Abstract
Water-flooded oil reservoirs have specific ecological environments due to continual water injection and oil production and water recycling. Using 16S rRNA gene clone library analysis, the microbial communities present in injected waters and produced waters from four typical water-flooded oil reservoirs with different in situ temperatures of 25°C, 40°C, 55°C and 70°C were examined. The results obtained showed that the higher the in situ temperatures of the oil reservoirs is, the less the effects of microorganisms in the injected waters on microbial community compositions in the produced waters is. In addition, microbes inhabiting in the produced waters of the four water-flooded oil reservoirs were varied but all dominated by Proteobacteria. Moreover, most of the detected microbes were not identified as indigenous. The objective of this study was to expand the pictures of the microbial ecosystem of water-flooded oil reservoirs.
Since 1859, the first oil well drilled near Titusville, Pennsylvania, oil reservoirs have been subjected to exploitation due to the demand of energy requirement saw a worldwide expansion. Multiple groups of microorganisms with diverse physiological characteristics and metabolic abilities and phylogenetic affiliations have gradually been recovered from oil reservoirs1,2,3. Magot3 identified sulfate reduction, fermentation, acetogenesis and methanogenesis as the major metabolic processes carried out by indigenous microbes in oil reservoirs. However, determining whether a microorganism is indigenous to an oil reservoir became increasingly difficult due to the introduction of allochthonous microorganisms and other sources of contaminations by enhanced oil recovery (EOR) processes. The major methods of improved oil recovery are water-flooding, gas injection, chemical flooding, and thermal recovery. All these techniques inevitably cause the change of physical and chemical characteristics and microbial compositions of oil reservoir ecosystems on short or long term bases2,4. Water-flooding, the least expensive and most widely used method for secondary oil recovery, is a technique where freshwater, seawater or recycled reservoir produced water after separation is reinjected into the reservoir formation of wells that have reduced production to displace/push some of the remaining oil toward producing wells in the same reservoir.
Recently, much attention has been paid to the potential applications of microbial enhanced oil recovery (MEOR) techniques, resulting in an increased interest in knowledge of microbial communities in subsurface oil reservoirs1,2,4. Most studies of microbial communities in oil reservoirs have for practical and economical reasons normally been carried out on samples obtained from the oil well production heads, and most of oil reservoirs under study are subjected to water-flooding5,6,7,8. Although the results of these studies, especially those based on culture-independent methods, broaden our knowledge of microbial diversity in oil reservoir's ecosystem and the detection of organisms however, they did not provide sufficient evidence whether these organisms were indigenous to the reservoirs. Water-flooding is believed to be the main contaminating factor to result in the continued reinoculation of reservoir with surface microorganisms as well as the alteration of the geochemistry of the formations9. Some microbes in injected water are thought to possess exceptional survival abilities and could be detected by various culture-dependent and culture-independent techniques long after being introduced to oil reservoirs2. However, detailed studies about the effects of the microbial communities present in injected water on the community in the oil reservoirs are limited. The main reason could be that most oil reservoirs, especially those in China, have been subjected to water-flooding for decades while no data about the microbial communities in these oil reservoirs before water-flooding were available. In order to discover the effects of microbes in injected water on microbial community in oil reservoirs, an alternative investigating strategy of comparison of microbial community compositions in injection and production well samples was applied in the study.
In general, the physical characteristics and chemical composition of oil reservoir environments are the main factors that determine the profile of the microbial communities in the ecosystems. Temperature appears to be the most important limiting factor for microbial growth in oil reservoirs1,3. For oil reservoirs, temperature increases generally with depth at a mean rate of 3°C per 100 m, therefore, different oil reservoirs have different in situ temperatures, which could have great effects on the possibility for the organisms in injected water to survive or thrive in oil reservoirs. In order to gain convictive investigating data of effects of microbes in injected water on microbial communities in oil reservoirs, samples from four water-flooded oil reservoirs with different in situ temperatures were selected in this study.
Microbial diversity assessment based on 16S rRNA gene clone library technique has yielded amazing insights into the community composition in a variety of environments. In this study, after comparing microbial communities in samples of injection and production wells from the four oil reservoirs with different in situ temperatures, microbial communities detected in samples of produced waters were also analyzed. We aim to expand knowledge of microbial ecosystem in oil reservoirs subjected to water-flooding and more importantly provide a significant referencing value for the performance of microbial enhanced oil recovery (MEOR) in theses oil fields.
Results
Microbial community compositions in samples of the injection and production wells from the oil reservoir with an in suit temperature of 25°C
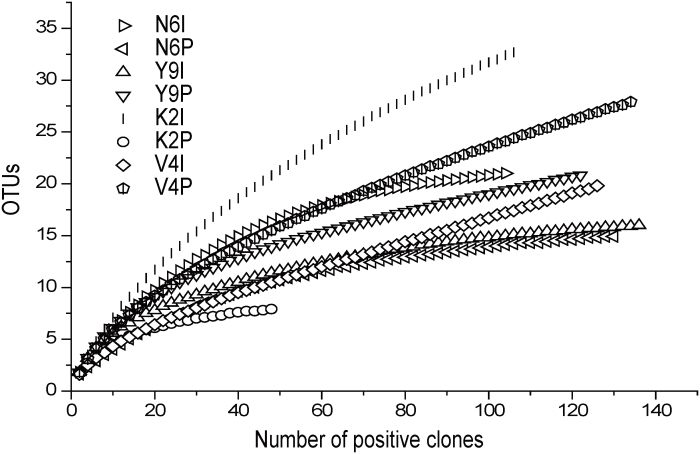
No. 6 oil reservoir, a typical representative of low-temperature oil reservoir with an in suit temperature of 25°C in Xinjiang oil field, was used in the study. Two 16S rRNA gene clone libraries, N6I and N6P, were constructed from wellhead samples of injection well of T6186 and production well of T6073. 21 and 15 OTUs were identified in each clone library according to ARDAR of 104 and 131 positive clones in N6I and N6P, respectively. Rarefaction curves of the two libraries tended to approach the saturation plateau indicating that positive clones in each library could well cover the diversity of bacteria (Fig. 1).
Figure 1. Rarefaction analysis of 16S rRNA gene positive clones for clone libraries of N6I, N6P, Y9I, Y9P, K2I, K2P, V4I and V4P using Analytic Refraction software.
