Abstract
CD8+ T-cell development in the thymus generates a predominant population of conventional naive cells, along with minor populations of “innate” T cells that resemble memory cells. Recent studies analyzing a variety of KO or knock-in mice have indicated that impairments in the T-cell receptor (TCR) signaling pathway produce increased numbers of innate CD8+ T cells, characterized by their high expression of CD44, CD122, CXCR3, and the transcription factor, Eomesodermin (Eomes). One component of this altered development is a non-CD8+ T cell-intrinsic role for IL-4. To determine whether reduced TCR signaling within the CD8+ T cells might also contribute to this pathway, we investigated the role of the transcription factor, IFN regulatory factor 4 (IRF4). IRF4 is up-regulated following TCR stimulation in WT T cells; further, this up-regulation is impaired in T cells treated with a small-molecule inhibitor of the Tec family tyrosine kinase, IL-2 inducible T-cell kinase (ITK). In contrast to WT cells, activation of IRF4-deficient CD8+ T cells leads to rapid and robust expression of Eomes, which is further enhanced by IL-4 stimulation. In addition, inhibition of ITK together with IL-4 increases Eomeso up-regulation. These data indicate that ITK signaling promotes IRF4 up-regulation following CD8+ T-cell activation and that this signaling pathway normally suppresses Eomes expression, thereby regulating the differentiation pathway of CD8+ T cells.
Keywords: T-cell receptor signal strength, T-cell signaling, ITK inhibitor
The immune system is composed of multiple lineages of T lymphocytes that differ in their antigen receptor specificity, their expression of effector cytokines, and their trafficking patterns in the body. The majority of circulating CD4+ and CD8+ T cells arise as naive lymphocytes, which require activation, expansion, and effector cell differentiation before participating in a protective immune response to infection. In contrast, a smaller subset of T cells completes thymic development as fully differentiated effector cells that are able to respond to activation signals rapidly and have transcription factor profiles and trafficking patterns of effector and/or memory T cells. This latter group, which includes invariant natural killer T (iNKT) cells, γδ T cells, and H2-M3–specific T cells, has been termed innate T cells (1).
The intracellular signals that direct developing T cells into conventional naive vs. innate cell lineages are not well understood. For CD1d-specific iNKT cells, the best studied of the innate T-cell lineages, factors regulating their development include the strength of T-cell receptor (TCR) signaling and differential use of costimulatory receptors, such as SLAM-family proteins (2). Additional insights into the development of innate T cells have come from studies of mice carrying deficiencies in T-cell signaling proteins or transcription factors. For example, in mice lacking the Tec family tyrosine kinase, IL-2 inducible T-cell kinase (ITK), thymocyte development produces a large population of innate CD8+ T cells instead of the normal subset of naive CD4−CD8+ (CD8SP) thymocytes. Specifically, Itk−/− CD8SP thymocytes have the hallmarks of antigen-experienced T cells because they are CD122+CD44hiCXCR3+, express high levels of the transcription factor Eomesodermin (Eomes), and produce IFN-γ within hours of ex vivo stimulation (3–5). A wealth of data indicates that alterations in TCR signaling pathways play a key role in promoting this innate T-cell development, because several lines of mice with defects in TCR signaling components, or downstream transcription factors regulated by TCR signaling, share this same phenotype; these include mice expressing a mutant version of SLP-76, in addition to mice deficient in KLF2, Creb-binding protein (CBP), or Id3 (6–10).
Recent studies have demonstrated that the development of innate memory-like CD8SP thymocytes is a complex process. One component of this altered development is a non-CD8+ T cell-intrinsic role for IL-4, which promotes the conversion of CD8SP thymocytes into memory-like T cells (8, 9). In addition to the environmental factors promoting this phenotype, reduced TCR signaling within the CD8+ T cell may contribute to the altered expression of lineage-determining transcription factors, such as Eomes.
