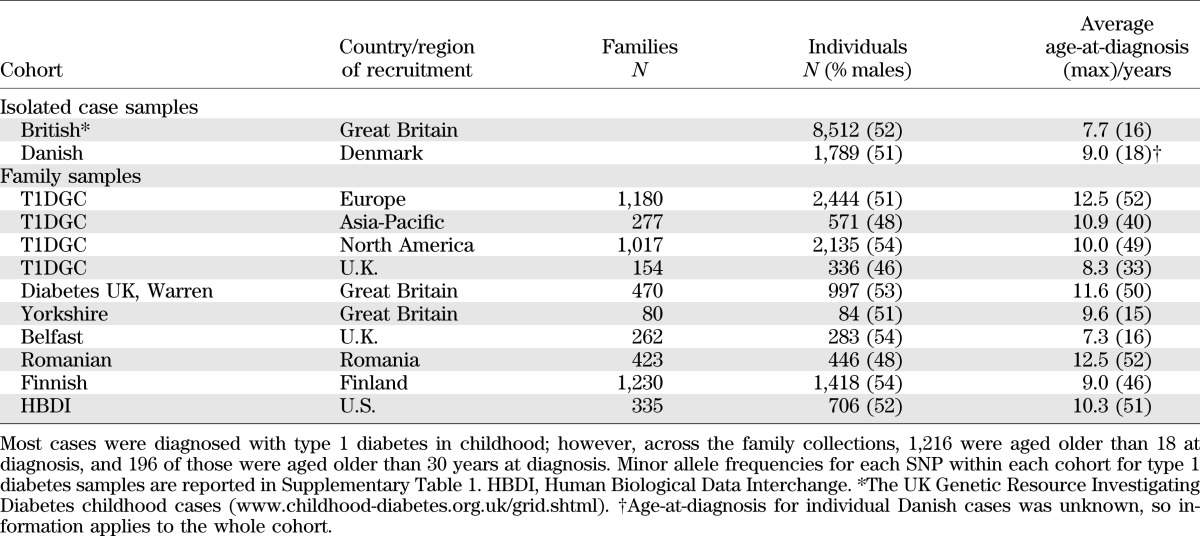
All subjects were of white European ancestry and are described in Table 1.
TABLE 1.
Description of the samples and cohorts genotyped and tested for interactions with age-at-diagnosis, sex, and HLA
Single nucleotide polymorphism genotyping.
Samples were genotyped using TaqMan assays or, where available, existing genotype data from two GWA studies in a subset of the British individuals was used and has been described elsewhere (1,2). All TaqMan genotyping was performed at the University of Cambridge, blinded to disease status, and double-scored. In this sample set, 18% of British case genotypes were common to both TaqMan and single nucleotide polymorphism (SNP) chip platforms. Concordance across genotypes was excellent (99.63%), and therefore, no further samples were double-genotyped. A total of 26.9% of genotypes in the British samples were from SNP chips that used a SNP call rate of 0.95, minor allele frequency of 0.01, and Hardy-Weinberg equilibrium cutoff of P < 5.7 × 10−8. All SNPs tested in the current study were in Hardy-Weinberg equilibrium in unaffected parents (P > 0.01) and controls (P ≥ 0.01).
Statistical methods.
Cases and affected offspring were both used to test for age-at-diagnosis effects in a regression model, with age-at-diagnosis as the outcome variable and genotype as the predictor, stratified by geographic region (cases) and collection (family samples). Robust variance estimates were used to account for nonindependence within families. Sex effects on genotype associations were tested in a similar manner using a logistic regression model with sex as the outcome variable. To test for differences in age-at-diagnosis by sex, age-at-diagnosis was used as the predictor and sex was a dependent variable in a logistic regression model. A genetic risk score was generated from the predictors of a logistic regression model, with disease status as the outcome variable and age-at-diagnosis associated loci (at P < 0.05, including DR3/DR4-DQB1*0302) as independent variables in the case–control data. We defined (statistical gene–gene) interaction as a deviation from a multiplicative interaction of the two test loci on the OR scale, equivalent to an additive model on the log odds scale (7).
