Abstract
Adipocyte renewal from preadipocytes has been shown to occur throughout life and to contribute to obesity, yet very little is known about the molecular circuits that control preadipocyte expansion. The soluble form of the preadipocyte factor (also known as pref-1) delta-like 1 homolog (DLK1S) is known to inhibit adipogenic differentiation; however, the impact of DLK1 isoforms on preadipocyte proliferation remains to be determined. We generated preadipocytes with different levels of DLK1 and examined differentially affected gene pathways, which were functionally tested in vitro and confirmed in vivo. Here, we demonstrate for the first time that only membrane-bound DLK1 (DLK1M) exhibits a substantial repression effect on preadipocyte proliferation. Thus, by independently manipulating DLK1 isoform levels, we established that DLK1M inhibits G1-to-S-phase cell cycle progression and thereby strongly inhibits preadipocyte proliferation in vitro. Adult DLK1-null mice exhibit higher fat amounts than wild-type controls, and our in vivo analysis demonstrates that this may be explained by a marked increase in preadipocyte replication. Together, these data imply a major dual inhibitory function of DLK1 on adipogenesis, which places DLK1 as a master regulator of preadipocyte homeostasis, suggesting that DLK1 manipulation may open new avenues in obesity treatment.
Obesity is a major risk factor for metabolic disorders such as type 2 diabetes and cardiovascular disease. Excessive dietary fat intake leads to expansion of adipose tissue, which, at the cellular level, is known to occur as a result of adipocyte hypertrophy; most recently it also has been demonstrated to be caused by an increase in the number of adipocytes during both childhood and adulthood (1–3). The latter certainly opens up new possibilities for developing novel strategies to treat or prevent obesity; however, the mechanisms responsible for preadipocyte proliferation are incompletely understood (4). Adipocytes originate from preadipocytes, for example, fat progenitors located in the stromal vascular fraction of adipose tissue. Little is known so far about the biology of these preadipocytes, but recent in vivo cell lineage tracing studies using peroxisome proliferator-activated receptor γ, a master regulator of adipogenesis, have suggested that preadipocytes in white adipose tissue are characterized by the expression of delta-like 1 homolog (Dlk1), GATA3, Wisp2, Smo, and Gli3 (4). Dlk1, also known as preadipocyte factor 1 (Pref-1), is a paternally expressed imprinted gene that encodes for a membrane protein member of the NOTCH receptor and ligand epidermal growth factor–like protein family (5,6). Dlk1 has been linked to the inhibition of adipogenesis, especially in studies using the preadipocyte cell line 3T3-L1, where Dlk1 is highly expressed during proliferation but downregulated upon adipogenic differentiation (7,8). Alternative splicing generates various forms of membrane-spanning DLK1 proteins that differ by in-frame deletions of an extracellular juxtamembrane protease recognition site (9). Thus, DLK1 isoforms lacking this region remain tethered to the membrane, whereas variants encompassing the protease recognition site can be processed to generate the large, active soluble DLK1S isoform (10) that is released into circulation (11) (Fig. 1A). Despite its well-known implication in adipogenesis, Dlk1’s mechanism of action still remains a large topic for discussion, and the interaction partner(s) still remains to be determined definitively (6,12–17). Thus, further insights into DLK1 signaling in preadipocytes will be important, and may serve to further unravel whether DLK1 itself or related interaction partners could serve as novel candidate targets in obesity therapy.
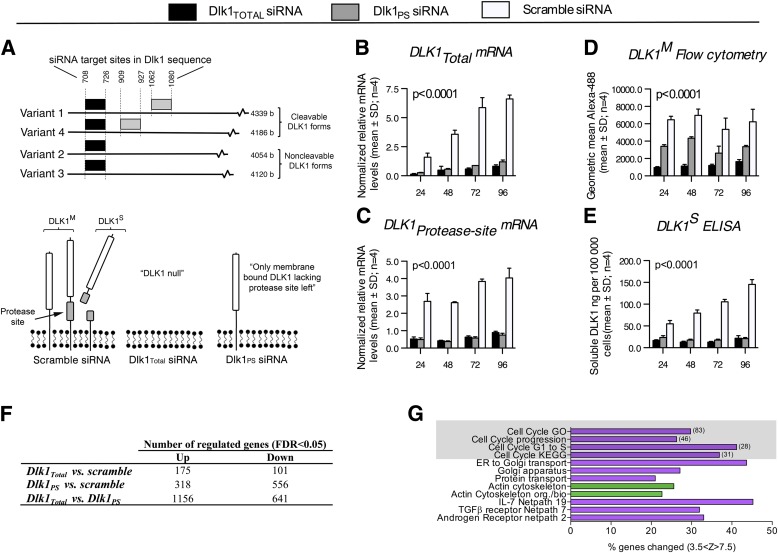
FIG. 1.
Cell cycle gene pathways are upregulated by knockdown of noncleavable Dlk1, but not cleavable Dlk1. A (top panel): The siRNA design. Three distinct siRNAs were designed to knock down all DLK1 isoforms independently (Dlk1Total, black boxes), only cleavable DLK1 (Dlk1PS, gray boxes), or none (scramble control), resulting in differential levels of DLK1 isoforms in 3T3-L1 preadipocytes, as indicated in the bottom panel of A, where the ideal scenario with 100% DLK1 knockdown is depicted. As such, DLK1M represents all membrane-bound DLK1, including those Dlk1 variants that contain the protease site but that still remain on the membrane and therefore are detected by flow cytometry. By contrast, DLK1S exclusively designates Dlk1 variants containing the protease site but that have been cleaved off the membrane and thus can be measured in the medium by ELISA. B–E: 3T3-L1 preadipocytes were transfected with either Dlk1Total, Dlk1PS, or scramble siRNA, and knockdown was verified by qRT-PCR (B, C), flow cytometry (D), and ELISA (E) at different time points after transfection. Note that DLK1S remains at the membrane until cleavage and, logically, Dlk1PS siRNA therefore also reduces DLK1M by 40–50% (D). Statistical significance was tested using a one-way ANOVA followed by a Bonferroni multiple comparisons test within each time point against scramble control. All tests resulted in P < 0.0001. F and G: 3T3-L1 preadipocytes transfected with Dlk1Total, Dlk1PS, or scramble siRNA were examined by global gene expression arrays (n = 4). Data were log2 transformed and analyzed by the Student t test, and the number of significantly regulated genes was examined by correcting for multiple testing (false discovery rate). Significantly (z > 0; P < 0.05, Westfall-Young adjustment for multiple testing) upregulated (magenta) and downregulated (green) gene pathways were determined using GenMAPP software. Numbers of genes regulated in cell cycle related pathways are listed in brackets. FDR, false discovery rate; TGFβ, transforming growth factor β; ER, endoplasmic reticulum.