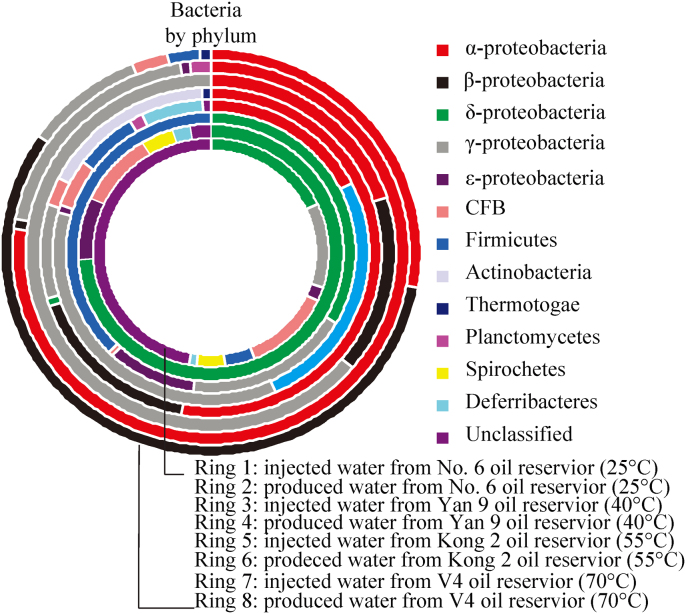
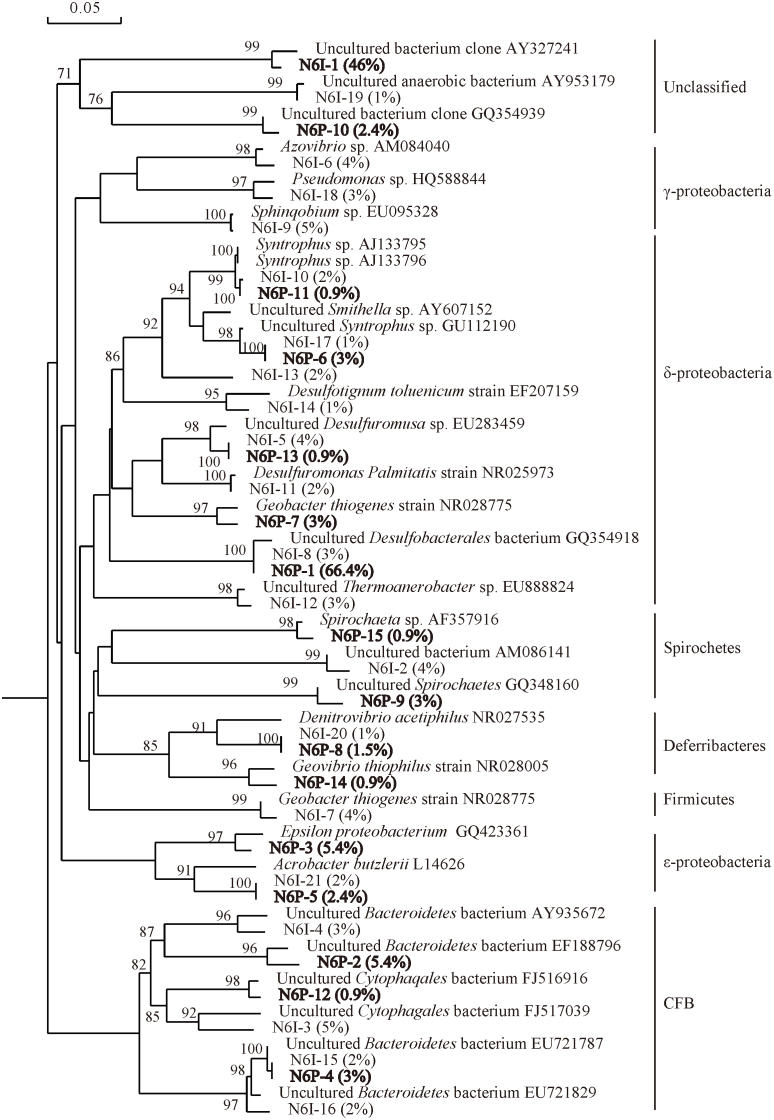
In N6I, 51% of the sequences were identified as unclassified bacteria, the remaining sequences were clustered within 4 phyla: Proteobacteria (32%), detailed in δ- (18%), γ- (12%), and ε- (2%), followed by Cytophaga-Flavobacter-Bacteroidetes (CFB) group (12%), Spirochetes (4%) and Deferribacteres (1%) (Fig. 2). Unlike N6I, only 2.4% sequences in N6P were unclassified, the predominant sequences were affiliated with δ-Proteobacteria (74%) which is followed by CFB (9.3%), Spirochetes (3.9%), Deferribacteres (2.4%) (Fig. 2). According to the results of phylogenetic affiliations obtained from algorithm of BLAST of nucleotide, 81% sequences in N6I and 94.6% sequences in N6P had a relatively low similarity (< 96%) to the sequences in GenBank database, which indicated that the oil reservoir harbours unique microbial communities comprising several unknown genera or species. In N6I (Fig. 3), the predominant group were the unknown bacterium (46%), and the other bacterial groups were uncultured CFB group (12%), Sphinqobium sp. (5%), Syntrophus sp. (5%), Desulfuromusa sp. (4%), Azovibrio sp. (4%), Geobacter sp. (4%), uncultured Desulfobacterales bacterium (3%), Pseudomonas sp. (3%), Desulfuromonas sp. (2%), Acrobacter sp. (2%), Denitrovibrio sp. (1%). In N6P, uncultured Desulfobacterales bacterium was predominant at 66.4%, followed by uncultured CFB group (9.3%), Syntrophus sp. (3.9%), Spirochaetes sp. (3%), Acrobacter sp. (2.4%) and Denitrovibrio sp. (1.5%) (Fig. 3). Obviously, the microbial communities in N6I and N6P appeared different, but even so, there were 5 common phyla, 5 common families, and 7 common existing OTUs in N6I and N6P (Table 1).
Figure 2. Bacterial population dynamics of the injected waters and produced waters collected from the four water-flooded oil reservoirs as revealed by 16S rDNA clone libraries.
Samples from inner to outer of the ring chart were injected water and produced water from the oil reservoir with high temperature (70°C) to the oil reservoir with low temperature (25°C).
Figure 3. Phylogenetic tree of bacteria 16S rDNA phylotypes of N6I and N6P (25°C).
The tree constructed was based on approximately 800bp 16S rRNA genes and their nearest clones retrieved from the GenBank database. The percentages behind the OTUs represent the relative abundance of each OTU. Bootstrap values are indicated at branch points. The scale bar represents 5% estimated sequence divergence.
