Abstract
We investigated the effects of obesity surgery-induced weight loss on transcription factor 7-like 2 gene (TCF7L2) alternative splicing in adipose tissue and liver. Furthermore, we determined the association of TCF7L2 splicing with the levels of plasma glucose and serum free fatty acids (FFAs) in three independent studies (n = 216). Expression of the short mRNA variant, lacking exons 12, 13, and 13a, decreased after weight loss in subcutaneous fat (n = 46) and liver (n = 11) and was more common in subcutaneous fat of subjects with type 2 diabetes than in subjects with normal glucose tolerance in obese individuals (n = 54) and a population-based sample (n = 49). Additionally, there was a positive correlation between this variant and the level of fasting glucose in nondiabetic individuals (n = 113). This association between TCF7L2 splicing and plasma glucose was independent of the TCF7L2 genotype. Finally, this variant was associated with high levels of serum FFAs during hyperinsulinemia, suggesting impaired insulin action in adipose tissue, whereas no association with insulin secretion or insulin-stimulated whole-body glucose uptake was observed. Our study shows that the short TCF7L2 mRNA variant in subcutaneous fat is regulated by weight loss and is associated with hyperglycemia and impaired insulin action in adipose tissue.
Variants in the transcription factor 7-like 2 gene (TCF7L2) increase the risk of type 2 diabetes by 30–40% (1). However, the molecular mechanisms by which intronic variations in TCF7L2 induce hyperglycemia remain unknown. TCF7L2 single nucleotide polymorphisms (SNPs) may regulate open chromatin (2) and transcription of TCF7L2 (3). However, increased mRNA expression of TCF7L2 in islets from individuals with type 2 diabetes suggests that impaired transcription is not the primary mechanism for increased diabetes risk (3,4). Intronic SNPs can regulate mRNA splicing, as shown with monogenic diseases (5). Thus, TCF7L2 SNPs could lead to impaired TCF7L2 function and increased diabetes risk by changing mRNA variant distribution (6–10). Alternatively, TCF7L2 splicing could be altered by obesity and hyperinsulinemia, as we have recently shown reduced expression of several mRNA processing genes regulating alternative splicing in obese individuals (11).
Human TCF7L2 consists of 18 exons and is subject to extensive alternative splicing. Alternative splicing of exons 12, 13, 13a, and 13b can lead to stop codons in exons 13a, 13b, or 14 and result in proteins with short, medium, or long reading frames (12). Only the protein encoded by the full exon 14 has binding sites for COOH-terminal protein binding protein, an inhibitor of the Wnt signaling pathway (13–15), and only variants including exon 13 encode a CRARF domain, which is needed for potent activation of the Wnt signaling cascade (14,16). Thus, changes in TCF7L2 splicing modify Wnt signaling and could alter diabetes risk.
We investigated the effect of weight loss induced by obesity surgery on the splicing of TCF7L2. We specifically focused on splicing variants that lead to impaired Wnt signaling (9,10) and therefore could contribute to increased adiposity. More specifically, we investigated tissue distribution of known TCF7L2 splice variants in subcutaneous and visceral fat tissue and in the liver and an impact of weight loss on TCF7L2 splicing in 95 individuals who had underwent obesity surgery. Finally, an association of adipose tissue TCF7L2 splice variants with the levels of plasma glucose and serum free fatty acids (FFAs) in three independent studies was investigated (n = 216). Our results demonstrate that a short 3′ end splice variant of TCF7L2 is regulated by weight loss and associates with glucose and fatty acid metabolism.