To identify additional components regulating the memory CD8+ T-cell phenotype seen in Itk−/− mice, we performed a gene expression microarray experiment and focused on transcription factors that were differentially expressed between WT and Itk−/− CD8SP thymocytes. This analysis revealed that IFN regulatory factor 4 (IRF4) was expressed more highly in TCR-β+CD8+ cells from thymi of WT vs. Itk−/− mice. IRF4 is up-regulated following antigen receptor stimulation in B cells and T cells (11, 12). In B cells, the strength of B-cell receptor signaling determines the level of IRF4 protein produced; in turn, this graded expression of IRF4 regulates memory B-cell vs. plasma cell lineage development (13, 14). IRF4 is also required for T-cell function and is essential for T helper (Th) 2, Th9, and Th17 CD4+ lineage development (15–23). In addition to the role of IRF4 in effector CD4+ T-cell differentiation, IRF4 is important in Foxp3+ regulatory T cells (T-regs), because Irf4−/− T-regs are unable to suppress spontaneous T-cell activation in vivo (24). However, the role of IRF4 in the development and function of CD8+ T cells has not been investigated.
To address the role of IRF4 in CD8+ T cells, we examined mice with a conditional allele of irf4 (Irf4fl/fl) (13) crossed to CD4-Cre+ transgenic mice, thereby deleting IRF4 in all αβ T cells. Although the innate memory-like phenotype seen with CD8SP thymocytes in Itk−/− mice was not seen in thymocytes from these conditional IRF4-deficient mice, peripheral CD8+ T cells in these mice showed a spontaneous conversion to a memory cell phenotype; specifically, Irf4fl/flCD4-Cre+ [hereafter referred to as Irf4−/−(T)] CD8+ T cells were CD44hiCXCR3+CD122+Eomes+ and produced IFN-γ on ex vivo stimulation. Further, as shown previously, impaired T-reg function in conditional IRF4-deficient mice leads to polyclonal activation of peripheral T cells (24). However, unlike IRF4-sufficient CD8+ T cells, IRF4-deficient CD8+ T cells activated in this environment up-regulate high levels of Eomes and acquire a memory cell phenotype. Furthermore, we show that IRF4 is required to suppress Eomes up-regulation following in vitro activation of naive CD8+ T cells and that IL-4 enhances Eomes expression in cells with impaired TCR signaling. Lastly, we show that a combination of weak T-cell stimulation due to impaired T-reg function, plus the absence of IRF4, induces the innate-like phenotype in Irf4−/−(T) CD8+ T cells in vivo. Together, these data demonstrate that IRF4 functions as a regulator of CD8+ T-cell differentiation via suppression of Eomes expression after TCR signaling via ITK.
Results
IRF4 Is Up-Regulated During Positive Selection in the Thymus.
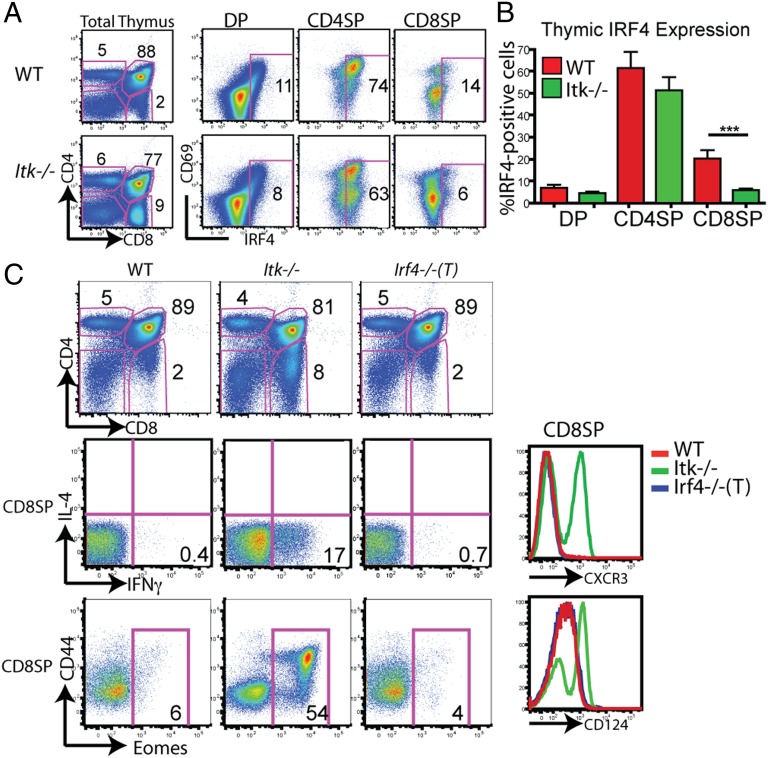
Using gene expression microarray analysis, we identified IRF4 as a gene more highly expressed in WT compared with Itk−/− CD8SP thymocytes. These data were confirmed using intracellular staining, followed by flow cytometry, to assess IRF4 protein levels (Fig. 1A). In WT thymocytes, IRF4 expression is detected at the CD4+8+ (DP) stage, and is predominantly restricted to the subset expressing high levels of CD69, representative of cells undergoing positive selection due to TCR signaling. IRF4 is also expressed in the majority of WT CD4+8− (CD4SP) thymocytes and, again, shows the highest correlation with CD69 expression. A smaller proportion of WT CD8SP thymocytes show detectable IRF4 expression. Overall, WT thymocytes had consistently higher levels of IRF4 protein expression directly ex vivo compared with Itk−/− thymocytes, but the largest difference was found in the CD8SP subset (Fig. 1 A and B). These data indicate that IRF4 is transiently up-regulated during thymic development as cells undergo positive selection and that ITK is required for optimal IRF4 expression.