Table 1. Co-existing families and the relative abundance of each family in the clone libraries created using the samples of injected water and produced water from each oil reservoir and the number of co-existing OTUs.
| Relative abundance (%) | ||||
|---|---|---|---|---|
| Co-existing Plylum | Co-existing Family | IW (OTUS) | PW (OTUS) | Co-existing OUT |
| N6 (25°C) | ||||
| Delta | Syntrophaceae | 5 (3) | 3.9 (2) | 2 |
| Desulfuromonadaceae | 6 (2) | 0.9 (1) | 1 | |
| Uncultured | 3 (1) | 66.4 (1) | 1 | |
| Desulfobacterales | ||||
| Epsilon | Campylobacteraceae | 2 (1) | 2.4 (1) | 1 |
| CFB | Cytophaqales | 5 (1) | 0.9 (1) | |
| Bacteroidetes | 7 (3) | 8.4 (2) | 1 | |
| Spirochetes | 4 (1) | 3.9 (2) | ||
| Deferribacteres | Deferribacteraceae | 2.4 (2) | 1 (1) | 1 |
| Unclassified | 47 (2) | 2.4 (1) | ||
| Total relative abundance (%) | 81.4 | 90.2 | 7 | |
| Y9 (40°C) | ||||
| Delta | Desulfuromonadaceae | 28.9 (1) | 0.9 (1) | |
| Gamma | Pseudomonadaceae | 15.5 (2) | 32.5 (1) | |
| Epsilon | ||||
| CFB | ||||
| Firmicutes | ||||
| Total relative abundance (%) | 44.5 | 33.4 | ||
| K2 (55°C) | ||||
| Alpha | Rhizobiaceae | 2.8 (1) | 6 (1) | |
| Beta | ||||
| Gamma | Xanthomonadaceae | 0.9 (1) | 2 (1) | |
| Pseudomonadaceae | 6.4 (5) | 62 (4) | ||
| Total relative abundance (%) | 10.1 | 70 | ||
| V4 (70°C) | ||||
| Alpha | Rhodobacteraceae | 0.8 (1) | 0.8 (1) | |
| Beta | ||||
| Gamma | Pseudomonadaceae | 0.8 (1) | 8.5 (7) | |
| Total relative abundance (%) | 1.6 | 9.3 | ||
Microbial community compositions in samples of the injection and production wells from the oil reservoir with an in suit temperature of 40°C
Water-oil samples from injection well of L28-45 and production well of L28-46 in Yan 9 oil reservoir with an in suit temperature of 40°C were used in the study. Two 16S rRNA gene clone libraries, Y9I and Y9P, were constructed. According to ARDAR of 137 and 124 positive clones, 15 and 21 OTUs were identified in Y9I and Y9P, respectively. Rarefaction curves of the two libraries showed that the positive clones in each library could well cover the diversity of bacteria (Fig. 1).
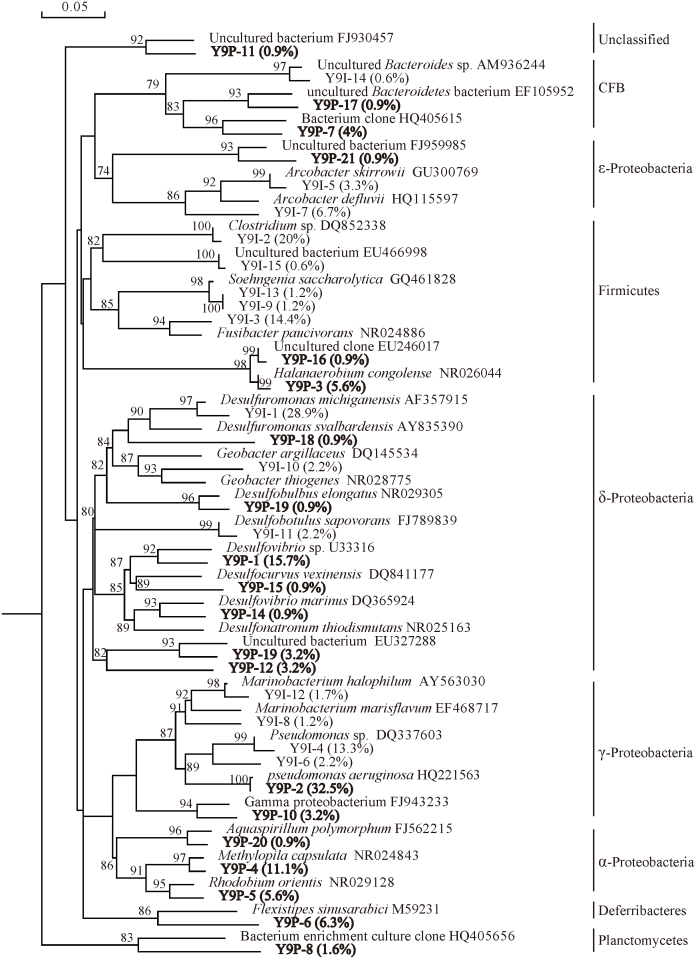
In Y9I, detected bacteria were clustered mainly within two phyla: Proteobacteria (61.7%) and Firmicutes (37.4%). Among Proteobacteria, δ-, γ-, and ε- Proteobacteria represented 33.3%, 18.4% and 10%, respectively. In Y9P, Proteobacteria predominated at 79.9%, including α- (17.6%), δ- (25.7%), γ- (35.7%), and ε- (0.9%), followed by Firmicutes (6.5%), Deferribacteres (6.3%), CFB (4.9%) and Planctomycetes (1.6%) (Fig. 4).
Figure 4. Phylogenetic tree of bacteria 16S rDNA phylotypes of Y9I and Y9P (40°C).
The tree constructed was based on approximately 800bp 16S rRNA genes and their nearest clones retrieved from the GenBank database. The percentages behind the OTUs represent the ralative abundance of each OTU. Bootstrap values are indicated at branch points. The scale bar represents 5% estimated sequence divergence.
55.6% sequences in Y9I and 50.8% sequences in Y9P were identified as unknown bacteria due to their low similarity (<96%) with reference sequences in GenBank database. The dominant bacteria groups in Y9I were affiliated with Desulfuromonas sp. (29.8%), Clostridium sp. (20%), Fusibacter sp. (14.4%) and Pseudomonas sp. (13.3%). However in Y9P, dominant bacteria were Pseudomonas aeruginosa (32.5%), Desulfovibrio sp. (15.7%), Methylopila sp. (11.1%), Flexistipes sp. (6.3%) (Figure 4). Although there were 5 co-existing phyla, microbial communities in wellhead samples of the injection well and the production well appeared divergent widely with only 2 common families, (Desulfuromonadaceae and Pseudomonadaceae) and no common OTU (Table 1).