To maximize power, interaction between the type 1 diabetes–associated SNPs and the HLA SNPs was tested in cases and affected offspring, which requires the SNPs to be conditionally independent in the general population (25). Two SNPs, rs2187668 and rs7454108, were used to tag the HLA-DRB1*03 (DR3) and HLA-DRB1*04 (not including HLA-DRB1*0403 and 0407; DR4) class II alleles (linkage disequilibrium r2 = 0.99 and 0.77 in cases, respectively). SNP coding was corrected to those of the classical genotypes where HLA-DRB1 classical genotypes were available (13,425 cases and affected offspring). Logistic regression was used to test for nonmultiplicative interaction, with the DR3/DR4-DQ302 genotype as the binary outcome variable, SNP genotype as the dependent variable, and geographic region or collection (for families and Danish cases) included as strata. Robust variance estimates, which relax the assumption of independent observations, were used for all gene–gene interaction tests to allow for nonindependence within families. SNP genotype was also regressed on class II genotypes using a linear regression model adjusted for geographic region or collection. Joint effects of SNP and HLA genotype were estimated using the method of Umbach and Weinberg (25) for case–control data, as detailed in Smyth et al. (20), and the method of Cordell et al. (26) for family data. Combined effects from family and case–control data were computed by the sum of effects (the vector of coefficients from the regression models), weighted by their variances, and divided by the sum of the weights (inverse of the variances). Quanto (http://hydra.usc.edu/GxE/) was used for power calculations. All other statistical analyses were performed in Stata (www.stata.com) or R (www.r-project.org) software. P < 0.0004 was considered significant, which equates to a Bonferroni correction for the number of loci and interactions tested.
RESULTS
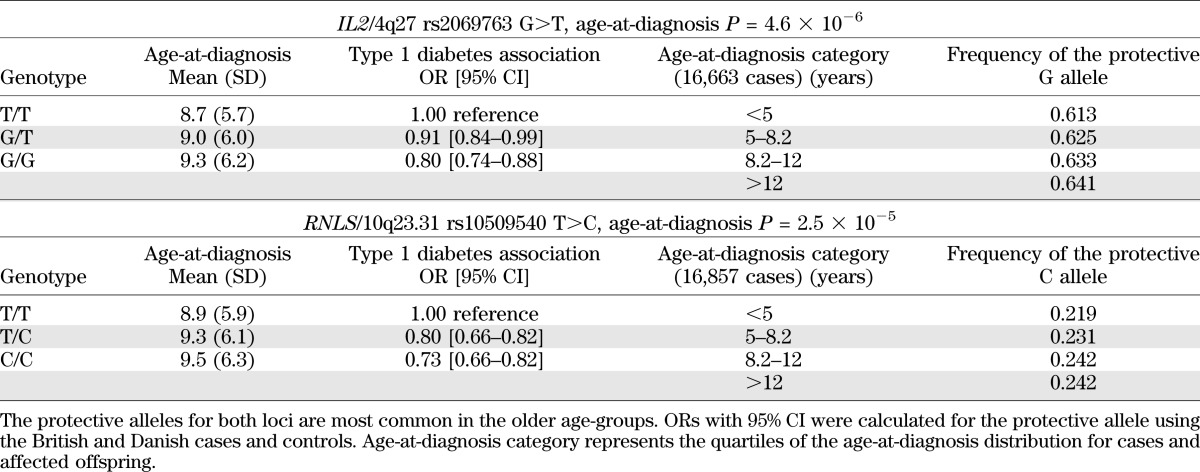
Two of the 38 susceptibility loci tested were associated with age-at-diagnosis, rs2069763 at interleukin 2 (IL2)/4q27 (P = 4.6 × 10−6) and rs10509540 at renalase, FAD-dependent amine oxidase (RNLS)/10q23.31 (P = 2.5 × 10−5; Supplementary Table 2). Consistent with a true biologically plausible age-at-diagnosis effect, the protective allele was more frequent with increasing age-at-diagnosis and the mean age-at-diagnosis increased with the number of protective alleles (Table 2). At rs2069763 (IL2/4q27) the major allele G confers protection from type 1 diabetes (www.t1dbase.org). The average age-at-diagnosis for G/G homozygotes was 9.3 years compared with 8.7 years for T/T homozygotes (7.2 months later). Furthermore, the frequency of the G allele was 2.8% higher in the upper quartile of the age-at-diagnosis distribution (64.1%) compared with the lower quartile (61.3%; Table 2). Similarly, for rs10509540 in RNLS/10q23.31, the minor allele C confers protection from type 1 diabetes (1), with the average age-at-diagnosis 7.2 months later for C/C homozygotes (9.5 years) compared with T/T homozygotes (8.9 years). The frequency of the C allele was also 2.3% higher in the upper quartile (24.2%) compared with the lower quartile (21.9%) of the age-at-diagnosis distribution (Table 2). The age-at-diagnosis effects at RNLS and IL2 were independent of each other (and also of the known DR3/DR4-DQ302 age-at-diagnosis effect), with the average age-at-diagnosis of individuals homozygous for the susceptible T/T genotype at both loci reduced to 8.4 years.