RESEARCH DESIGN AND METHODS
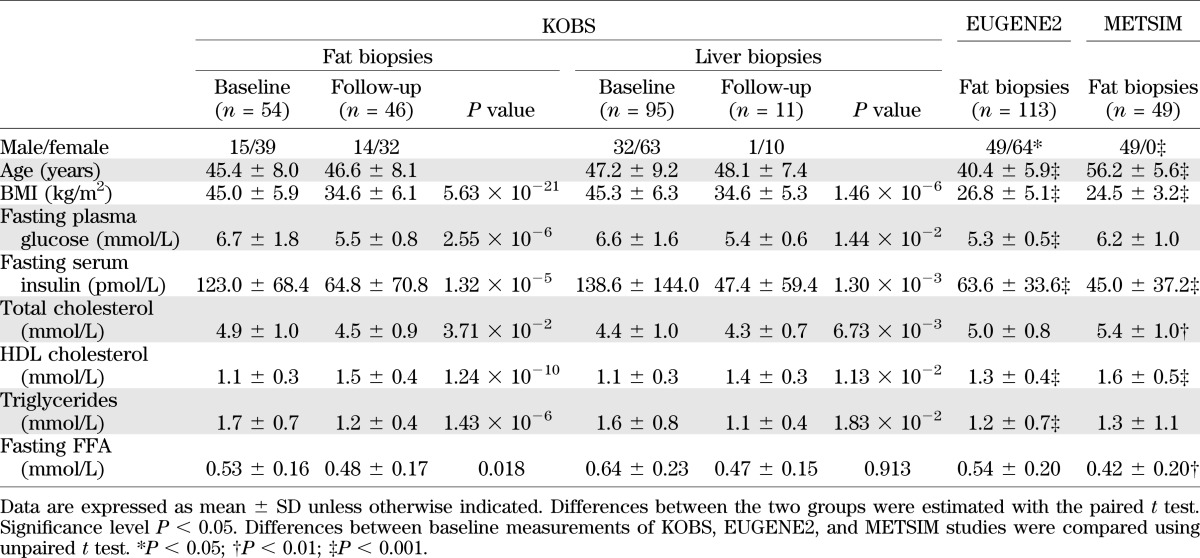
A total of 95 morbidly obese individuals participated in the ongoing Kuopio Obesity Surgery Study (KOBS) (17). All subjects undergoing the Roux-en-Y gastric bypass surgery at Kuopio University Hospital were recruited to this study (>90% participated). Liver, subcutaneous, and visceral adipose tissue biopsies were collected during the operation. Subcutaneous fat and liver biopsies were taken a year after surgery. Clinical parameters were assessed prior to the surgery (at baseline) and at 1-year postsurgery (follow-up).
Two independent study groups were used for replication of the results related to mRNA variants in subcutaneous adipose tissue. A total of 49 men (22 with type 2 diabetes and 27 with normal glucose tolerance) were included from the population-based Metabolic Syndrome in Men (METSIM) study (18), and 113 nondiabetic individuals were included from the European Network on Functional Genomics of Type 2 Diabetes (EUGENE2) study (19). Baseline characteristics of the study groups are shown in Table 1.
TABLE 1.
Characteristics of the study groups
The study protocol was approved by the Ethics Committee of Northern Savo Hospital District and carried out in the accordance with Helsinki Declaration. Informed written consent was obtained from all participants.
Analytical methods used in human studies.
Plasma glucose, insulin, and serum lipids and lipoproteins (total cholesterol, HDL cholesterol, and triglycerides) and FFAs were measured from fasting venous blood samples. Plasma glucose was measured by enzymatic hexokinase photometric assay (Konelab Systems Reagents; Thermo Fischer Scientific, Vantaa, Finland). Plasma insulin was determined by immunoassay (ADVIA Centaur Insulin IRI, number 02230141; Siemens Medical Solutions Diagnostics, Tarrytown, NY). An oral glucose tolerance test (OGTT) was carried after a 12-h fasting period. Serum FFAs were assayed with an enzymatic colorimetric method (Wako NEFA C test kit; Wako Chemicals, Neuss, Germany). Glucose, insulin, and FFA levels were determined at 0, 30, and 120 min in an OGTT.
Insulin secretion and insulin action.
In the EUGENE2 study, an intravenous glucose tolerance test (IVGTT) and hyperinsulinemic euglycemic clamp were performed after a 12-h fast. In order to determine the first-phase insulin release, an IVGTT was performed. A bolus of glucose (300 mg/kg in a 50% solution) was given within 30 s into the antecubital vein. Samples for the measurement of blood glucose and plasma insulin were drawn at −5, 0, 2, 4, 6, 8, 10, 20, 30, 40, 50, and 60 min. Whole-body insulin sensitivity was assessed with the euglycemic hyperinsulinemic clamp technique (insulin infusion rate of 40 mU ×min−1 × m−2 body surface area), as described (20). Blood glucose was clamped at 5.0 mmol/L for the next 120 min by infusion of 20% glucose at various rates according to blood glucose measurements made every 5 min. The mean amount of glucose infused during the last hour was used to calculate the rates of whole-body glucose uptake. Adipose tissue insulin sensitivity was estimated from the levels of serum FFAs during the clamp.
Cell-culture experiments.