Fig. 1.
IRF4 is up-regulated during thymic selection but is not required for normal T-cell development. (A) Thymocytes from WT or Itk−/− mice were isolated and surface-stained for CD4, CD8, and CD69, and then for intracellular IRF4 protein. Thymocyte subsets (DP, CD4SP, and CD8SP) were gated on, and IRF4 vs. CD69 is displayed. Numbers on dot-plots represent the percentage of cells expressing IRF4, based on comparison with an isotype control. Data are representative of six or more independent experiments. (B) Compilation of the percentage of IRF4-expressing cells for each thymocyte subset (n = 15; ***P < 0.001 based on the Mann–Whitney test). (C) Thymocytes from WT, Itk−/−, and Irf4−/−(T) mice were stained with antibodies to CD4, CD8, CD44, intracellular Eomes, CXCR3, and CD124; stimulated for 4 h with phorbol 12-myristate 13-acetone (PMA) and ionomycin; and stained for intracellular IL-4 and IFN-γ. CD4 vs. CD8 profiles are shown (Top) along with gated CD8SP cells (Middle and Bottom).
IRF4-Deficient T Cells Show Normal Thymic Development.
To determine whether IRF4 is a component of the pathway regulating CD8+ T-cell development or differentiation, we analyzed the thymic and peripheral T cells in conditional IRF4-deficient mice. For these experiments, Irf4fl/fl mice (13) were crossed to CD4-Cre+ transgenic mice [hereafter referred to as Irf4−/−(T)], thereby initiating Irf4 deletion at the DP stage in the thymus. In these mice, deletion of the floxed sequences in the Irf4 locus is accompanied by expression of GFP; as shown, 70% of DP thymocytes and nearly 100% of CD4SP and CD8SP thymocytes are GFP+ (Fig. S1A). To confirm that deletion of the Irf4 locus occurred on both alleles of Irf4, CD8SP thymocytes and CD8+ peripheral T cells were isolated, stimulated in vitro with αCD3/αCD28 stimulation, and analyzed by Western blotting. Although abundant IRF4 protein is detected in WT cells, IRF4 is virtually absent in cells from the Irf4−/−(T) mice (Fig. S1A). These data demonstrate that Irf4 is efficiently deleted by the CD8SP stage of thymocyte maturation.
Analysis of thymocytes from Irf4−/−(T) mice indicated that IRF4 is not required for normal T-cell development. Compared with WT thymocytes, Irf4−/−(T) thymocytes showed normal CD4/CD8 ratios, as has been previously reported for germline Irf4−/− mice (15) and unlike that seen in the absence of ITK (Fig. 1C). Additionally, there were no differences in the cell surface phenotype of CD8SP cells; thus, expression of CXCR3 and CD124 in Irf4−/−(T) CD8SP cells appeared identical to that in WT cells (Fig. 1C). Irf4−/−(T) CD8SP thymocytes also did not produce IFN-γ following ex vivo stimulation, consistent with their lack of expression of Eomes, a transcription factor found at abundant levels in the Itk−/− CD8SP cells (Fig. 1C).