DLK1M and soluble DLK1 isoforms both have been shown to be involved in adipogenesis; however, their significance and actual roles in this process are still debated (7,10,18), and little attention has been given to DLK1’s role in preadipocyte proliferation. Interestingly, Dlk1-null mice display considerably increased fat mass compared with wild-type controls, but this was concluded previously, reflecting adipocyte hypertrophy rather than hyperplasia (19). Vice versa, reduced fat mass due to decreased cell size has been reported for mice that overexpress DLK1S (20). DLK1 thus has been shown to be a major inhibitor of adipogenesis, and although questioned, this inhibitory function has been ascribed solely to the large soluble form of DLK1 interfering with adipocyte differentiation (10). However, neither of these studies investigated the impact of DLK1 on preadipocyte proliferation. This led us to test the effect of DLK1 on preadipocyte proliferation in vitro and in vivo and specifically whether differential roles exist for DLK1M and soluble DLK1 isoforms.
RESEARCH DESIGN AND METHODS
Animals.
Dlk1−/− and Dlk1+/+ C57BL/6 mice (21) were backcrossed to C57BL/6, and obtained heterozygotes were intercrossed to generate homozygotes. Tail or ear DNA was isolated using a DNeasy kit (Qiagen), and genotype analysis was performed by PCR amplification using the following primers: 5′Dlk1_F, CCAAATTGTCTATAGTCTCCCTC; 5′Dlk1_R, CTGTATGAAGAGGACCAAGG; 5′Neo_F, TTGAACAAGATGGATTGCACGCAGG; 5′Neo_R,GGCTGGCGCGAGCCCCTGATGCTCT, and a Taq DNA polymerase (Invitrogen). Animals were housed in plastic cages with a 12-/12-h light/dark cycle, and fed ad libitum with normal chow (10% fat, 20% protein, 70% carbohydrate). Specific characteristics of the animals have been described previously (21) and are in agreement with others (19). For all animal experiments we used age- and sex-matched animals as indicated, and all procedures were approved by the Danish Council for Supervision with Experimental Animals (no. 2011/561–1966).
In vitro cell culture.
The preadipocyte cell line 3T3-L1 was kept as previously described (22). Briefly, cells were plated at 600 cells/cm2 3 days before small interfering RNA (siRNA) transfection at day 3 with media replaced every 24 h. Cell culture medium consisted of Dulbecco’s modified Eagle’s medium (Lonza) supplemented with 10% calf serum (CS; Sigma-Aldrich) and 1% penicillin-streptomycin (PS; Lonza).
Cell number and size.
Cultured cells were detached gently with 0.25% trypsin-ethylenediaminetetraacetic acid (Gibco, Invitrogen), pelleted and resuspended in Hanks' balanced salt solution (HBSS, Lonza)/10% CS/1% PS. Cell number and cell diameter were determined using a Beckman Coulter Counter Z2 (Ramcon) fitted with a 100 μm aperture. The size range of particles counted was set at 11–27 μm, and counting was performed in the indicated number of independent experiments, each comprising triplicate measurements.
Flow cytometry to determine DLK1M.
Cells were detached, washed twice in HBSS/10% CS/1% PS, and fixed for 30 min on ice in 1% normal buffered formaldehyde. Fixed cells were washed three times and stored at 4°C in HBSS/5% CS/1% PS/0.05% NaN3 until analysis. Cells were immunostained with rabbit α-mouse DLK1 antibody (0.45 μg/mL) generated in house (23,24) or with rabbit immunoglobulin (Ig) G (control; Santa Cruz Biotechnology) for 30 min on ice. After washing twice, samples were incubated with Alexa 488-conjugated donkey α-rabbit IgG (1:200; Molecular Probes, Invitrogen) for 30 min on ice followed by two washes before flow cytometric analysis using a BD FACSCalibur Instrument. The BD FACSDiva software (version 5.0.1; BD Biosciences, San Jose, CA) was used to analyze flow cytometric results. Debris was excluded from the analysis by gating in the forward and side scatter as previously described (25–27). The relative DLK1M levels were calculated as fold geometric Alexa-488 fluorescence (sample/control) of all live cells on a given day. In parallel, the percentage of DLK1M–positive cells in this live cell gate was measured by gating using the control.
ELISA to determine DLK1S.
Quantification of soluble mouse DLK1S in the culture medium of Dlk1- and scramble siRNA transfected cells was performed by enzyme-linked immunosorbant assay (ELISA), as previously described (24). The amount (ng/mL) of soluble DLK1 per 105 cells was calculated using cell numbers as quantified by Coulter counting.
MTT proliferation assay.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction was evaluated 48 h after Dlk1Total−, Dlk1PS−, and scramble siRNA transfection. Growth medium was replaced with Dulbecco’s modified Eagle’s medium (without phenol red) supplemented with 10% CS, 2 mM l-glutamine, and 1 mM/L pyruvate 24 h before addition of MTT to a final concentration of 0.5 mg/mL. After 4 h of incubation, reduced MTT formazan crystals were solubilized overnight in a 5% sodium dodecyl sulfate/0.005 mol/L hydrogen chloride solution, and absorbance was red at 570–690 nm.
Relative quantitative real-time PCR.
Total RNA was extracted from cell cultures or gonadal fat tissue using the semiautomated 6100 Nucleic Acid Prep Station system according to the manufacturer’s instructions (Applied Biosystems). For cDNA synthesis, 0.3–0.4 μg of total RNA was reverse transcribed with a High Capacity cDNA RT kit (no. 4368813; Applied Biosystems), and quantitative real-time PCR (qRT-PCR) reactions were performed in technical triplicates with 1.5–2 ng of cDNA and 3 pM of forward and reverse primer in a 10 μL reaction mixture using Power SYBRGreen RCR mastermix (no. 4367659; Applied Biosystems). The PCR was run on a 7900HT Fast Real-time PCR system (Applied Biosystems) using 95°C for 10 min and the following PCR cycling (×40) conditions: denaturation at 94°C for 15 s; annealing at 57°C for 30 s; and elongation at 72°C for 30 s. As recommended by others, robust and valid qRT-PCR data were obtained by normalizing the raw data against multiple stably expressed endogenous control genes as determined by the qBase Plus platform (Biogazelle) (28,29).
siRNA-mediated knockdown.