TABLE 2.
Age-at-diagnosis effects at IL2/4q27 and RNLS/10q23.31
No convincing association with sex was obtained at any of the 38 loci (minimum P ≥ 0.01; Supplementary Table 2). However, there was suggestive evidence that females had a lower mean age-at-diagnosis (8.9 years) than males (9.2 years, P = 7.29 × 10−4), consistent with the literature for adult-onset autoimmune type 1 diabetes (10).
HLA-SNP interaction results.
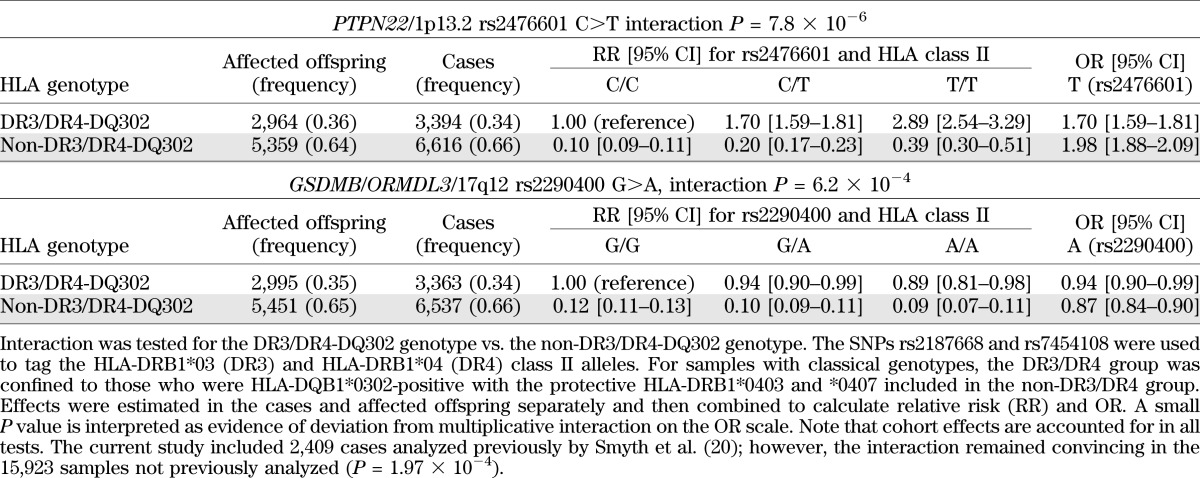
HLA was modeled using the SNPs, rs2187668 and rs7454108, to capture the DR3 and DR4 (not HLA-DRB1*0403 or HLA-DRB1*0407) haplotypes, respectively, in a maximum of 18,548 type 1 diabetes individuals and affected offspring (the minimum number of samples used was 16,336, for rs689 at the insulin [INS] gene; Supplementary Table 2). We obtained evidence of a statistical interaction (deviation from a multiplicative interaction on the OR scale) between the DR3/DR4-DQB1*0302 genotype and the PTPN22 SNP, rs2476601 (P = 7.82 × 10−6). We confirm previous reports (20–23) that the effect of the PTPN22 SNP was to increase susceptibility in those who are DR3/DR4-DQ302–negative compared with those who are DR3/DR4-DQ302–positive. The OR at rs2476601 for the T allele was 1.70 in the DR3/4-DQ302–positive group and 1.98 in the DR3/4-DQ302–negative group (Table 3). Importantly, DR3/DR4-DQ302 individuals remain significantly more susceptible than the non–DR3/DR4-DQ302 individuals, regardless of PTPN22 genotype (Table 3). Some evidence of