More detailed analysis of minor thymocyte populations indicated that Irf4−/−(T) mice had reduced numbers of CD1d/αgalcer-binding iNKT cells and had no expansion of CD4+ IL-4–producing γδ T cells (Fig. S1B). In addition, the thymus of Irf4−/−(T) mice lacked the abundant promyelocytic leukemia zinc finger-positive population of innate CD4+ T cells seen in Itk−/− mice, and shown to be essential in the conversion of Itk−/− CD8SP thymocytes to the innate phenotype (8) (Fig. S1C). Surprisingly, although reduced numbers of CD1d/αgalcer-binding iNKT cells were found in the thymus of Irf4−/−(T) mice, spleens and livers of these mice did not show a reduction in the proportions of iNKT cells relative to WT (Fig. S2). Moreover, splenic Irf4−/−(T) CD4+ T cells showed a trend toward enhanced IL-4 production compared with WT cells, but this difference was not statistically significant (Fig. S2). Together, these data indicate that the Irf4−/−(T) mice do not contain an obvious IL-4–producing subset that would promote Eomes expression in CD8SP thymocytes or T cells.
IRF4-Deficient Peripheral CD8+ T Cells Acquire a Memory Phenotype.
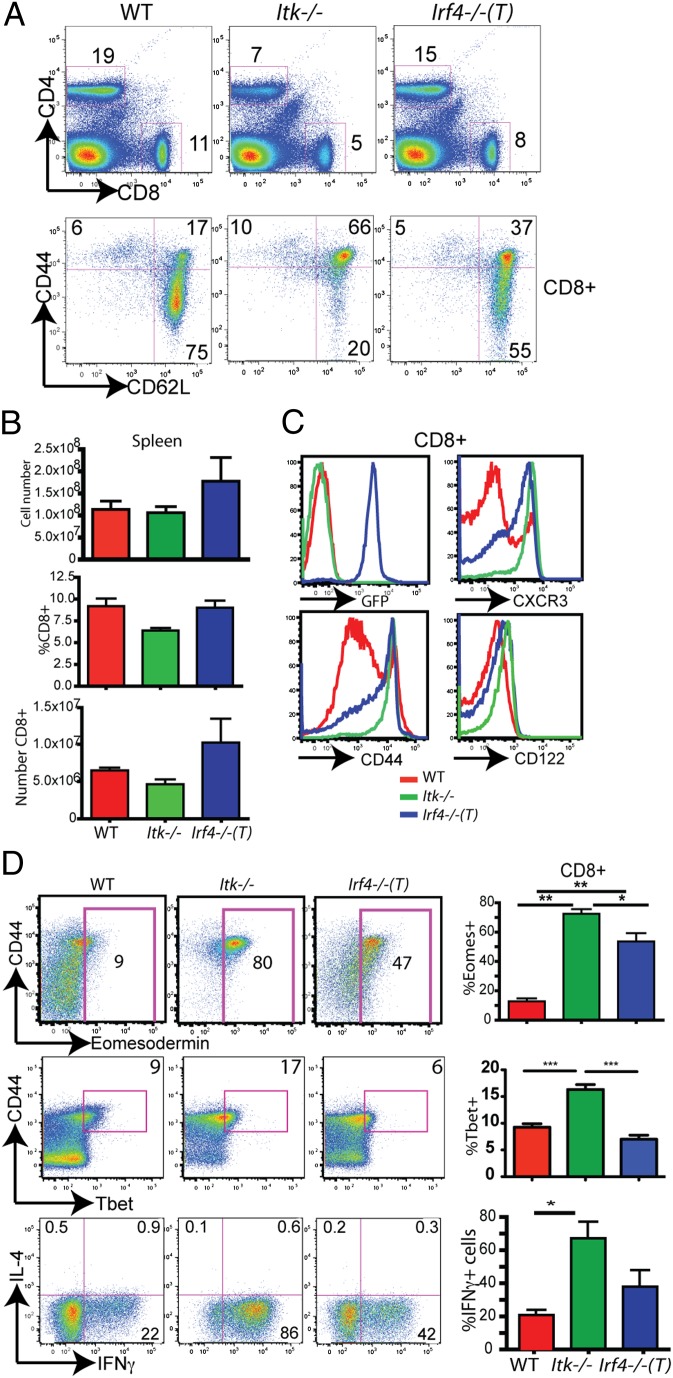
In contrast to thymocytes, peripheral CD8+ T cells in the Irf4−/−(T) mice showed a spontaneous conversion to an innate/memory phenotype (25), with increased proportions of CD44hiCD62Lhi cells relative to WT CD8+ T cells (Fig. 2A). As has been reported previously for the germline Irf4-deficient mice (15), Irf4−/−(T) mice had a slight increase in the absolute numbers of cells in their spleens (Fig. 2B). Cell surface analysis indicated additional differences between Irf4−/−(T) and WT T cells. In particular, the Irf4−/−(T) CD8+ T cells had increased levels of CXCR3, CD44, and CD122 compared with WT T cells, similar to those seen on Itk−/− T cells (Fig. 2C). These data indicate that Irf4−/−(T) CD8+ T cells undergo a spontaneous conversion from naive to memory-like T cells following their migration into the periphery.