Microbial community compositions in samples of the injection and production wells from the oil reservoir with an in suit temperature of 55°C
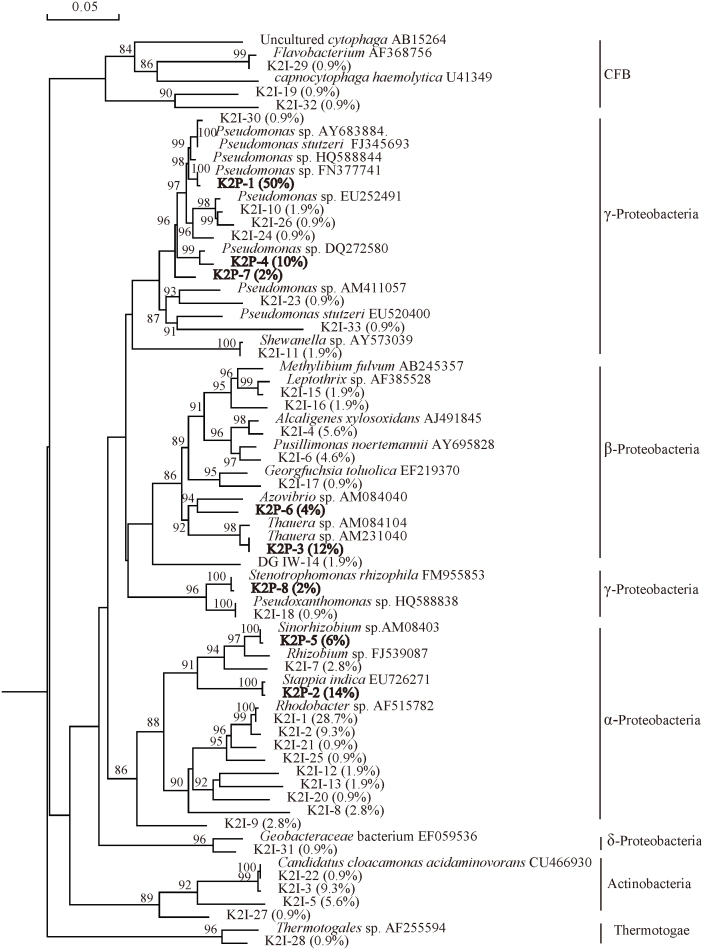
Water-oil samples from injection well of 1098-8 and production well of 1017-7 in Kong 2 oil reservoir with an in suit temperature of 55°C were used in the study. According to ARDAR of 108 and 50 positive clones in clone libraries of K2I and K2P, 33 and 8 OTUs were identified in each clone library. Rarefaction curves of the two libraries indicated that the positive clones in each library could well cover the diversity of bacteria (Fig. 1). The predominant phylum in K2I was Proteobacteria predominated at 79.8%, encompassing α- (52.9%), β- (16.8%), δ- (0.9%), γ- (9.2%). The other phyla in K2I were Actinobacteria (16.7%), CFB (2.7%) and Thermotogae (0.9%). In K2P there was only one phylum of Proteobacteria, including α- (20%), β- (16%), and γ- (64%) (Fig. 2).
92% and 91.8% sequences in K2I and K2P were not affiliated with specific genera. The dominant populations in K2I were Rhodobacter sp. (47.3%), followed by Candidatus sp. (16.7%), Alcaligenes sp. (5.6%), Pseudomonas sp. (6.4%), Pusillimonas sp. (4.6%). In K2P, Pseudomonas sp. covered a major percentage of 62%, which was followed by Stappia sp. (14%), Thauera sp. (12%), Sinorhizobium sp. (6%), Azovibrio sp. (4%) and Stenotrophomonas sp. (2%) (Fig. 5). The results revealed that the microbial communities in wellhead samples from the injection well and the production well were different. In addition, the co-existing common phyla were α-, β- and γ- Proteobacteria, and the co-existing common families were Rhizobiaceae, Xanthomonadaceae and Pseudomonadaceae. There were no co-existing common OTU (Table 1).
Figure 5. Phylogenetic tree of bacteria 16S rDNA phylotypes of K2I and K2 (55°C).
The tree constructed was based on approximately 800bp 16S rRNA genes and their nearest clones retrieved from the GenBank database. The percentages behind the OTUs represent the ralative abundance of each OTU. Bootstrap values are indicated at branch points. The scale bar represents 5% estimated sequence divergence.
Microbial community compositions in samples of the injection and production wells from the oil reservoir with an in suit temperature of 70°C
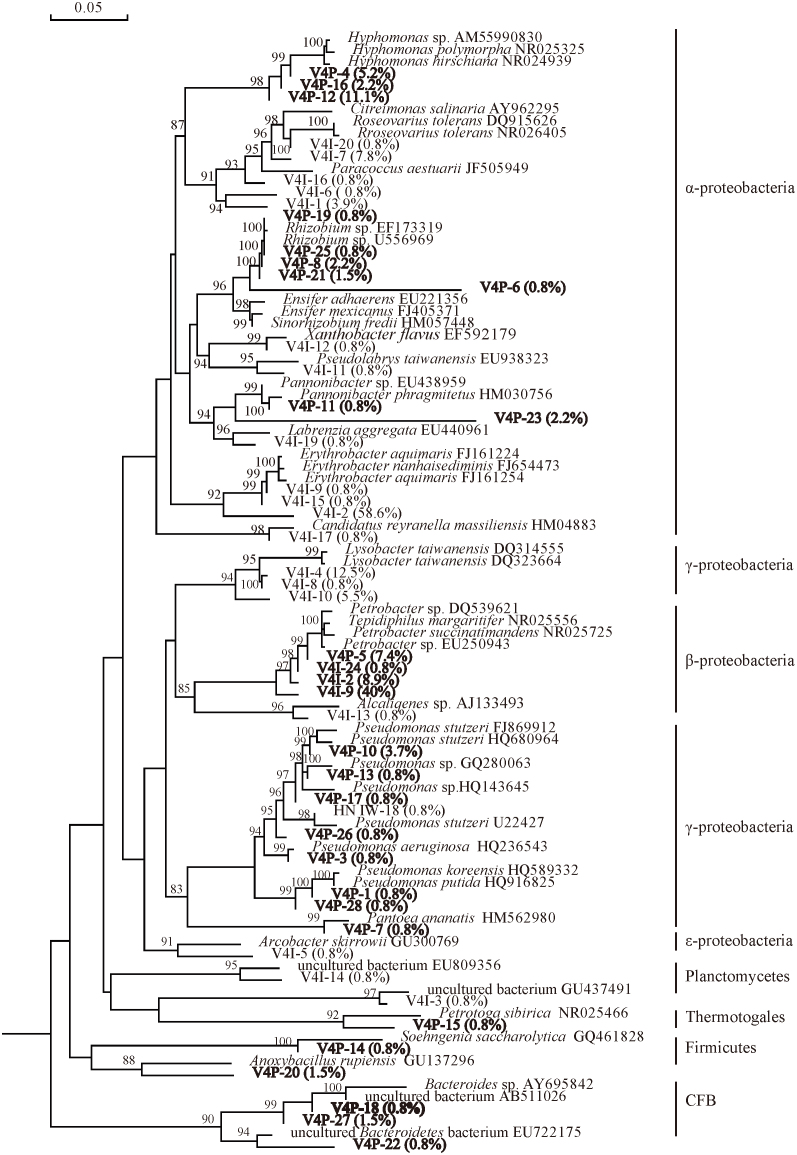
Water-oil samples from injection well of V187 and production well of V149 in V4 oil reservoir with an in suit temperature of 70°C were used in the study. According to ARDAR of 128 and 135 positive clones, two 16S rRNA gene clone libraries, V4I and V4P were constructed with 20 and 28 OTUs in each clone library. Rarefaction curves of the two libraries also indicated that the positive clones in each library could well cover the diversity of bacteria (Fig. 1). The predominant phylum in V4I was Proteobacteria representing 98.7% including α- (77.5%), β- (0.8%), γ- (19.6%) and ε- (0.8%). Another phylum in V4I was Planctomycetes (1.6%). In V4P, Proteobacteria also were predominant at 94.8% including α- (27.6%), β- (57.1%), γ- (9.3%). The other phyla detected were CFB (3.1%), Firmicutes (2.3%) and Thermotogae (0.8%) (Fig. 2).