deviation from multiplicative interaction on the OR scale (although not at P ≤ 4 × 10−4) was also obtained between the 17q12 region, containing the candidate genes gasdermin B (GSDMB) and ORM1-like 3 (orosomucoid 1-like 3, ORMDL3) (rs2290400 G>A) and with the DR3/DR4-DQ302 genotype (P = 6.24 × 10−4). The interaction between HLA and rs2290400 manifested as increased protection in the non–DR3/DR4-DQ302 protective group (OR 0.87 for the A allele at rs2290400) with little association of the allele in the susceptible DR3/DR4-DQ302 group (OR 0.94 for the minor A allele at rs2290400; Table 3). All other regions had P > 0.007 for statistical interaction with the DR3/DR4-DQB1*0302 genotype (Supplementary Table 2).
TABLE 3.
Joint effects of HLA class II genotypes, DR3/DR4-DQ302 and rs2476601 at PTPN22 and rs2290400 at GSDMB/ORMDL3
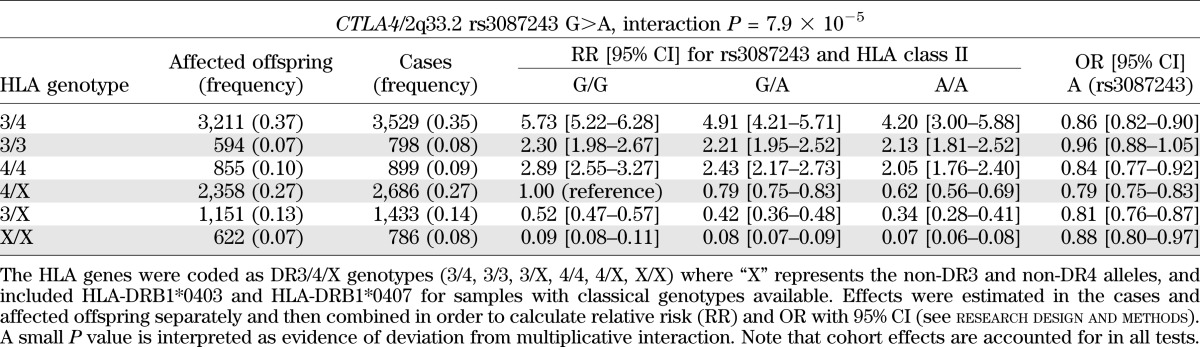
When SNPs were used to encode DR3/4/X genotypes rather than as presence or absence of the DR3/DR4-DQ302 genotype (research design and methods), PTPN22-DR3/4/X deviated from multiplicative interaction on the OR scale (P = 4.87 × 10−4). In addition, there was evidence of nonmultiplicative statistical interaction on the OR scale with the cytotoxic T-lymphocyte antigen 4 (CTLA4) SNP, rs3087243 (P = 7.90 × 10−5). The protective rs3087243 T allele conferred greatest protection in those who were DR4/X and DR3/X, but there was little effect in those with the DR3/DR3 genotype (Table 4). All other regions had P > 0.006.
TABLE 4.