Human Simpson-Golabi-Behmel syndrome (SGBS) preadipocytes, which are characterized by a high capacity for adipogenic differentiation (21), were morphologically differentiated into mature adipocytes. Preadipocytes were cultured in Dulbecco’s modified Eagle’s medium/F12 Nut mix supplemented with 33 μM biotin, 17 μM pantothenate, 1% penicillin/streptomycin, and 10% FBS until reaching confluence. To induce differentiation, preadipocytes at day 0 were washed three times with PBS and cultured in serum-free 3FC medium (Dulbecco’s modified Eagle’s medium/F12 nut mix, 33 μM biotin, 17 μM pantothenate, 1% penicillin/streptomycin, 0.01 mg/mL transferrin, 20 nmol/L insulin, 0.1 μM hydrocortisone, and 0.2 nmol/L T3) with 25 nmol/L dexamethasone, 0.5 μM isobutylmethylxanthine, and 2 μM rosiglitazone. At day 4 of postdifferentiation, the medium was changed to 3FC medium with 25 nmol/L dexamethasone and 0.5 μM isobutylmethylxanthine. At day 7 of postdifferentiation, the medium was changed to 3FC medium, and after that, the medium was replenished twice a week with 3FC medium.
RNA isolation, reverse transcription, and PCR.
Total RNA was extracted and purified using the miRNeasy Mini Kit (Qiagen, Hilden, Germany). Total RNA was converted to cDNA with random primers using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. PCR reaction was performed using DyNAzyme II Hot Start DNA Polymerase (Finnzymes, Espoo, Finland) and a suitable primer pair flanking an alternatively spliced region resulting in PCR products that vary in size depending on splicing. The primers used in this study are shown in Supplementary Table 1.
Analysis of alternative splicing and sequencing.
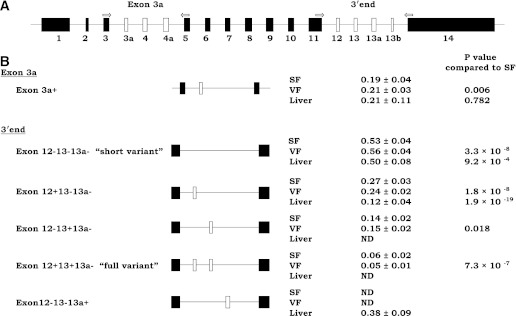
In order to determine the relative ratio of gene splice variants, we implemented the PCR-capillary electrophoresis method for efficient screening of published alternatively spliced exons 3a, 12, 13, 13a, and 13b (6–8,22). Our method, based on a single PCR (for exon 3a and exons in the 3′ end separately), was optimized to give reliable information on the isoform distribution and on changes of the isoform distribution due to alternative splicing. We were primarily not interested in absolute quantification of the variants. The cDNA equivalent of 2 ng total RNA was amplified by PCR in a thermocycler (ABI PRISM 2720; Applied Biosystems) and run on the ABI 3100 DNA Genetic Analyzer (Applied Biosystems). The location of the primers spanning the regions around alternatively spliced exons 3a, 4, and 4a and exons 12, 13, 13a, and 13b is shown in Fig. 1A (6–8,22). The results were analyzed using Peak Scanner Software v1.0 (Applied Biosystems). Peak areas were used to calculate the relative quantities of PCR products. PCR products were initially gel-purified and sequenced in order to determine exon incorporation and ensure the specificity of amplification using BigDye Terminator v1.1 (Applied Biosystems).
FIG. 1.
Distribution of TCF7L2 splice variants in subcutaneous fat (SF), visceral fat (VF), and liver. A: Structure of the TCF7L2 gene. Exon lengths are drawn approximately to scale. Arrows indicate the position and orientation of the primer pairs used in capillary electrophoresis. Examined exons that are subject to alternative splicing are white. B: Schematic overview and relative expression level (mean ± SD) of the existing TCF7L2 mRNA variants at baseline. The + and − in variant names indicate existence of this exon.
Total gene expression analysis.
Quantitative RT-PCRs were conducted in a 7500 Real-Time PCR System (Applied Biosystems) using cDNA as template (equivalent of 3 ng total RNA) and gene-specific primers localized in the exon 4–exon 5 junction and in the exon 5 that were not subject to alternative splicing. PCR products were detected using SYBR Green (KAPA SYBR FAST qPCR Kit; Kapa Biosystems, Woburn, MA). Gene expression was normalized to RPLP0 (ribosomal protein, large, P0). Primers for SYBR Green PCR were designed using Primer3 v. 0.4.0. Primer information is presented in Supplementary Table 1.
Statistical analysis.
Data analyses were conducted with the SPSS/Win programs (SPSS Inc., Chicago, IL). Data before and after surgery were compared with paired t tests and between study groups with unpaired two-tailed t tests. Correlations were assessed with nonparametric Spearman correlations. A P value <0.05 was considered statistically significant. Insulin and FFA levels were logarithmically transformed to obtain normal distribution. The main effects of BMI, genotype, fasting glucose level, fasting insulin level, sex, age, and study group on splicing pattern was evaluated by general linear model. Descriptive statistics are presented as mean ± SDs.