Two specific siRNAs were designed (Fig. 1A) to differentially target either all Dlk1 mRNA splice variants (Dlk1Total: AGAUCGUAGCCGCAACCAATT; UUGGUUGCGGCUACGAUCUCA; Ambion no. 4390771) or only Dlk1 mRNA splice variants containing coding sequences for the protease site for extracellular cleavage (Dlk1PS: UCCUGAAGGUGUCCAUGAATT; UUCAUGGACACCUUCAGGATG; Ambion no. 4399665). Silencer Select Negative control (no. 4390846; Ambion) was used as control, and transfections were performed for 4 h using Lipofectamine 2000 (Invitrogen) as previously described (22). To achieve the highest possible Dlk1 knockdown and avoid off-target effects due to high concentrations of double-stranded RNA in the cells (30,31), we tested three slightly different siRNAs at 1–30 μM for each knockdown design as well as varying cell plating densities (data not shown), and thereby established robust and reproducible Dlk1 knockdown conditions.
mRNA microarray expression profiles.
RNA was reverse transcribed using the MessageAmp II Enhanced kit (Applied Biosystems) and hybridized to Affymetrix GeneChips (Mouse Genome 430 2.0 Array) that were run using the GeneChip Scanner 3000 (Affymetrix, Santa Clara, CA). Data analysis was performed using the R software (Bioconductor); expression indexes were calculated using RMA and data were normalized using the quantile method. Differentially expressed genes were analyzed by the Student t test with false discovery rates to adjust for multiple tests. Gene signaling pathway analysis was performed by the GenMAPP/MAPPFinder 2.0 software (www.genmapp.org; Gladstone Institute, University of California San Francisco) (32,33). Search criterions were set at a log twofold change higher than 1.1 or lower than 0.9 and a significance filter of unadjusted P values less than 0.05. Genes meeting those criteria were assigned to the local MappFinder pathways and gene families (Mapps) as well as to the gene sets of the Gene Ontology database (Gene Ontology terms). Mapps or Gene Ontology terms were considered significantly enriched when the z scores were above zero and the adjusted P values were less than 0.05 (Westfall-Young adjustment for multiple testing). Gene sets that comprised fewer than 5 or more than 100 genes changed were excluded from the analysis because they were considered either too specific or too general, respectively.
In vitro and in vivo 5-ethynyl-2′-deoxyuridine incorporation and detection.
For in vitro S-phase detection and quantification, 5-ethynyl-2′-deoxyuridine (EdU) was added to Dlk1Total−, Dlk1PS−, and scramble siRNA transfected cells to a final concentration of 10 μM and incubated for 1 h before detachment and ethanol fixation. For each siRNA treatment, cells without EdU added were included as negative controls. The Click-iT EdU Alexa Fluor 647 Cell Proliferation Assay (Molecular Probes, Invitrogen) was used for detection of EdU incorporation according to the manufacturer’s instructions. Samples were analyzed using a BD LSR II Flow cytometer (BD Biosciences) with acquisition of 10,000 cells in a live mononuclear gate. Debris was excluded by gating in the forward and side scatter as previously described (26). Cell doublet discrimination was performed by gating the single-cell population in a width versus area plot of the propidium iodide signal (34,35).
For in vivo determination of proliferation rate, EdU was injected intraperitoneally to Dlk1−/− and Dlk1+/+ female mice (n = 6) at a concentration of 50 μg EdU/g body weight. One week after injection, the stromal vascular fraction (SVF) was isolated from the gonadal fat pads as previously described (36) and was used for EdU analysis. Briefly, gonadal fat pads were excised from mice, weighed, washed in HBSS/100 IU/mL 1% PS, and minced extensively. Tissue was digested with 0.35% type II collagenase (Worthington, U.K.) at 37°C for 60 min, and cells were released by gentle trituration in 37°C growth medium. A single cell suspension was obtained by filtering the cell samples through 100-μm cell strainers. Erythrocytes were lysed and isolated cells were fixed in 2% paraformaldehyde followed by permeabilization using a saponin-based buffer as described by the manufacturer. Animals injected with PBS were included as EdU-negative controls. EdU detection was performed as described earlier, and SVF cells were co-stained with Rat-α-mouse-CD45 (BD Pharmingen; 1:100) to distinguish preadipocytes present in the nonhematopoietic (CD45-negative) fraction of the SVF. Samples were analyzed using a BD LSR II flow cytometer (BD Biosciences) with acquisition of 10,000 cells in a live mononuclear gate. Flow data were analyzed by FACSDiva Software (version 5.0.1, BD Biosciences) with debris and cell doublets being excluded from the analysis by gating in the forward and side scatter, as previously described (26).
Statistical analysis.
For cell culture, independent experiments (n) represent cells with at least two intervening passages. All analyses comprised 4–8 independent experiments, and one- or two-way ANOVA and two-tailed t tests were performed as indicated (GraphPad Prism software, 5.0a Mac version) to test significance (P < 0.05). For animal experiments, 5–12 animals were used within each group.
RESULTS
Cell cycle signaling pathways are differentially regulated by DLK1 isoforms in preadipocytes.
DLK1 exists in four major isoforms in the mouse (9) (Fig. 1A), whereas only one cleavable and one noncleavable isoform is present in humans (37). We designed two different siRNAs, one specifically targeting the protease encoding site, only present in mRNA encoding for DLK1 forms (siRNA Dlk1PS) that may be cleaved, and another targeting an mRNA sequence common to all Dlk1 mRNA variants (siRNA Dlk1Total) (Fig. 1A). To our knowledge, this is the first study examining the function of DLK1 isoforms by specifically reducing the endogenous level of specific DLK1 variants in cells that naturally express the protein.
When we knocked down the DLK1 isoforms in proliferating preadipocytes, we achieved 85 and 80% reductions of Dlk1Total and Dlk1PS mRNA levels, respectively, compared with scramble transfected cells. This highly efficient knockdown was maintained for at least 96 h (Fig. 1B and C) and successfully confirmed at the protein level (Fig. 1D and E).