Joint effects of HLA class II genotypes and rs3087243 at CTLA4
DISCUSSION
We report convincing evidence of age-at-diagnosis effects outside of the HLA region in childhood-onset type 1 diabetes. The evidence for genetic effects on age-at-diagnosis involved SNPs in the IL-2 and RNLS genes. The focus here was on childhood-onset individuals, but with an emphasis on adult-onset type 1 diabetes, more loci may yet be identified. Indeed, a recent study of 1,384 individuals with autoimmune diabetes aged between 3 and 89 years at diagnosis also found suggestive evidence of age-at-diagnosis effects at IL2 (P = 0.026) and RNLS (P = 0.033) (10). The same study reported a suggestive association at interleukin 2 receptor α (IL2RA)/10p15.1/ rs2104286 (P = 0.027), a locus that was close to significant in the present sample set (P = 9.8 × 10−4). Regulator of G-protein signaling 1 (RGS1)/1q31.2, GLIS family zinc finger 3 (GLIS3)/9p24.2, approached significance in our study, and a further seven SNPs had P < 0.05 (Supplementary Table 2), but no support for RGS1 or GLIS3 was obtained previously (10).
The type 1 diabetes risk alleles at all 11 loci with P < 0.05 for age-at-diagnosis effects were associated with a younger age-at-diagnosis. The genetic risk score was strongly correlated with age-at-diagnosis, accounting for ∼1% of the variance in age-at-diagnosis (P = 9.2 × 10−18). Stratifying the risk score into quintiles demonstrated a trend toward an earlier age-at-diagnosis with increasing risk: average age-at-diagnosis from lowest risk to highest risk was 8.2, 8.2, 7.8, 7.4, and 7.0 years. Children carrying a higher dose of the earlier age-at-diagnosis alleles will probably have an earlier diagnosis of disease compared with individuals who do not carry these risk alleles, presumably due to a more rapid development of autoimmunity and/or progression from autoimmunity, as detected by being autoantibody-positive (10). Another possibility is that as the immune system matures and ages, alterations in its functions may nullify the effects of certain susceptibility alleles, for example, of the genes in the IL-2 pathway, and so cause these age-at-diagnosis–dependent associations.
The IL-2 pathway is recognized as being important for the pathogenesis of type 1 diabetes. IL2 SNP genotypes are correlated with IL-2 gene expression (J. Yang, J.A.T., unpublished data), supporting IL2 as the causal gene in the chromosome 4q27 region for type 1 diabetes. Two IL-2 receptor genes, IL2RA and IL2RB, are both associated with type 1 diabetes susceptibility (1). In the NOD mouse model, the major non-major histocompatibility complex locus, insulin dependent diabetes susceptibility 3 (Idd3), is the IL2 gene and its effect, via polymorphic gene expression, is age-dependent (27). The RNLS gene encodes renalase, a FAD-dependent amine oxidase secreted by the kidney that circulates in blood and modulates cardiac function and systemic blood pressure, perhaps through its ability to metabolize catecholamines (28). Little is known about its function, if any, in the immune system. Renalase RNA is expressed in monocytes (http://dil.t1dbase.org/page/HaemAtlasView). We have found a correlation between the type 1 diabetes risk SNPs and SNPs associated with RNA levels of the RNLS gene in monocytes, implicating it as the causal gene in the chromosome 10q23.31 region (29).
We tested HLA*non-HLA gene interactions because the HLA class II genes have the largest effects on type 1 diabetes in the genome. Hence, we expect the HLA to have the highest prior probability of showing a nonmultiplicative interaction on the OR scale with a non-HLA locus. The PTPN22-HLA class II genotype interaction is most convincing, probably because the main effect at PTPN22 is large compared with other non-HLA genes in type 1 diabetes with a genotype OR ≥ 3.5. The biological interpretation of a statistical interaction is difficult, but for PTPN22 and HLA class II, one can hypothesize that their coexpression and hence biological interaction in certain cell types critical for type 1 diabetes, such as T lymphocytes and antigen-presenting cells, could contribute to our observed statistical result. The CTLA4*HLA finding requires confirmation in future studies; however, the result is not surprising given the key role of HLA class II molecules and CTLA-4 in autoantigen presentation and autoreactive T-cell activation. Further insights into these molecules and their role in disease require detailed laboratory-based investigations.
Our findings here illustrate that statistical gene–gene interactions can be detected, and we can anticipate that evidence for many more interactions may be found (and those we report confirmed) with larger numbers of samples, and with the use of non-European samples. However, in keeping with another report (4), our data suggest that for common variants with ORs < 2, statistical interactions are unlikely to contribute substantially to the “missing heritability” in type 1 diabetes.