RESULTS
Clinical characteristics of obese subjects.
Ninety-five individuals participated in the KOBS study and were examined after a 1-year follow-up. At 12 months, the subjects had lost 24 ± 9% of their initial weight (P = 4 ×10−37). Significant improvement in the levels of fasting glucose and insulin was also observed (Table 1).
Distribution of splicing variants in adipose tissue and liver.
We implemented the PCR-capillary electrophoresis method to investigate distribution of TCF7L2 splicing variants. Distribution of splicing variants in adipose tissue and liver was concordant with previously published data (Fig. 1B) (7,8). Exon 13 was predominant in subcutaneous and visceral adipose tissue, whereas exon 13a was detected only in the liver. Two common mRNA splice variants were found in the middle of the gene (with and without exon 3a) and five common mRNA splice variants at the 3′ end, as expected (7,8). The following variants in the 3′ end were observed: the short Ex12−13−13a− mRNA spice variant (negative for exons 12, 13, and 13a; GenBank accession numbers FJ010171 and FJ010172), variant Ex12+13−13a− (GenBank FJ010170), variant Ex12−13+13a− (GenBank FJ010174), the full adipose tissue Ex12+13+13a− mRNA variant (GenBank FJ010173), and the liver-specific variant Ex12−13−13a+ (GenBank FJ010167). We also detected additional rare splice variants, a variant excluding exon 12 and including exons 13 and 13a (Ex12−13+13a+), and a variant positive for exons 12, 13, and 13a (Ex12+13+13a+) in the adipose tissue as well as the variants Ex12−13+13a+ and Ex12+13−13a+ in the liver. The proportion of these variants was very low (≤2%), and therefore, they were excluded from statistical analysis.
Change in the splicing pattern in response to obesity surgery.
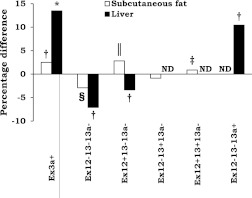
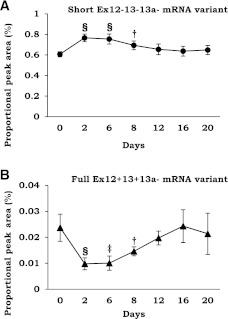
Next, we analyzed whether distribution of mRNA variants changes in response to obesity surgery (Fig. 2) using PCR-capillary electrophoresis. No change in total TCF7L2 expression was observed using quantitative PCR. The relative proportion of the short Ex12−13−13a− mRNA variant, related to impaired activation of Wnt signaling (9,10), was reduced by 3% in subcutaneous fat (P = 1 × 10−4) and by 7% in the liver (P = 0.008). Reciprocal increases of 1 and 3% were observed in the proportion of the full adipose tissue-specific Ex12+13+13a− mRNA variant (P = 0.001) and in the Ex12+13−13a− variant (P = 3 × 10−6) in subcutaneous fat, respectively. In the liver, an 11% increase in the relative amount of liver-specific variant Ex12−13−13a+ in response to obesity surgery was seen (P = 0.005). We also observed an increase in Ex3a+ variant both in subcutaneous fat (3%; P = 0.004) and in the liver (14%; P = 0.012). Because Ex3a+ was not associated with glucose and FFA levels (Supplementary Table 2), we concentrated on the short 3′ end variant in the following analyses.
FIG. 2.
Change in expression level of TCF7L2 splice variants during the year after gastric bypass surgery in subcutaneous fat and liver. *P < 0.05; †P < 0.01; ‡P < 0.001; §P < 1 × 10−4; ‖P < 1 × 10−5, paired t test. ND, not detected.
The short Ex12−13−13a− mRNA variant associates with high levels of plasma glucose.