To identify pathways that were differentially regulated by cleavable and noncleavable DLK1 variants in proliferating preadipocytes, we next performed global gene expression profiling and found a large number of significantly (false discovery rate, <0.05) regulated genes among the three groups (Fig. 1F). Interestingly, the highest number of significantly regulated genes (1,156 up- and 641 downregulated) was obtained by comparing the Dlk1Total- and Dlk1PS siRNA-treated cell populations with each other (Fig. 1F), indicating that cleavable and noncleavable DLK1 isoforms act differently, as previously suggested (7,38). Gene signaling pathway analysis revealed that 4 of the 12 significantly regulated pathways between Dlk1Total and Dlk1PS siRNA-transfected cells (Fig. 1G) concerned cell cycle control, especially in relation to the G1- to S-phases (Fig. 2A).
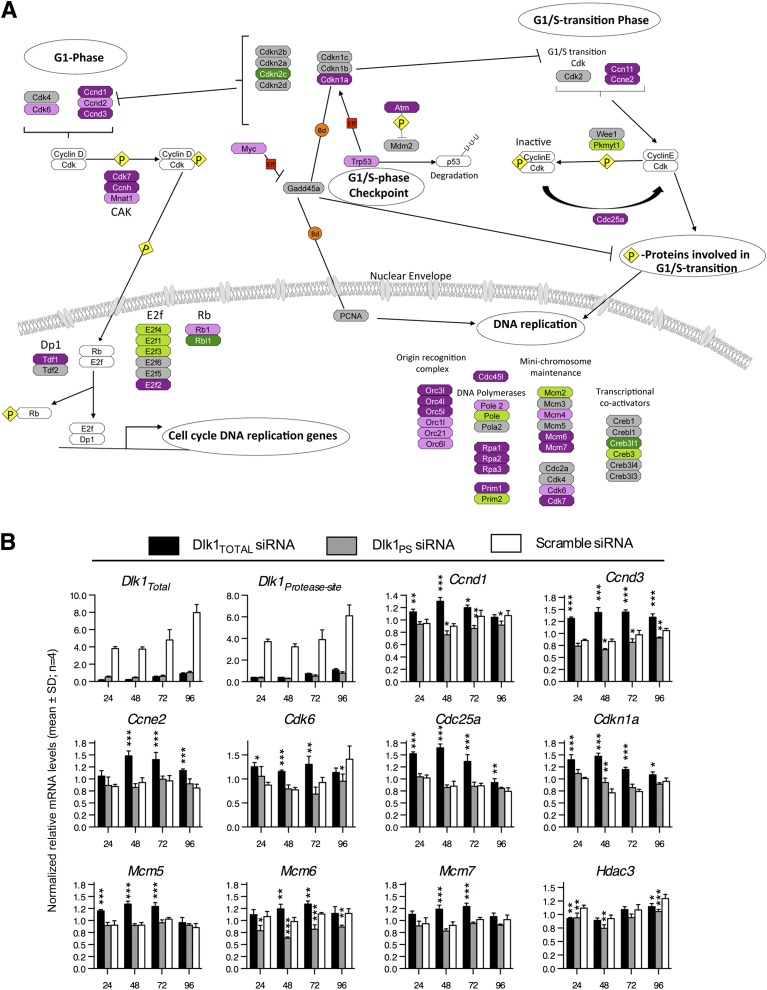
FIG. 2.
Noncleavable Dlk1 regulates numerous genes during the G1-to-S-phase transition. A: Relative variation of gene expression in Dlk1Total versus Dlk1PS siRNA transfected 3T3-L1 preadipocytes at 48 h, as determined using GenMAPP software. Upregulated genes are magenta (P < 0.05) and purple (0.05 < P < 0.2), and downregulated genes are dark green (P < 0.05) and green (0.05 < P < 0.2). P values have been assigned to each gene after log2 transformation and a Student t test. B: Relative qRT-PCR of cell cycle–associated genes at 24–96 h after transfection of 3T3-L1 preadipocytes with DLK1Total, DLK1PS, or scramble siRNA. Significance was tested by a one-way ANOVA followed by a Bonferroni multiple comparisons test within each time point against scramble control. *P < 0.05; **P < 0.01; ***P < 0.0001. Raw data were normalized against multiple stably expressed reference genes (data not shown).
Relative qRT-PCR supported our microarray data and showed that genes known to promote cell cycle progression (Ccnd1, Ccnd3, Ccne2, Cdk6, Cdc25a,and Mcm5–7) were all significantly upregulated ∼27–94% (P < 0.001–0.01) in Dlk1Total knockdown cells, whereas all these genes except Cdkn1a were unaffected or even significantly downregulated in Dlk1PS siRNA-transfected cells (Fig. 2B). Cdkn1a is known to be decreased in DLK1 over-expressing liver cells (39), which is in agreement with our results.
Thus, simultaneous knockdown of cleavable and noncleavable DLK1 variants, but not cleavable DLK1 alone, results in enhanced expression of genes that are known to promote cell cycle progression, suggesting that only DLK1M affects preadipocyte proliferation. However, it is also possible that cleavable and noncleavable DLK1 variants exert similar effects and that the amounts of noncleavable DLK1 remaining on the membrane when only cleavable DLK1 forms are knocked down are sufficient to prevent an increase in proliferation-associated events. To test for this possibility, we restored the level of soluble DLK1S in Dlk1Total knocked down cells by adding different amounts of conditioned preadipocyte medium containing soluble DLK1 or purified human soluble DLK1 to the culture medium and examined the effect on cyclin expression levels. However, no apparent difference in the levels of Ccnd1, Ccnd3, and Ccne2 were found (Fig. 3), suggesting that only noncleavable DLK1 isoforms exert an inhibitory function on preadipocyte proliferation.
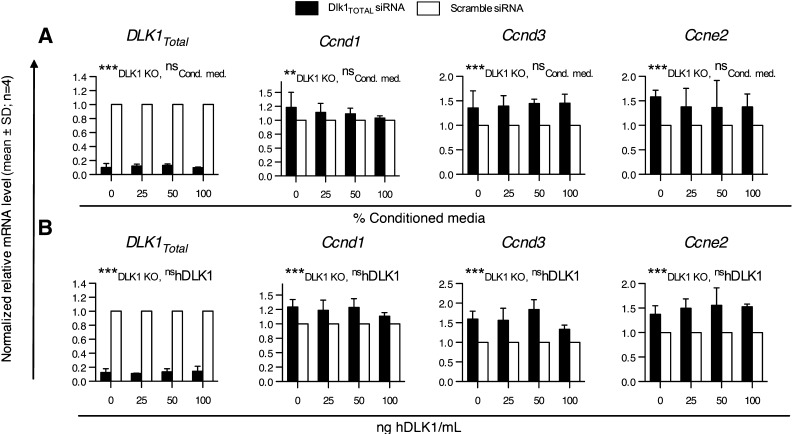
FIG. 3.