ACKNOWLEDGMENTS
This research uses resources provided by the T1DGC, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development, and Juvenile Diabetes Research Foundation International (JDRF) and is supported by U01 DK-062418. This work was funded by the JDRF, the Wellcome Trust (WT), under award 076113, and by the National Institute for Health Research Cambridge Biomedical Centre. The Cambridge Institute for Medical Research (CIMR) is in receipt of a WT Strategic Award (079895). T1DGC supplied samples.
No potential conflicts of interest relevant to this article were reported.
J.M.M.H. analyzed and interpreted the data, and wrote, reviewed, and edited the manuscript. J.D.C. researched data and reviewed and edited the manuscript. D.J.S. performed TaqMan genotyping and reviewed and edited the manuscript. N.M.W. managed the data and reviewed and edited the manuscript. H.S. managed DNA samples and reviewed and edited the manuscript. J.-X.S., G.S.E., M.R., B.A., P.C., H.A.E., C.J., G.M., J.N., C.N., and S.S.R. reviewed and edited the manuscript. J.A.T. interpreted the data, and wrote, reviewed, and edited the manuscript. F.P. contributed Danish DNA samples and reviewed and edited the manuscript. J.M.M.H. is the guarantor of this work and as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank the participation of all the patients, control subjects, and family members. The authors thank David Dunger and Barry Widmer of the University of Cambridge, and the British Society for Paediatric Endocrinology and Diabetes for the type 1 diabetes case collection. The authors acknowledge use of DNA from the Human Biological Data Interchange and Diabetes UK for the USA and UK multiplex families, respectively, David Savage of the Belfast Health and Social Care Trust, Chris Patterson and Dennis Carson of Queen's University Belfast, and Peter Maxwell of Belfast City Hospital for the Northern Irish families, the Genetics of Type 1 Diabetes in Finland (GET1FIN, Jaakko Tuomilehto, Leena Kinnunen, Eva Tuomilehto-Wolf, Valma Harjutsalo, and Timmo Valle of the National Public Health Institute, Helsinki) for the Finnish families, and Christian Guja and Constantin Ionescu-Tirgoviste of the Institute of Diabetes “N Paulescu,” Romania for the Romanian families. Danish subjects were from the Danish Society of Childhood Diabetes (DSBD) and from the Inter99 study cohort. This study makes use of data generated by the WT Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk/. The authors also thank Pamela Clarke, Gillian Coleman, Simon Duley, Deborah Harrison, Steve Hawkins, Meeta Maisuria, Trupti Mistry, and Niall Taylor from the JDRF/WT Diabetes and Inflammation Laboratory, University of Cambridge, for preparation of DNA samples, and Heather Cordell from Newcastle University and David Clayton from the JDRF/WT Diabetes and Inflammation Laboratory for useful discussions.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1694/-/DC1.
A full list of members appears in the Supplementary Data online.