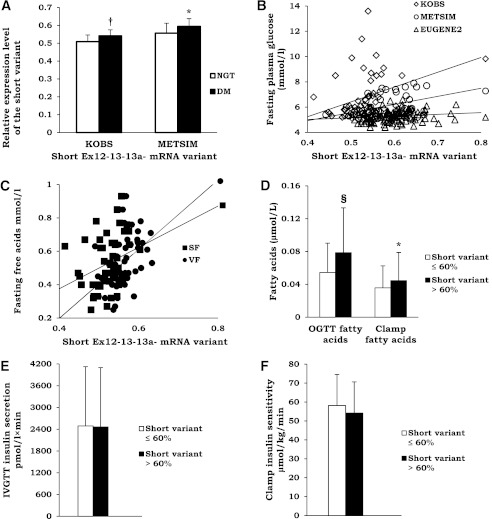
In pooled data of 54 participants from the KOBS study, 49 men from the population-based METSIM study and 113 nondiabetic subjects from the EUGENE2 study (n = 216), the short Ex12−13−13a− mRNA variant was associated with high BMI (P = 0.003), high glucose level (P = 0.008), and the study group (participants of the METSIM and EUGENE2 studies were leaner than participants of the KOBS study; Table 1), but not with fasting insulin level, sex, age, and the rs7903146 genotype (Table 2). We also detected higher levels of this mRNA variant in subcutaneous adipose tissue (P = 0.002) and liver (P = 0.045) in individuals with type 2 diabetes compared with nondiabetic individuals in the KOBS study (Fig. 3A). Similarly, a higher adipose tissue expression of the short Ex12−13−13a− mRNA variant was observed in patients with type 2 diabetes than in normoglycemic individuals in the METSIM study (P = 0.031). Finally, there was a positive correlation between fasting glucose and proportion of the short Ex12−13−13a− mRNA variant in nondiabetic individuals (r = 0.218; P = 0.026, EUGENE2 study) (Fig. 3B and Supplementary Table 2).
TABLE 2.
Multivariate analysis investigating determinants of the subcutaneous adipose tissue expression of Ex3a+ and Ex12−13−13a− mRNA variants
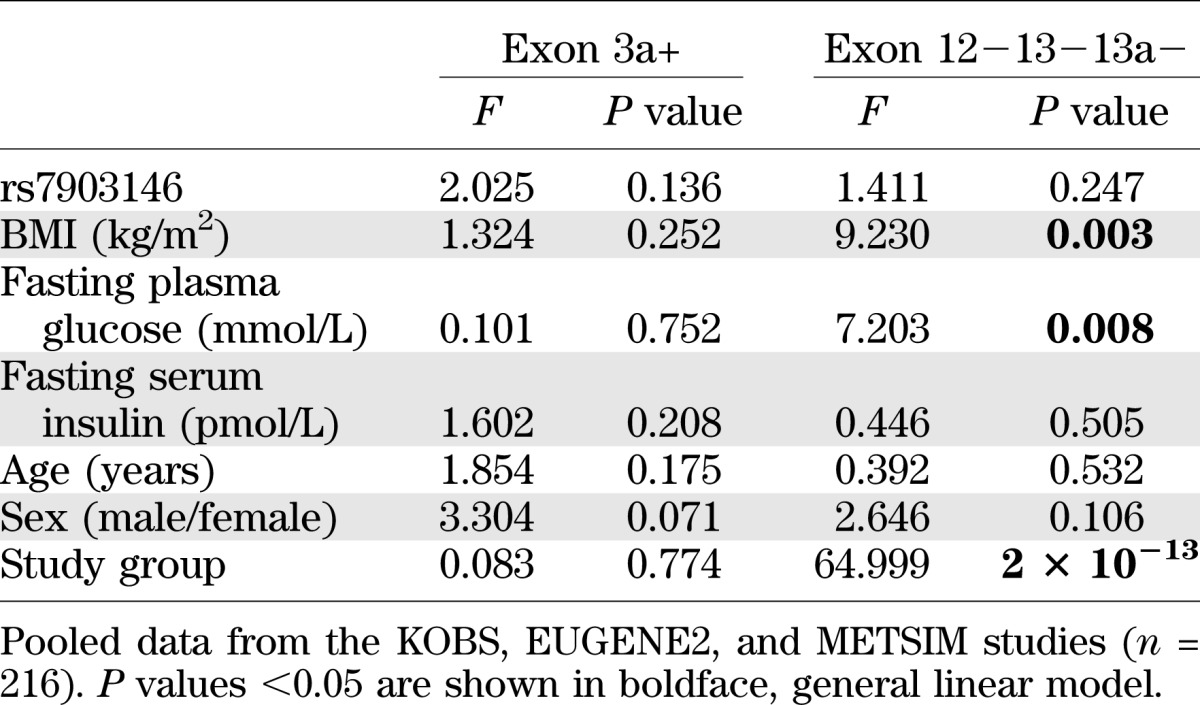
FIG. 3.