Restoration of cleaved DLK1 levels in the medium of Dlk1Total knocked down 3T3-L1 cells. 3T3-L1 cells were transfected at 0 h with either Dlk1Total or scramble siRNA and grown in either 3T3-L1–conditioned medium (Cond. med.) (A) or a medium containing known concentrations of purified human DLK1 (hDLK1) (B). mRNA levels of cell cycle–related genes were determined by qRT-PCR 48 h after transfection. Significance was tested by a two-way ANOVA (**P < 0.01; ***P < 0.0001), and qRT-PCR raw data were normalized against multiple stably expressed endogenous controls (data not shown).
Membrane-tethered DLK1 inhibits preadipocyte proliferation in vitro by lowering the number of cells in the S phase of the cell cycle.
We next performed an EdU cell cycle distribution assay, which is a new and far more robust method to detect S-phase cells than conventional 5-bromo-2-deoxyuridine (BrdU) assays (40). As expected, we found that 40.1 ± 1.6% (P = 0.0006) and 34.0 ± 1.4% (P = 0.02) of Dlk1Total− and Dlk1PS siRNA-treated cells were present in the S phase at 48 h, whereas only 30.7 ± 1.6% of scramble control cells were replicating (Fig. 4A and B).
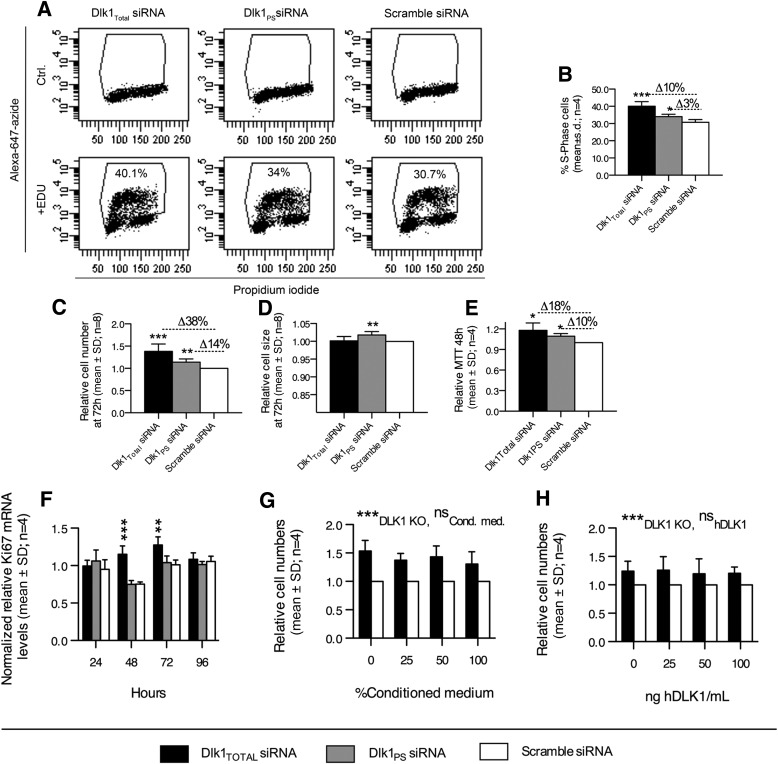
FIG. 4.
Membrane-bound DLK1 restricts in vitro preadipocyte proliferation by repressing G1-to-S-phase transition. A, B: The number of 3T3-L1 preadipocytes in the S phase of the cell cycle was quantified by EdU incorporation 48 h after transfection with Dlk1Total, Dlk1PS, or scramble siRNA. Cell doublets and debris were excluded from the flow cytometric analysis, and EdU+ gating was established within each siRNA treatment from negative controls that did not receive any EdU. C–F: Proliferation capacity was examined in Dlk1Total, Dlk1PS, or scramble siRNA-treated cells using Coulter counting (C, D), MTT (E), and qRT-PCR (F). G, H: Dlk1Total and scramble siRNA-transfected 3T3-L1 preadipocytes were cultured in the presence of different percentages of (G) conditioned medium (Cond. med.) from proliferating 3T3-L1 preadipocytes or (H) purified human DLK1 (ng/mL). Cell numbers were then examined by Coulter counting at 72 h. Significance was tested by Student t test (A–E), a one-way ANOVA followed by a Bonferroni multiple comparisons test within each time point against scramble control (F), and two-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.0001) (G, H). qRT-PCR raw data in (F) were normalized against multiple stably expressed reference genes (data not shown).
We next determined the proliferative capacity of Dlk1Total, Dlk1PS, and scramble siRNA-transfected cells. Cell numbers were significantly elevated (38%; P < 0.001, one-sample t test) in Dlk1Total transfected cells compared with scramble control cells (Fig. 4C). This effect was much less pronounced but still significant (14%; P < 0.05, one-sample t test) in Dlk1PS knocked down cells (Fig. 4C). Previous studies have suggested that the cleavable form of DLK1 has a hypotrophic effect on adipocytes (19,20). In agreement, we found a small but significant increase in cell size (1.8%; P < 0.001) in only Dlk1PS siRNA-transfected cells (Fig. 4D). However, this minor effect may be insignificant in a physiological sense. Nevertheless, the enhanced proliferation was confirmed by higher MTT levels (Fig. 4E) and an increased expression of the proliferation marker Ki-67 (43%; P < 0.001) (Fig. 4F). Finally, we restored the level of cleaved DLK1 in Dlk1Total knocked down cells and found that cleaved DLK1 alone did not reduce cell numbers (Fig. 4G and H), which is in agreement with the lack of a suppressive effect on cyclin gene expression, as described earlier (Fig. 3).
Adipose tissue expansion in Dlk1−/− mice due to increased preadipocyte proliferation.