REFERENCES
- 1.Barrett JC, Clayton DG, Concannon P, et al. Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd JA, Walker NM, Cooper JD, et al. Genetics of Type 1 Diabetes in Finland. Wellcome Trust Case Control Consortium Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007;39:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet 2009;5:e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet 2011;88:294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol 2011;35:310–317 [DOI] [PubMed] [Google Scholar]
- 7.Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet 2002;11:2463–2468 [DOI] [PubMed] [Google Scholar]
- 8.Howson JM, Stevens H, Smyth DJ, et al. Evidence that HLA class I and II associations with type 1 diabetes, autoantibodies to GAD and autoantibodies to IA-2, are distinct. Diabetes 2011;60:2635–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caillat-Zucman S, Garchon HJ, Timsit J, et al. Age-dependent HLA genetic heterogeneity of type 1 insulin-dependent diabetes mellitus. J Clin Invest 1992;90:2242–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howson JM, Rosinger S, Smyth DJ, Boehm BO, Todd JA, ADBW-END Study Group Genetic analysis of adult-onset autoimmune diabetes. Diabetes 2011;60:2645–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, Howson JM, Walker NM, et al. Wellcome Trust Case Control Consortium Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 2007;450:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howson JM, Walker NM, Clayton D, Todd JA, Type 1 Diabetes Genetics Consortium Confirmation of HLA class II independent type 1 diabetes associations in the major histocompatibility complex including HLA-B and HLA-A. Diabetes Obes Metab 2009;11(Suppl. 1):31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espino-Paisan L, de la Calle H, Fernández-Arquero M, et al. Polymorphisms in chromosome region 12q13 and their influence on age at onset of type 1 diabetes. Diabetologia 2011;54:2033–2037 [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Jin Y, Reddy MV, et al. Genetically dependent ERBB3 expression modulates antigen presenting cell function and type 1 diabetes risk. PLoS ONE 2010;5:e11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espino-Paisan L, de la Calle H, Fernández-Arquero M, et al. A polymorphism in PTPN2 gene is associated with an earlier onset of type 1 diabetes. Immunogenetics 2011;63:255–258 [DOI] [PubMed] [Google Scholar]
- 16.Payne F, Cooper JD, Walker NM, et al. Interaction analysis of the CBLB and CTLA4 genes in type 1 diabetes. J Leukoc Biol 2007;81:581–583 [DOI] [PubMed] [Google Scholar]
- 17.Bergholdt R, Taxvig C, Eising S, Nerup J, Pociot F. CBLB variants in type 1 diabetes and their genetic interaction with CTLA4. J Leukoc Biol 2005;77:579–585 [DOI] [PubMed] [Google Scholar]
- 18.Maier LM, Chapman J, Howson JM, et al. No evidence of association or interaction between the IL4RA, IL4, and IL13 genes in type 1 diabetes. Am J Hum Genet 2005;76:517–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bugawan TL, Mirel DB, Valdes AM, Panelo A, Pozzilli P, Erlich HA. Association and interaction of the IL4R, IL4, and IL13 loci with type 1 diabetes among Filipinos. Am J Hum Genet 2003;72:1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth DJ, Cooper JD, Howson JM, et al. PTPN22 Trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with HLA class II genotypes. Diabetes 2008;57:1730–1737 [DOI] [PubMed] [Google Scholar]
- 21.Bjørnvold M, Undlien DE, Joner G, et al. Joint effects of HLA, INS, PTPN22 and CTLA4 genes on the risk of type 1 diabetes. Diabetologia 2008;51:589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermann R, Lipponen K, Kiviniemi M, et al. Lymphoid tyrosine phosphatase (LYP/PTPN22) Arg620Trp variant regulates insulin autoimmunity and progression to type 1 diabetes. Diabetologia 2006;49:1198–1208 [DOI] [PubMed] [Google Scholar]
- 23.Steck AK, Liu SY, McFann K, et al. Association of the PTPN22/LYP gene with type 1 diabetes. Pediatr Diabetes 2006;7:274–278 [DOI] [PubMed] [Google Scholar]
- 24.Nielsen C, Hansen D, Husby S, Lillevang ST. Sex-specific association of the human PTPN22 1858T-allele with type 1 diabetes. Int J Immunogenet 2007;34:469–473 [DOI] [PubMed] [Google Scholar]
- 25.Umbach DM, Weinberg CR. Designing and analysing case-control studies to exploit independence of genotype and exposure. Stat Med 1997;16:1731–1743 [DOI] [PubMed] [Google Scholar]
- 26.Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol 2004;26:167–185 [DOI] [PubMed] [Google Scholar]
- 27.Todd JA, Aitman TJ, Cornall RJ, et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature 1991;351:542–547 [DOI] [PubMed] [Google Scholar]
- 28.Malyszko J, Zbroch E, Malyszko JS, Koc-Zorawska E, Mysliwiec M. Renalase, a novel regulator of blood pressure, is predicted by kidney function in renal transplant recipients. Transplant Proc 2011;43:3004–3007 [DOI] [PubMed] [Google Scholar]
- 29.Wallace C, Rotival M, Cooper JD, et al. The Cardiogenics Consortium Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum Mol Genet 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]