A: Prevalence of the short TCF7L2 variant (see Fig. 1) in subcutaneous fat in individuals with and without type 2 diabetes in the KOBS and METSIM studies. White bars, normal glucose tolerance (NGT) subjects; black bars, type 2 diabetes (DM) subjects. Mean ± SD shown. *P < 0.05; †P < 0.01. B: Scatter plot demonstrating correlation between the short mRNA TCF7L2 variant expression (see Fig. 1) and fasting plasma glucose in the KOBS, EUGENE2, and METSIM studies in subcutaneous fat. White diamonds, KOBS study; white circles, METSIM study; white triangles, EUGENE2 study (KOBS, r = 0.335, P = 0.016; METSIM, r = 0.382, P = 0.011; EUGENE2, r = 0.218, P = 0.026). C: Scatter plot demonstrating correlation between the expression of the short TCF7L2 variant in subcutaneous fat (SF; black squares) (r = 0.350; P = 0.014) and visceral fat (VF; black circles) (r = 0.581; P = 1.8 × 10−5) and fasting plasma glucose and serum FFAs in the KOBS study. D: Insulin action in adipose tissue measured as serum FFA levels at 120 min after an oral glucose load (OGTT) (n = 216, all studies combined) or as serum FFA levels at the end of 40 mU euglycemic clamp (n = 113, EUGENE2 study) in individuals with low (white bars) or high (black bars) prevalence of the TCF7L2 short mRNA variant (divided by the median value). Mean ± SD shown. *P < 0.05; §P < 1 × 10−4. Insulin secretion measured during IVGTT (E) and insulin sensitivity measured as whole-body glucose uptake during the hyperinsulinemic euglycemic clamp (F) in the EUGENE2 study in individuals with low (white bars) or high (black bars) prevalence of the TCF7L2 short mRNA variant (divided by the median value).
The short Ex12−13−13a− mRNA variant associates with FFAs and adipose tissue insulin action.
Interestingly, we found a positive correlation between fasting FFAs and the short TCF7L2 mRNA variant in both subcutaneous (r = 0.350; P = 0.014) and visceral adipose tissue (r = 0.581; P = 2 × 10−5) (Fig. 3C), whereas the correlation with fasting plasma glucose was observed only with the short variant in subcutaneous adipose tissue. Furthermore, individuals with higher level of the short Ex12−13−13a− mRNA variant (divided by the median value) had higher levels of FFAs at 120 min during an OGTT (pooled data of 216 individuals from the KOBS, METSIM, and EUGENE2 studies) and higher levels of FFAs during the hyperinsulinemic clamp in 113 individuals from the EUGENE2 study (Fig. 3D). In contrast, no association of TCF7L2 splicing with insulin release in an IVGTT or insulin sensitivity during the hyperinsulinemic euglycemic clamp in the EUGENE2 study was observed (Fig. 3E and F). These results suggest that adipose tissue splicing of TCF7L2 associates primarily with fatty acid metabolism and insulin action in adipose tissue. The potential significance of this regulation in adipocytes is also supported by the changes in the splicing pattern during adipocyte differentiation. We observed that the expression of the short Ex12−13−13a− mRNA variant, which was associated with high levels of glucose and FFAs, increased during differentiation of SGBS adipocyte cell strain. This was linked with a decrease in the amount of the full Ex12+13+13a− mRNA variant (Fig. 4).
FIG. 4.
Proportion of the short Ex12−13−13a− mRNA variant (black circles) (A) and the full Ex12+13+13a− mRNA variant (black triangles) (B) during differentiation of SGBS cell strain. †P < 0.01; ‡P < 0.001; §P < 1 × 10−4, t test.
DISCUSSION
In this study, we demonstrate that weight loss regulates alternative splicing of TCF7L2 (Fig. 2). Importantly, this regulation of splicing in the adipose tissue associates with obesity and the levels of fasting plasma glucose and serum FFAs (Fig. 3A–C). Finally, the association between splicing variants and serum FFA levels during hyperinsulinemia suggests an effect on insulin action in adipose tissue (Fig. 3 D). Because weight loss (Fig. 2) and obesity (Table 2) associated with TCF7L2 splicing but the rs7903146 genotype of TCF7L2 did not, these findings reveal an important regulation of TCF7L2 independent of the genotype.
The first novel finding in our study was that weight loss, induced by obesity surgery, led to a change in TCF7L2 mRNA variant distribution, investigated using PCR-capillary electrophoresis. Both in subcutaneous fat and the liver, the relative proportion of the short Ex12−13−13a− mRNA variant was reduced in response to weight loss (Fig. 2). No change in total gene expression, determined by quantitative PCR, of TCF7L2 was observed, suggesting that the effect on splicing is independent of transcriptional regulation. An explanation for the altered TCF7L2 splicing in obesity could be related to insulin resistance. Splicing factors have been shown to be regulated by insulin (11,23), and their expression is lower in obesity (11). The exact pathways responsible for the change in TCF7L2 splicing after obesity surgery should be investigated in experimental studies.