It has been suggested that the Dlk1 knockout mouse possesses increased amounts of adipose tissue (19,21) as a sole consequence of DLK1 inhibiting adipocyte hypertrophy (19,20), which previously was the general view of how fat expansion occurred. Recently, it has become clearer that adipocyte numbers also determine fat content (1–3), and we therefore hypothesized that obesity in the Dlk1 knockout mouse also could be explained by an increased proliferation of preadipocytes residing in vivo, as our in vitro results suggest. Thus, to support our in vitro data, we performed qRT-PCR on gonadal adipose tissue from Dlk1−/− and Dlk1+/+ mice and found substantially increased (26–67%; P < 0.05–0.001) levels of cell cycle–promoting genes (Ccnd1, Ccne2, and Ccnd3, Mcm5–6) (Fig. 5A). No difference was observed between sexes (data not shown), which is in agreement with a previous study (19) showing that both female and male Dlk1 knockouts possess higher amounts of fat compared with their respective wild-type littermates. No Dlk1 expression was present in adipose tissue from the Dlk1−/− mice (Fig. 5A), confirming the lack of Dlk1 in the knockout mice (21).
FIG. 5.
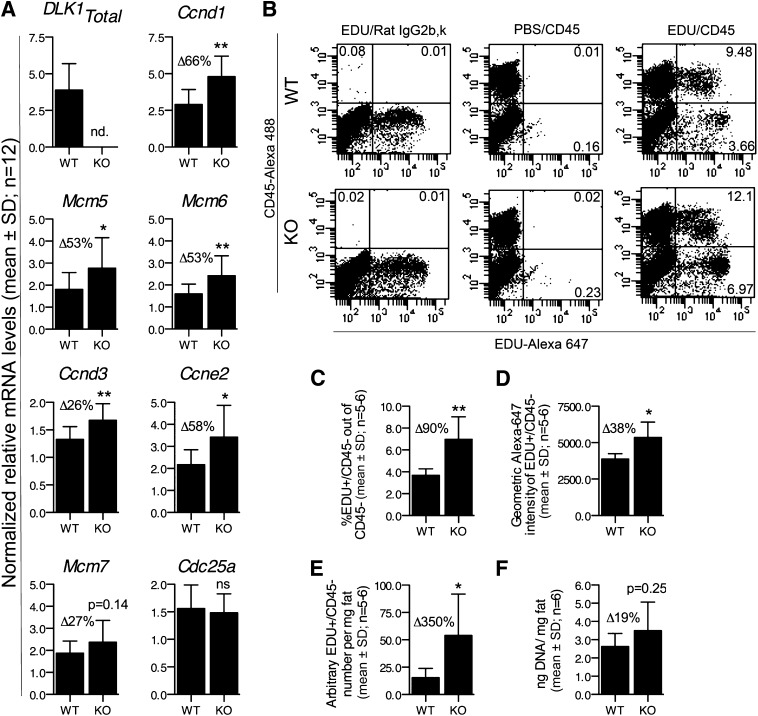
DLK1 represses cell cycle gene expression and proliferation of preadipocytes in vivo. A: Expression analysis by qRT-PCR of several cell cycle genes in gonadal fat tissue from Dlk1+/+ or Dlk1−/− mice (n = 12; 6 males and 6 females within each group). B–E: Flow cytometric analysis of EdU/CD45 in SVF cells isolated from gonadal fat tissue of Dlk1+/+ or Dlk1−/− mice injected with 50 mg/mL/kg of EdU 1 week before SVF isolation and analysis. The number of SVF cells isolated per milligram of fat was counted by Coulter counting. F: The DNA content per milligram of gonadal fat tissue in Dlk1+/+ and Dlk1−/− mice. Significance was tested by a Student t test (*P < 0.05; **P < 0.01), and qRT-PCR raw data in (A) were normalized against multiple stably expressed reference genes (data not shown).
To test our novel results on a functional level in vivo, we next quantified the proliferative capacity of preadipocytes in young mice, when the gonadal adipose tissue is supposed to be established (1,3). In general, there is a major lack of knowledge about the phenotype and identity of preadipocytes in vivo (4). Because we intended to confirm only our in vitro data and not to exclude any preadipocytes by selecting the wrong preadipocyte markers, we chose to analyze the adipose-derived, CD45-negative portion of the stromal vascular fraction because this is generally accepted to comprise all preadipocytes. We therefore administered the nucleoside analog EdU into 5-week-old Dlk1−/− and Dlk1+/+ mice and determined its incorporation into CD45−SVF cells after 1 week (Fig. 5B and E). In wild-type mice we found that 3.7% (mean, n = 6) of the CD45−SVF had been proliferating (Fig. 5B and C), a figure highly similar to what was reported recently for preadipocytes in 6-week-old mice (2) and which confirms our experimental setup. For comparison, 7.0% (mean, n = 5) of the CD45−SVF in the Dlk1−/− mice were EdU positive (Fig. 5B–C), suggesting that nearly twice as many preadipocytes are proliferating in the absence of DLK1. By measuring the relative total amount of incorporated EdU, we also established that Dlk1−/− preadipocytes contained 38% more EdU than their wild-type littermates (Fig. 5D), indicating that the Dlk1−/− preadipocytes not only are higher in numbers, but they also have progressed through the cell cycle more times than Dlk1+/+ preadipocytes. Overall, we found that Dlk1−/− mice possessed 3.5 times more proliferating preadipocytes per milligram of fat than their wild-type littermates (Fig. 5E), which was confirmed by a tendency of Dlk1−/− fat to contain more DNA per milligram of fat than Dlk1−/− mice (Fig. 5F). Thus, our in vivo results confirm our in vitro data that Dlk1 inhibits preadipocyte proliferation.
DISCUSSION
Adipose tissue expansion is known to occur through proliferation and differentiation of preadipocytes; however, the mechanisms especially responsible for preadipocyte proliferation are incompletely understood (4). Numerous data suggest that only cleaved DLK1 inhibits adipogenesis by interfering with the differentiation machinery (8), but our results invoke an additional explanation suggesting that DLK1M slows preadipocyte proliferation by regulating numerous components in the G1-S phase of the cell cycle.