The microbial communities in the samples of the injection well and production well of the oil reservoir were also different. In the sample of the injection well, Erythrobacter sp. predomenated at 60.2%, which was followed by Lysobacter sp. (18.8%), Roseovarius sp. (8.6%). In the sample of the production well, the dominant bacterial populations included Petrobacter sp. (57.1%), Hyphomonas sp. (18.5%), Pseudomonas sp. (8.5%), Rhizobium sp. (5.9%) (Fig. 6). In V4I and V4P, 96.8% and 90% sequences were not affiliated with a specific genera, the common co-existing phyla were α-, β- and γ- Proteobacteria while the common co-existing families were Rhodocyclaceae and Pseudomonadaceae and no common co-existing OTU were detected (Table 1).
Figure 6. Phylogenetic tree of bacteria 16S rDNA phylotypes of V4I and V4P (70°C).
The tree constructed was based on approximately 800bp 16S rRNA genes and their nearest clones retrieved from the GenBank database. The percentages behind the OTUs represent the ralative abundance of each OTU. Bootstrap values are indicated at branch points. The scale bar represents 5% estimated sequence divergence.
Discussion
Water-flooding procedure is always regarded as an important source affecting native-microbial communities in oil reservoirs2,10. However, surveys investigating how microbial communities in production wells are affected by microorganisms in injected water are limited. In this study, microbial communities in samples of the injection wells and their adjacent production wells in four typical oil reservoirs with different in situ temperatures were investigated to explore this topic systematically. The results showed that microbial communities in samples of the injection well and the production well in each oil reservoir, under study, were different. The increase of in situ temperature of the oil reservoir exacerbated the difference in microbial communities in samples of the injection and production wells. A reduction in the number of common co-existing families and OTUs in the samples collected from the injection and production wells of the oil reservoirs with higher temperatures was observed.
Several previous studies on microbial communities of oil reservoirs also pointed out significant difference between the microbial community structure in samples of injection and production wells. Ren et al.11 reported that only 2 co-existing OTUs appeared in the samples of the injection and production wells of a high-temperature oil reservoir (69°C). In our study, analogously, there were no co-existing OTU in samples of injection and production wells of oil reservoirs (K2 and V4) with high temperatures; and at family level, only 9.3% and 1.6% sequences in clone libraries of the injection wells appeared in the clone libraries of the production wells in oil reservoirs of K2 (55°C) and V4 (70°C), respectively (Table 1). In the low-temperature oil reservoir (25°C) although microbial communities were different, there were 5 co-existing families, and 7 co-existing OTUs in the samples of the injection well and the production well. At family level, 81.4% sequences in the sample of the injection well appeared in the sample of the production well (Table 1). She et al.12 reported similarities between an injection well and two production wells in a low-temperature oil reservoir of 30% and 20%, respectively, using Unweighted Pair Group Method Clustering (UPGMC).
For the mesothermal Yan 9 oil reservoir (40°C) in our study, there was no co-existing OTU in samples of the injection and production well, but 44.5% sequences, at family level, in the sample of the injection well appeared in the sample of the production well (Table 1). Therefore, although microbial communities in injection wells appeared different to those in the production wells, the effects of the microorganisms in injected water on microbial community compositions in oil reservoirs remained ; and the in situ temperatures of oil reservoirs was a crucial factor for the degrees of effect, the higher the in situ temperature, the less such effects were.
The results show that bacterial communities in the samples of produced water were different to those detected in samples of the injection wells in the four oil reservoirs. However, great care has to be taken when drawing inferences concerning in situ microbes of oil reservoirs in produced water. An investigation carried out by Dahle et al.10 indicated that microorganisms in produced water might only to a small extent reflect the indigenous microorganisms in the respective oil formations, even when the formations are not water-flooded. Considering the difficulty of judging the indigenous nature of encountered bacteria and the specific environments of water-flooded oil reservoirs, we classified the detected bacteria in the produced water samples of the study and compared them with those previously reported in samples from oil reservoirs to expose microbial communities inhabiting oil reservoirs subjected to water-flooding in China.
Alphaproteobacteria appeared in samples of production wells from Yan 9 (40°C), Kong 2 (55°C) and V4 (70°C) oil reservoirs at a predominance level of 17.6%, 20% and 27.8%, respectively (Fig. 2). Among all the encountered members belonging to Alphaproteobacteria, sequences of one OTU detected in the sample from the Kong 2 oil reservoir were affiliated with the genus of Stappia. The Stappia-related clones were reported to be detected in oil phase separated from produced oil-water sample from a mesothermic oil field13, while the remaining Alphaproteobacteria were never reported to be isolated or detected in oil reservoirs. Collectively, members belonging to the order of Rhizobiales (Methylopila sp., Rhodobium sp., Sinorhizobium sp. and Rhizobium sp.) appeared in the three produced water samples and indicating that Rhizobiales might be a common Alphaproteobacteria inhabiting water-flooded oil reservoirs. The second dominant microbial population affiliated with the Hyphomonas sp., in the produced water sample from V4 oil reservoir was often reported in marine environments14,15 and seldom detected in oil reservoirs.