It is important to understand that although splicing factors and alternative splicing can be dysregulated in obesity, they can also contribute to the metabolic phenotype, as shown for the splicing factor SFRS10 and its alternatively spliced target exon 6 of LPIN1 (11). Based on this study, we propose that TCF7L2 splicing affects FFA levels by regulating adipocyte function for the following reasons. We observed an association of TCF7L2 splicing in both subcutaneous and visceral fat with fasting FFAs (Fig. 3C), as opposed to glucose that was only related to TCF7L2 splicing in subcutaneous adipose tissue. Moreover, we detected that subjects with high expression of the short Ex12−13−13a− TCF7L2 mRNA variant had higher FFAs during hyperinsulinemia, suggesting impaired insulin action in adipose tissue (Fig. 3D). Finally, the amount of the short Ex12−13−13a− mRNA variant increased during adipocyte differentiation in an SGBS cell strain.
TCF7L2 splicing can contribute to cell metabolism by modifying Wnt signaling (9,10). Our results are concordant with a predicted lack of the short Ex12−13−13a− mRNA variant to induce Wnt signaling (9,10). The short Ex12−13−13a− mRNA variant lacks exon 13 and 13a, which encode complete auxiliary DNA interaction motif C-clamp (CRARF and CRALF motifs) (14,16) needed for the activation of Wnt/β-catenin target promoters (10). In contrast, the lack of exon 12 in this variant may also be crucial because the variant Ex12+13−13a−, which has exon 12 but lacks exon 13, was negatively correlated with fasting glucose level (Supplementary Table 2). The importance of exon 12 is supported by a recent study demonstrating that overexpression of the short Ex12−13−13a− mRNA variant induced β-cell apoptosis and glucagon-like peptide 1-stimulated insulin secretion, whereas variant containing exon 12, and not exon 13 or 13a, has a protective effect on β-cell survival (9). Thus, it is possible that the lack of both exons 12 and 13 in the short Ex12−13−13a− mRNA variant, and resulting change in Wnt signaling, could explain the association with adipose tissue insulin resistance.
We recognize that we had a small sample size for a genetic study. An intronic TCF7L2 SNP rs7903146 that has been consistently associated with type 2 diabetes (3,24–26) has been associated with TCF7L2 splicing in one previous study (6). Although we did not observe any association between this genotype and TCF7L2 splicing, we cannot exclude the possibility of an association with the current sample size. However, paired tissue samples taken before and a year after bariatric surgery provided enough power to demonstrate statistically highly significant changes in splicing in response to weight loss. In addition, we presented data from three independent studies to replicate the association of fasting glucose and FFA levels with TCF7L2 splicing showing a genotype-independent association between TCF7L2 splicing and metabolism. Despite different methodology, our results with respect to distribution of mRNA variants in adipose tissue and liver are in agreement with the earlier studies (6,8). The benefit of our method, PCR-capillary electrophoresis, is the efficient screening of even subtle changes in splicing and direct comparison of variant proportions across the different tissue samples without a need for a separate control gene. Ultimately, studies using whole-transcriptome RNA sequencing will be needed to detect all splicing variants.
In conclusion, we demonstrated that TCF7L2 splicing is regulated by weight loss and associated with obesity and high levels of plasma glucose and serum FFAs. These findings could be explained by impaired ability of the short mRNA TCF7L2 variant to activate Wnt signaling in adipose tissue. Our results suggest that the regulation of TCF7L2 splicing in adipose tissue by weight loss may be important in adipocyte biology, leading to increased diabetes risk.
ACKNOWLEDGMENTS
This study was supported by a grant from the Academy of Finland (contracts 124243 to M.L. and 120979 and 138006 to J.P.), the Emil Aaltonen Foundation (to J.P.), the Finnish Diabetes Research Foundation (to M.L. and J.P.), the Finnish Cultural Foundation (to J.P.), the Finnish Heart Foundation (to M.L.), Tekes–the Finnish Funding Agency for Technology and Innovation (contract 1510/31/06 to M.L. and 40058/07 to M.V. and L.P.), Commission of the European Community (contract LSHM-CT-2004-512013 EUGENE2 to M.L.), and the Sigrid Juselius Foundation (to M.V. and L.P.).
No potential conflicts of interest relevant to this article were reported.
D.K. researched data and wrote the manuscript. T.K. researched data and contributed to the discussion. S.V. and P.K. conducted clinical studies. M.V. carried out the SGBS cell experiment. L.P. was responsible for the SGBS cell experiment. M.P. and H.G. conducted clinical studies. M.L. contributed to the discussion and reviewed the manuscript. J.P. designed the study, contributed to the discussion, and reviewed and edited the manuscript. J.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Tiina Sistonen (Department of Medicine, University of Eastern Finland) and Päivi Turunen (Institute of Public Health and Clinical Nutrition, University of Eastern Finland) for assistance with tissue biopsies and laboratory analyses.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db12-0239/-/DC1.