As such, we have examined the effect on preadipocyte proliferation of cleavable and noncleavable DLK1 isoforms by specifically reducing the endogenous level of specific DLK1 variants in cells that naturally express the protein. Although one study previously used antisense RNA molecules to study DLK1’s function in adipocyte differentiation (7), the role of DLK1 has only been assessed by over-expressing the protein in cell types lacking endogenous Dlk1 (13,41) or expressing different endogenous DLK1 levels (10,39,42). Consequently, these previous studies have introduced a concentration variable that has been suggested to be an important factor for DLK1’s function (14). We believe, therefore, that this study provides a novel setup that mimics biology more closely than those used in the previous studies described earlier. Furthermore, this study is one of a few (13,15) that have focused on investigating whether different functions exist for DLK1M and soluble DLK1. In this study, we used mRNA arrays and identified four cell cycle–related signaling pathways that were regulated differentially by cleavable and noncleavable DLK1 isoforms. However, many of the genes only differed by 20–50% in the qRT-PCR verification. Such a small statistically significant change may not be biologically relevant per se, but in our case we chose the genes to be validated at random and therefore cannot rule out that other cell cycle genes may have differed more. Furthermore, we clearly showed that many components, and not merely a few genes, in the cell cycle all are affected at different time points in the same direction of having more proliferation in the treated cells. Most importantly, however, we saw a substantial functional change in the proliferation rate of the treated cells, and we therefore believe that our results are very strong indeed. Accordingly, we found a 0, 14, and 38% increase in the proliferative rate of scramble, Dlk1PS and Dlk1Total siRNA-treated preadipocytes, respectively, and, bearing in mind that these cells exhibit 100, 45, and 15% membrane DLK1, it seems highly likely that the lower levels of DLK1M increase preadipocyte proliferation. This is in agreement with another study, which showed that ectopic expression of only full-length membrane spanning, and not soluble DLK1, diminishes proliferation of leukemic cells with a slower progression through the transition from the G1 to the S phase (41). However, this phenomenon may be cell type/tissue specific because DLK1 over-expression in some cancer cell lines with a small but significant level of endogenous DLK1 reveals that both cleavable and noncleavable DLK1 isoforms promote S-phase cell cycle progression (39,42). Our results showing that cleavable DLK1 does not exert an effect on preadipocyte proliferation is in line with a previous study showing that DLK1 mutants encoding only the cleavable DLK1 isoform do not have an impact on hematopoietic cell proliferation (41). By contrast, Bray et al. (13) showed that Drosophila mutants expressing either cleavable or noncleavable DLK1 result in fewer and higher cell numbers, respectively, indicating that both DLK1 isoforms in some way may be important for cell proliferation and likely have opposite or interfering actions. We also cannot rule out that much higher nonphysiological concentrations of cleaved DLK1 may have a suppressive effect on cell proliferation or, as recently reported (43), that DLK1 isoforms exert their effects with different potencies, with the DLK1M being far more effective. However, our in vivo results clearly support our in vitro data, firmly demonstrating that DLK1 in vivo inhibits preadipocyte proliferation and not only induces adipocyte hypertrophy, as previously assumed (19,20). Yet it should be noted that our in vivo data do not allow us to distinguish between the cleavable and noncleavable DLK1 isoforms. Nevertheless, the data presented here together firmly establish that DLK1 inhibits preadipocyte proliferation in its membrane-bound but not soluble form, which instead exerts its inhibitory adipogenic effect on adipocyte differentiation, as shown by others (7,10,18–20,44).
Conclusively, we believe that this novel and dual function of DLK1 firmly places this molecule as a major regulator of adipogenesis, and further insights into DLK1 signaling will be important and may serve to unravel novel candidate targets to treat obesity.
ACKNOWLEDGMENTS
This work was supported by The Danish National Research Council, The Lundbeck Foundation, Hertha Christensens Foundation, Eva and Henry Frænkels Foundation, A.P. Møller Foundation for the Advancement of Medical Science, and the Department of Clinical Biochemistry and Pharmacology, Odense University Hospital.
No potential conflicts of interest relevant to this article were reported.
S.B.M. and C.H.J. contributed to collection, analysis, and interpretation of data as well as manuscript preparation. M.S. contributed to the study design. M.T. and T.A.K. contributed to analysis and interpretation of data. J.L. and S.P.S. contributed to interpretation of data and approval of the manuscript. D.C.A. contributed to the conception and design of this study; collection, analysis, and interpretation of data; and manuscript preparation and approval as well as funding. D.C.A. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Charlotte Nielsen and Tonja L. Jørgensen (LMCC, Odense University Hospital) for technical assistance on qRT-PCR.
REFERENCES
- 1.Spalding KL, Arner E, Westermark PO, et al. Dynamics of fat cell turnover in humans. Nature 2008;453:783–787 [DOI] [PubMed] [Google Scholar]
- 2.Rigamonti A, Brennand K, Lau F, Cowan CA. Rapid cellular turnover in adipose tissue. PLoS One 2011;6:e17637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arner E, Westermark PO, Spalding KL, et al. Adipocyte turnover: relevance to human adipose tissue morphology. Diabetes 2010;59:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park KW, Halperin DS, Tontonoz P. Before they were fat: adipocyte progenitors. Cell Metab 2008;8:454–457 [DOI] [PubMed] [Google Scholar]
- 5.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993;73:725–734 [DOI] [PubMed] [Google Scholar]
- 6.Baladrón V, Ruiz-Hidalgo MJ, Nueda ML, et al. Dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res 2005;303:343–359 [DOI] [PubMed] [Google Scholar]
- 7.Garcés C, Ruiz-Hidalgo MJ, Bonvini E, Goldstein J, Laborda J. Adipocyte differentiation is modulated by secreted delta-like (dlk) variants and requires the expression of membrane-associated dlk. Differentiation 1999;64:103–114 [DOI] [PubMed] [Google Scholar]
- 8.Sul HS. Minireview: Pref-1: role in adipogenesis and mesenchymal cell fate. Mol Endocrinol 2009:23:1717–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smas CM, Green D, Sul HS. Structural characterization and alternate splicing of the gene encoding the preadipocyte EGF-like protein pref-1. Biochemistry 1994;33:9257–9265 [DOI] [PubMed] [Google Scholar]