Betaproteobacteria appeared in samples of production wells from Kong 2 (55°C) and V4 (70°C) oil reservoirs. All the detected groups of Betaproteobacteria were members belonging to the family of Rhodocyclaceae genera of Thauera and Azovibrio in the sample from Kong 2 oil reservoir and members related to Petrobacter sp. in the sample from V4 oil reservoir. Thauera sp. was seldom found in samples from oil reservoirs, recently, Thauera –related phylotype was only detected in oil fields injected with nitrate16,17. Petrobacter-affiliated populations were predominant at 57.1% in the produced water from V4 oil reservoir. This genus was isolated and detected in samples from higher temperature oil reservoirs18,19, suggesting that they may belong to the natural community in higher temperature oil-bearing formations.
Microbial groups belonging to Deltaproteobacteria covered an overwhelming percentage of 74.2% in the lower temperature oil reservoir No. 6, and 25.7% in the mesothermic oil reservoir of Yan 9. The detected sequence affiliated with uncultured Desulfobacterales bacterium, that is a common order detected in oil reservoirs2, were predominant at 66.4% abundance in the produced water from oil reservoir of No. 6. Moreover, a co-existing OTU of the uncultured Desulfobacterales bacterium appeared in the sample of the injected water. In addition, bacteria related to Syntrophus sp. were also found in the in the produced water from oil reservoir of No. 6. The genus of Syntrophus was recently demonstrated to be important in processes that are happening in oil reservoirs20,21 and members of this genus were also identified in enrichments derived from an oil reservoir in Canada22. However they could not be considered indigenous because of a co-existing OTU detected in the sample of the injected water.
Members of the genus of Desulfovibrio appeared in produced water samples from mesothermic oil reservoir of Yang 9 (40°C). Desulfovibrio was reported to be a major mesophilic genus that is recovered frequently from oil fields23,24,25. Although no clone affiliated to the genus was found in the injected water sample, the indigenous nature of the detected Desulfovibrio-related bacteria remained unidentified without further culture-dependent assessment.
Members represented by Geobacter-related and Desulfuromonas-related phylotypes, belonging to the order of Desulfuromonadales, were found in both produced and injected water samples in both oil reservoirs. Notably, in the samples of the produced water, these bacteria were prevailing, suggesting that bacteria affiliated with Desulfuromoas sp. and Geobacter sp. were exogenous in the both oil reservoirs No. 6 and Yan 9.
Bacteria affiliated with Pseudomonas sp. phylum Gammaproteobacteria were often detected in oil reservoirs7,8,26. We detected these bacteria in all reservoirs, except for the lower temperature of No. 6 oil reservoir. . In samples of produced water from oil reservoirs of Yan 9 and Kong 2, Pseudomonas sp. was at 32.5% and 62%. Pseudomonas, however were not believed to originate from oil reservoirs. The fact that these mesophilies were dominant at high temperature oil reservoir Kong 2 and the higher temperature oil reservoir of V4 implied that water flooded oil reservoirs were open and recycling systems and some microbial communities detected in produced water might come from the cooler portions near wellbores7,8.
Two OTUs, accounting for 7.8% of the clone library constructed from the produced water from low temperature oil reservoir No. 6 (25°C), were clustered within the Epsilonproteobacteria. Sequences of one OTU were related to uncultivated species and sequences of the other OTU similar to Arcobacter sp. which have been previously reported in clone libraries from oil field produced water27, and to be the dominant microbial group in non-water-flooded low-temperature oil reservoir5. However, with the appearance of the Arcobacter-related sequences in injected water samples, the bacteria involved in situ were still unknown.
CFB-affiliated sequences, detected in the produced sample from low temperature oil reservoir No. 6, were generally distantly related to known cultivated species, the results were similar to that reported by Grabowski et al.5. However, most of the sequences were close to sequences that were also retrieved from a mesothermic petroleum reservoir28. In the water samples of the mesothermic oil reservoir Kong 2 and the higher temperature oil reservoir V4, bacteria belonging to CFB group, accounted to 2.7% and 3.1%, respectively. Similarity, most of them were uncultured but related to environmental sequences associated with oil reservoirs26,28,29. Although members of CFB group were detected widely in samples collected from oil reservoirs, their ecological significance and indigenous nature are not clear due to their uncultured characteristics, which underlined the need to isolate and characterize pure cultures of these microorganisms.
Bacteria affiliated with Halanaerobium sp. detected in produced water sample of Yan 9 oil reservoir were closely related to strains isolated from an African oil field30. Interestingly, although non Halanaerobium-related sequence was found in the injected water sample of Yan 9 oil reservoir, sequences belonging to other genera of Firmicutes (Clostridium, Fusibacter and Soehngenia) occupied a dominant ralative abundance (37.4%) in the injected sample, which indicated that the subsurface strata might have an anaerobic high-salt condition.
Sequences of the genera of Denitrovibrio and Geovibrio, covered limited percentages of 1.5% and 0.9% the clone library of produced water from the low temperature oil reservoir No. 6. Denitrovibrio-related bacteria isolated from reservoirs appeared the ability to reduce nitrate by denitrification17,31. In the produced water sample from the mesochermic oil reservoir Yan 9, harboured Flexistipes-related bacteria that were closely similar with the isolate originated from a multiply extreme environment with high temperature, high salinity, and high concentrations of heavy metals32, which further confirmed that the Yan 9 oil reservoir might be anaerobic and high insalt content.
Sequences affiliated with the phylum of Spirochetes were only detected at low temperature oil reservoir of No. 6. One OTU was closely similar to the phylotype present in the production waters of a low-temperature biodegraded oil reservoir5.
Among the 28 OTUs in clone library of the sample from the higher-temperature V4 oil reservoir, only one OTU (V4P-15), occupying only 0.8% ralative abundance, is relatively distantly related to Petrotoga sibirica (92%). Petrotoga species are considered as typically indigenous since it has been isolated only from oil reservoirs so far33,34.
In conclusion, bacteria inhabiting in the produced waters of the four water-flooded oil reservoirs in China were dominated by Proteobacteria. At low-temperature oil reservoir No 6, δ-Proteobacteria was prevailing while at high-temperature oil reservoirs Kong 2 and V 4, α-, β-, γ-Proteobacteria were dominant. In mesothermic oil reservoir Yan 9, α-, δ- and γ-Proteobacteria were foremost. Most detected bacteria could not be identified as indigenous. In the study, we just focused on bacteria inhabiting in oil reservoirs without involving archaea that will be investigated in our following study. Apparently, water-flooded oil reservoirs are changing environments, harboring groups of surviving bacteria indigenous and exogenous that form new ecological systems different to the original ones. The identification of microbes present in produced waters within water-flooded oil reservoirs reflects the inhabiting microbial communities which are of great importance to ecological study of the specific environments of water-flooded oil reservoirs.