See accompanying commentary, p. 2657.
REFERENCES
- 1.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323 [DOI] [PubMed] [Google Scholar]
- 2.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet 2010;42:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 2007;117:2155–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu L, Matveyenko AV, Kerr-Conte J, Cho JH, McIntosh CH, Maedler K. Decreased TCF7L2 protein levels in type 2 diabetes mellitus correlate with downregulation of GIP- and GLP-1 receptors and impaired beta-cell function. Hum Mol Genet 2009;18:2388–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev 2003;17:419–437 [DOI] [PubMed] [Google Scholar]
- 6.Mondal AK, Das SK, Baldini G, et al. Genotype and tissue-specific effects on alternative splicing of the transcription factor 7-like 2 gene in humans. J Clin Endocrinol Metab 2010;95:1450–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osmark P, Hansson O, Jonsson A, Rönn T, Groop L, Renström E. Unique splicing pattern of the TCF7L2 gene in human pancreatic islets. Diabetologia 2009;52:850–854 [DOI] [PubMed] [Google Scholar]
- 8.Prokunina-Olsson L, Welch C, Hansson O, et al. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet 2009;18:3795–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Bacquer O, Shu L, Marchand M, et al. TCF7L2 splice variants have distinct effects on beta-cell turnover and function. Hum Mol Genet 2011;20:1906–1915 [DOI] [PubMed] [Google Scholar]
- 10.Weise A, Bruser K, Elfert S, et al. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucleic Acids Res 2010;38:1964–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pihlajamäki J, Lerin C, Itkonen P, et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab 2011;14:208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiina H, Igawa M, Breault J, et al. The human T-cell factor-4 gene splicing isoforms, Wnt signal pathway, and apoptosis in renal cell carcinoma. Clin Cancer Res 2003;9:2121–2132 [PubMed] [Google Scholar]
- 13.Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 2006;25:2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atcha FA, Munguia JE, Li TW, Hovanes K, Waterman ML. A new beta-catenin-dependent activation domain in T cell factor. J Biol Chem 2003;278:16169–16175 [DOI] [PubMed] [Google Scholar]
- 15.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci USA 2008;105:9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atcha FA, Syed A, Wu B, et al. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol 2007;27:8352–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pihlajamäki J, Kuulasmaa T, Kaminska D, et al. Serum interleukin 1 receptor antagonist as an independent marker of non-alcoholic steatohepatitis in humans. J Hepatol 2012;56:663–670 [DOI] [PubMed] [Google Scholar]
- 18.Stancáková A, Javorský M, Kuulasmaa T, Haffner SM, Kuusisto J, Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes 2009;58:1212–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmenniemi U, Ruotsalainen E, Pihlajamäki J, et al. Multiple abnormalities in glucose and energy metabolism and coordinated changes in levels of adiponectin, cytokines, and adhesion molecules in subjects with metabolic syndrome. Circulation 2004;110:3842–3848 [DOI] [PubMed] [Google Scholar]
- 20.Vauhkonen I, Niskanen L, Vanninen E, Kainulainen S, Uusitupa M, Laakso M. Defects in insulin secretion and insulin action in non-insulin-dependent diabetes mellitus are inherited. Metabolic studies on offspring of diabetic probands. J Clin Invest 1998;101:86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wabitsch M, Brenner RE, Melzner I, et al. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int J Obes Relat Metab Disord 2001;25:8–15 [DOI] [PubMed] [Google Scholar]
- 22.Prokunina-Olsson L, Kaplan LM, Schadt EE, Collins FS. Alternative splicing of TCF7L2 gene in omental and subcutaneous adipose tissue and risk of type 2 diabetes. PLoS ONE 2009;4:e7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel NA, Chalfant CE, Watson JE, et al. Insulin regulates alternative splicing of protein kinase C beta II through a phosphatidylinositol 3-kinase-dependent pathway involving the nuclear serine/arginine-rich splicing factor, SRp40, in skeletal muscle cells. J Biol Chem 2001;276:22648–22654 [DOI] [PubMed] [Google Scholar]
- 24.Helgason A, Pálsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 2007;39:218–225 [DOI] [PubMed] [Google Scholar]
- 25.Groves CJ, Zeggini E, Minton J, et al. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes 2006;55:2640–2644 [DOI] [PubMed] [Google Scholar]
- 26.Florez JC, Jablonski KA, Bayley N, et al. Diabetes Prevention Program Research Group TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]