- 10.Mei B, Zhao L, Chen L, Sul HS. Only the large soluble form of preadipocyte factor-1 (Pref-1), but not the small soluble and membrane forms, inhibits adipocyte differentiation: role of alternative splicing. Biochem J 2002;364:137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen CH, Krogh TN, Hojrup P, et al. Protein structure of fetal antigen 1 (FA1). A novel circulating human epidermal-growth-factor-like protein expressed in neuroendocrine tumors and its relation to the gene products of dlk and pG2. Eur J Biochem 1994;225:83–92 [DOI] [PubMed] [Google Scholar]
- 12.Nueda ML, García-Ramírez JJ, Laborda J, Baladrón V. dlk1 specifically interacts with insulin-like growth factor binding protein 1 to modulate adipogenesis of 3T3-L1 cells. J Mol Biol 2008;379:428–442 [DOI] [PubMed] [Google Scholar]
- 13.Bray SJ, Takada S, Harrison E, Shen SC, Ferguson-Smith AC. The atypical mammalian ligand Delta-like homologue 1 (Dlk1) can regulate Notch signalling in Drosophila. BMC Dev Biol 2008;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baladrón V, Ruiz-Hidalgo MJ, Bonvini E, Gubina E, Notario V, Laborda J. The EGF-like homeotic protein dlk affects cell growth and interacts with growth-modulating molecules in the yeast two-hybrid system. Biochem Biophys Res Commun 2002;291:193–204 [DOI] [PubMed] [Google Scholar]
- 15.Baladrón V, Ruiz-Hidalgo MJ, Gubina E, Bonvini E, Laborda J. Specific regions of the extracellular domain of dlk, an EGF-like homeotic protein involved in differentiation, participate in intramolecular interactions. Front Biosci 2001;6:A25–A32 [DOI] [PubMed] [Google Scholar]
- 16.Miyaoka Y, Tanaka M, Imamura T, Takada S, Miyajima A. A novel regulatory mechanism for Fgf18 signaling involving cysteine-rich FGF receptor (Cfr) and delta-like protein (Dlk). Development 2010;137:159–167 [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Zhao L, Smas C, Sul HS. Pref-1 interacts with fibronectin to inhibit adipocyte differentiation. Mol Cell Biol 2010;30:3480–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nueda ML, Baladrón V, Sánchez-Solana B, Ballesteros MA, Laborda J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J Mol Biol 2007;367:1281–1293 [DOI] [PubMed] [Google Scholar]
- 19.Moon YS, Smas CM, Lee K, et al. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol 2002;22:5585–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K, Villena JA, Moon YS, et al. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J Clin Invest 2003;111:453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghunandan R, Ruiz-Hidalgo M, Jia Y, et al. Dlk1 influences differentiation and function of B lymphocytes. Stem Cells Dev 2008;17:495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen DC, Jensen CH, Schneider M, et al. MicroRNA-15a fine-tunes the level of Delta-like 1 homolog (DLK1) in proliferating 3T3-L1 preadipocytes. Exp Cell Res 2010;316:1681–1691 [DOI] [PubMed] [Google Scholar]
- 23.Jensen CH, Jauho EI, Santoni-Rugiu E, et al. Transit-amplifying ductular (oval) cells and their hepatocytic progeny are characterized by a novel and distinctive expression of delta-like protein/preadipocyte factor 1/fetal antigen 1. Am J Pathol 2004;164:1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachmann E, Krogh TN, Højrup P, Skjødt K, Teisner B. Mouse fetal antigen 1 (mFA1), the circulating gene product of mdlk, pref-1 and SCP-1: isolation, characterization and biology. J Reprod Fertil 1996;107:279–285 [DOI] [PubMed] [Google Scholar]
- 25.Andersen DC, Andersen P, Schneider M, Jensen HB, Sheikh SP. Murine “cardiospheres” are not a source of stem cells with cardiomyogenic potential. Stem Cells 2009;27:1571–1581 [DOI] [PubMed] [Google Scholar]
- 26.Andersen DC, Petersson SJ, Jørgensen LH, et al. Characterization of DLK1+ cells emerging during skeletal muscle remodeling in response to myositis, myopathies, and acute injury. Stem Cells 2009;27:898–908 [DOI] [PubMed] [Google Scholar]
- 27.Andersen DC, Schrøder HD, Jensen CH. Non-cultured adipose-derived CD45- side population cells are enriched for progenitors that give rise to myofibres in vivo. Exp Cell Res 2008;314:2951–2964 [DOI] [PubMed] [Google Scholar]
- 28.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007;8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 2004;10:12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds A, Anderson EM, Vermeulen A, et al. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA 2006;12:988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol 2003;4:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salomonis N, Hanspers K, Zambon AC, et al. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics 2007;8:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ormerod MG. Flow Cytometry. New York, : Oxford University Press, 2000 [Google Scholar]
- 35.Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G. Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A 2009;75:535–546 [DOI] [PubMed] [Google Scholar]
- 36.Andersen DC, Jensen L, Schrøder HD, Jensen CH. “The preadipocyte factor” DLK1 marks adult mouse adipose tissue residing vascular cells that lack in vitro adipogenic differentiation potential. FEBS Lett 2009;583:2947–2953 [DOI] [PubMed] [Google Scholar]
- 37.Lee YL, Helman L, Hoffman T, Laborda J. dlk, pG2 and Pref-1 mRNAs encode similar proteins belonging to the EGF-like superfamily. Identification of polymorphic variants of this RNA. Biochim Biophys Acta 1995;1261:223–232 [DOI] [PubMed] [Google Scholar]
- 38.Ferrón SR, Charalambous M, Radford E, et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 2011;475:381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu F, Hao X, Zhao H, et al. Delta-like 1 contributes to cell growth by increasing the interferon-inducible protein 16 expression in hepatocellular carcinoma. Liver Int 2010;30:703–714 [DOI] [PubMed] [Google Scholar]
- 40.Li K, Lee LA, Lu X, Wang Q. Fluorogenic “click” reaction for labeling and detection of DNA in proliferating cells. Biotechniques 2010;49:525–527 [DOI] [PubMed] [Google Scholar]
- 41.Li L, Forman SJ, Bhatia R. Expression of DLK1 in hematopoietic cells results in inhibition of differentiation and proliferation. Oncogene 2005;24:4472–4476 [DOI] [PubMed] [Google Scholar]
- 42.Yin D, Xie D, Sakajiri S, et al. DLK1: increased expression in gliomas and associated with oncogenic activities. Oncogene 2006;25:1852–1861 [DOI] [PubMed] [Google Scholar]
- 43.Sánchez-Solana B, Nueda ML, Ruvira MD, et al. The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other’s activities. Biochim Biophys Acta 2011;1813:1153–1164 [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Hudak C, Sul HS. Role of preadipocyte factor 1 in adipocyte differentiation. Clin Lipidol 2010;5:109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]