Methods
Sample collection
We obtained permission from the Institute of Oil Recovery Research of Xinjiang, Changqing, Dagang and Hennan Oil Field Ltd. for data acquisition and field studies which did not involve endangered and protected species. Eight wellhead samples of injection wells and production wells from the four oil reservoirs were collected. All the oil reservoirs under study are subjected to water-flooding. The detail information about the oil reservoirs and sampling is shown in Table 2. The water-oil samples were retrieved from wellheads and stored in sterile plastic bottles and transported to the laboratory as soon as possible for cultural and molecular analyses.
Table 2. The locations and characteristics of injection and production wells from the four oil fields in China used in this study.
| Oil Field | Location | Oil Reservior | Wells | Waterfloodig Date | Sampling Date | Depth (m) | In situ Temperature |
|---|---|---|---|---|---|---|---|
| Xinjiang | Junggar basin | No. 6 | T6186 | 1974 | 10/09/2009 | 800 | 25°C |
| T6073 | |||||||
| Changqing | Ordos Basin | Yan 9 | L28-45 | 1999 | 03/05/2010 | 1289 | 40°C |
| L28-46 | |||||||
| Dagang | Bohai Sea coast | Kong 2 | 1098-8 | 1979 | 06/08/2004 | 1400 | 55°C |
| 1017-7 | |||||||
| Henan | Henan Province | V4 | V187 | 1977 | 05/09/2010 | 1355 | 70°C |
| V149 |
16S rRNA gene amplification, library construction and sequencing
For water-oil samples retrieved from wellheads, in the study, 100 ml water sample is an optimal amount for cell collection (10,000 g, 10 min). DNA in the pellet cells were extracted following the manufacturer's protocol for the FastDNA Spin Kit for Soil (Qbiogen, Carlsbad, CA. USA). Obtained DNA was detected by agarose gel electrophoresis.
The bacterial 16S rDNA in the bulk DNA was amplified using the universal bacteria-specific primers, 27F and 1492R35. The 25μl PCR reacting system contains a 2.5μl of 10×PCR buffer (Mg2+ plus), 10 nmol of deoxynucleotide triphosphates, 1 U Taq DNA polymerase (TakaRa), 10 pmol of each primer, and 1μl of template DNA. The thermal cycling conditions were as follows: an initial denaturation at 94°C for 5 min, 40 cycles of 94°C for 30 sec, 56°C for 60 sec, 72°C for 90 sec, and a final extension step of 72°C for 10 min26,36. Amplified fragments were approximately 1450 bp.
After purified with an Agarose Gel DNA Purification Kit, amplicons of 16S rRNA genes were (TianGen Biotech, Beijing, China) cloned into Trans1-T1 competent cells (TransGen Biotech, Beijing, China) using PGEM-T Easy Vector (Promega, Madison, WI, USA) as instructed by the suppliers. Randomly chosen one hundred and fifty putative clones (white) from each plate were transferred to another labelled LB plate with ampicillin (100μg/ml) and cultured at 37°C overnight. A re-amplification, with sets of vector-specific primers T7/SP6, was taken to determine positive clones. During the re-amplification procedure, a small quantity of cells of putative clones was used as template DNA in the reaction mixtures. PCR products of positive clones were classified into different operational taxonomic units (OTUs) using amplified ribosomal DNA restriction analysis (ARDRA) with HinfI and HaeIII (TaKaRa)26,36. Representative clones were selected for sequencing which was performed using an ABI PRISM 3730 DNA sequencer (SinoGenoMax Co., Ltd., Beijing, China). Clone libraries were statistically evaluated by rarefaction analysis using Analytic Rarefaction 1.3 (http://www.uga.edu/strata/software/Software.html) software.
Taxonomic classification and phylogenetic analysis of 16S rRNA gene sequences
The obtained sequences were manually trimmed and edited using DNAMAN version 5.2.2.0. The obtained sequences were submitted to the GenBank database of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) employing the Basic Local Alignment Search Tool (BLAST) algorithm of nucleotide to determine their phylogenetic affiliations. The sequences being the nearest neighbours of the submitted sequences, included cultured and uncultured sequences, were cited from the GenBank. Sequences, sequenced and cited, were aligned using DNAMAN software to construct phylogenetic trees with maximum likelihood.
Nucleotide sequence accession numbers
16S rRNA gene sequences, submitted to the GenBank, have been assigned accession numbers: JQ519726-JQ519747 (clone library of N6I), JQ519802-JQ519816 (clone library of N6P), JQ519787-JQ519801 (clone library of Y9I), JQ519748-JQ519767 (clone library of Y9P), AY770941-AY770937 (clone library of K2I), AY770933-AY770940 (clone library of K2P), JQ519768-JQ519786 (clone library of V4I) and JQ519699-JQ519726 (clone library of V4P).
Author Contributions
Experiments were conceived and designed by Y.H.S., F.Z. and D.J.H. and performed by F.Z., L.J.C. and X.T.Z. Data were analyzed by F.Z., L.J.C., I.M.B., X.T.Z., F.C.S., Z.L.W. and L.J.Y. The manuscript was written by F.Z.
Acknowledgments
This study was sponsored by the 863 Program (No. 2008AA06Z204) of the Ministry of Science and Technology and the National Natural Science Foundation (No. 50974022 and No. 51074029) in P. R. China. The authors wish to thank the technical personnel in the oil field under study, for kindly collecting samples and providing geological information of the oil reservoirs.
References
- Magot M. Indigenous microbial communities in oil fields. In: Ollivier B., and Magot M., eds. , editors. Petroleum microbiology. ASM, Washington, DC. pp. 21–34 (2005).
- Youssef N., Elshahed M. S. & McInerney M. J. Microbial process in oil field: culprits, problems, and opportunities. Adv Appl Microbiol 66, 141–251 (2009). [DOI] [PubMed] [Google Scholar]
- Magot M., Ollivier B. & Patel B. K. C. Microbiology of petroleum reservoirs. Antonie van Leeuwenhoek 77, 103–116 (2000). [DOI] [PubMed] [Google Scholar]
- Basso O., Lascourreges J. F., Jarry M. & Magot M. The effect of cleaning and disinfecting the sampling well on the microbial communities of deep subsurface water samples. Environ Microbiol 7, 13–21 (2005). [DOI] [PubMed] [Google Scholar]
- Grabowski A., Nercessian O., Fayolle F., Blanchet D. & Jeanthon C. Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol Ecol 54, 427–443 (2005). [DOI] [PubMed] [Google Scholar]
- Kaster K. M., Bonaunet K., Berland H., Kjeilen-Eilertsen G. & Brakstad O. G. Characterisation of culture-independent and –dependent microbial communities in a high-temperature offshore chalk petroleum reservoir. Antonie van Leeuwenhoek 96, 423–39 (2009). [DOI] [PubMed] [Google Scholar]
- Li H., Yang S-Z., Mu B-Z., Rong Z-F. & Zhang J. Molecular analysis of the bacterial community in a continental high-temperature and water-fooded petroleum reservoir. FEMS Microbiol Lett 257, 92–98 (2006). [DOI] [PubMed] [Google Scholar]
- Orphan V. J., Taylor L. T., Hafenbradl D. & Delong E. F. Culture-dependent and culture-independent characterization of microbial assemblages associated with high temperature petroleum reservoirs. Appl Environ. Microbiol 66, 700–711 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance I. & Thrasher D. R. In: B. Ollivier and M. Magot, editors. Petroleum Microbiology. Reservoir souring: Mechanisms and prevention. ASM, Washington, DC. pp. 103–142 (2005).
- Dahle H., Garshol F., Madsen M. & Birkeland N. K. Microbial community structure analysis of produced water from a high-temperature North Sea oil-field. Antonie van Leeuwenhoek 93, 37–49 (2008). [DOI] [PubMed] [Google Scholar]
- Ren H. Y. et al. Comparison of Microbial Community Compositions of Injection and Production Well Samples in a Long-Term Water-Flooded Petroleum Reservoir. PloS One 6(8), e23258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Y. H., Zhang F., Xiang T. S., Liu B. B. & Zhao, L. P. Microbial diversity in petroleum reservoirs analyzed by PCR-DGGE. Acta Ecologica Sinica 25, 237–242 (2005). [Google Scholar]
- Kryachko Y., Dong X., Sensen C. W. & Voordouw G. Compositions of microbial communities associated with oil and water in a mesothermic oil field. Antonie Van Leeuwenhoek 101, 493–506 (2010). [DOI] [PubMed] [Google Scholar]
- Kang H. S. & Lee S. D. Ponticaulis koreensis gen. nov., sp. nov., a new member of the family Hyphomonadaceae isolated from seawater. Int J Syst Evol Microbiol 59, 2951–2955 (2009). [DOI] [PubMed] [Google Scholar]
- Maeda R., Nagashima H., Widada J., Iwata K. & Omori T. Novel marine carbazole-degrading bacteria. FEMS Microbiol Lett 292, 203–209 (2009). [DOI] [PubMed] [Google Scholar]
- Hubert C. & Voordouw G. Oil field souring control by nitrate-reducing Sulfurospirillum spp. that outcompete sulfate-reducing bacteria for organic electron donors. Appl Environ. Microbiol 73, 2644–2652 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shartau S. L. et al. Ammonium concentrations in produced waters from a mesothermic oil field subjected to nitrate injection decrease through formation of denitrifying biomass and anammox activity. Appl Environ Microbiol 76, 4977–4987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas M. B. et al. Petrobacter succinatimandens gen. nov., sp. nov., a moderately thermophilic, nitrate-reducing bacterium isolated from an Australian oil well. Int J Syst Evol Microbiol 54, 645–649 (2004). [DOI] [PubMed] [Google Scholar]
- Yamane K. et al. Diversity and similarity of microbial communities in petroleum crude oils produced in Asia. Biosci Biotechnol Biochem 72, 2831–2839 (2008). [DOI] [PubMed] [Google Scholar]
- Jones D. M. et al. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 451, 176–180 (2008). [DOI] [PubMed] [Google Scholar]
- Gray N. D. et al. The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ Microbiol 13, 2957–2975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski A., Blanchet D. & Jeanthon C. Characterization of long-chain fattyacid- degrading syntrophic associations from a biodegraded oil reservoir. Res Microbiol 156, 814–821 (2005). [DOI] [PubMed] [Google Scholar]
- Birkeland N. K. Sulfate-reducing bacteria and archaea. In “Petroleum Microbiology” (B. Ollivier and M. Magot, Eds.), pp. 35–54. ASM, Washington, DC (2005).
- Magot M. et al. Desulfovibrio longus sp. nov., a sulfate-reducing bacterium isolated from an oil-producing well. Int J Syst Bacteriol 42, 398–403 (1992). [DOI] [PubMed] [Google Scholar]
- Magot M., Basso O., Tardy-Jacquenod C. & Caumette P. Desulfovibrio bastinii sp. nov. and Desulfovibrio gracilis sp. nov., moderately halophilic, sulfate-reducing bacteria isolated from deep subsurface oilfield water. Int J Syst Evol Microbiol 54, 1693–1697 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang F. et al. Impact of an indigenous microbial enhanced oil recovery field trial on microbial community structure in a high pour-point oil reservoir. Appl Microbiol Biotechnol 95, 811–821 (2012). [DOI] [PubMed] [Google Scholar]
- Voordouw G. et al. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl Environ. Microbiol 62, 1623–1629 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V. D. et al. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11, 176–87 (2009). [DOI] [PubMed] [Google Scholar]
- Shimizu S., Ueno A. & Ishijima Y. Microbial communities associated with acetate-rich gas-petroleum reservoir surface facilities. Biosci Biotechnol Biochem 75, 1835–1837 (2011). [DOI] [PubMed] [Google Scholar]
- Ravot G. et al. Haloanaerobium congolense sp. nov., an anaerobic, moderately halophilic, thiosulfate- and sulfur-reducing bacterium from an African oil field. FEMS Microbiol. Lett 147, 81–88 (1997). [DOI] [PubMed] [Google Scholar]
- Myhr S. & Torsvik T. Denitrovibrio acetiphilus, a novel genus and species of dissimilatory nitrate-reducing bacterium isolated from an oil reservoir model column. Int J Syst Evol Microbiol 50, 1611–1619 (2000). [DOI] [PubMed] [Google Scholar]
- Lapidus A. et al. Genome sequence of the moderately thermophilic halophile Flexistipes sinusarabici strain (MAS10). Stand Genomic Sci 15, 86–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Tello E. et al. Petrotoga mexicana sp. nov., a novel thermophilic, anaerobic and xylanolytic bacterium isolated from an oil-producing well in the Gulf of Mexico. Int J Syst Evol Microbiol 54, 169–174 (2004). [DOI] [PubMed] [Google Scholar]
- Miranda-Tello E. et al. Petrotoga halophila sp. nov., a thermophilic, moderately halophilic, fermentative bacterium isolated from an offshore oil well in Congo. Int J Syst Evol Microbiol 57, 40–44 (2007). [DOI] [PubMed] [Google Scholar]
- Lane D. J. 16S/23S rRNA sequencing. In: E. Stackebrandt, M. Goodfellow, eds. Nucleic Acid Techniques in Bacterial Systematics, John Wiley and Sons, New York. pp 5–175 (1991).
- Zhang F. et al. Response of microbial community structure to microbial plugging in a mesothermic petroleum reservoir in China. Appl Microbiol Biotechnol 88, 1413–1422 (2010). [DOI] [PubMed] [Google Scholar]