Fig. 2.
Peripheral IRF4-deficient T cells share the innate phenotype of Itk−/− CD8+ T cells. (A) Splenocytes were isolated from WT, Itk−/−, and Irf4−/−(T) mice, and they were then stained with antibodies to CD4, CD8, CD44, and CD62L. CD4 vs. CD8 staining of total splenocytes (Upper) and CD44 vs. CD62L staining on gated CD8+ T cells (Lower) are shown. (B) Compilations of the total splenocyte numbers, percentages of CD8+ T cells, and absolute numbers of CD8+ T cells in the spleens are shown (n = 8). (C) CD8+ splenic T cells from WT, Itk−/−, and Irf4−/−(T) mice were analyzed for GFP, CXCR3, CD44, and CD122 expression. (D) CD8+ splenic T cells from WT, Itk−/−, and Irf4−/−(T) mice stained with antibodies to CD44, Eomes, and Tbet or were stimulated for 4 h with phorbol 12-myristate 13-acetone and ionomycin, and they were then analyzed for IL-4 vs. IFN-γ expression. Compilations of the data show the percentages of Eomes+ cells (n = 12), Tbet+ cells (n = 6–10), and IFN-γ+ cells (n = 10). Statistically significant differences are indicated by *(P < 0.05), **(P < 0.001), or ***(P < 0.0001) based on the Mann–Whitney test.
Because Itk−/− CD8+ T cells express high levels of Eomes (3), a transcription factor responsible for promoting IFN-γ transcription in effector and memory T cells (26, 27), we assayed for Eomes expression in splenic Irf4−/−(T) T cells. WT CD8+ T cells expressed modest amounts of Eomes protein, and this expression was largely restricted to the CD44hi subset. The proportions of Eomes+ cells among the peripheral CD8+ T cells in Itk−/− and Irf4−/−(T) mice were increased dramatically compared with WT mice (Fig. 2D). We also examined the expression of the related transcription factor, Tbet, in splenic Irf4−/−(T) T cells. Peripheral WT and Irf4−/−(T) CD8+ T-cell populations contained similar proportions of Tbet+ cells, whereas Itk−/− CD8+ T cells had a significant increase in Tbet expression (Fig. 2D). As for Eomes, the expression of Tbet was limited to the CD44hi subset. This increased Eomes expression correlated with enhanced IFN-γ production following ex vivo stimulation, although the difference between WT and Irf4−/−(T) CD8+ T cells did not achieve statistical significance (Fig. 2D).
Loss of Peripheral Tolerance in Irf4−/−(T) Mice.
IRF4 has been shown to be required for the function of FoxP3+ T-regs (24). Therefore, we considered whether defects in peripheral tolerance might account for the spontaneous activation of CD8+ T cells in Irf4−/−(T) mice. To test this possibility, we generated mixed bone marrow chimeras in which Irf4−/−(T) bone marrow was mixed with WT bone marrow and then used to reconstitute irradiated congenic Rag2−/− (CD45.1+) hosts. When analyzed at 7.5 wk after reconstitution, we found that CD8+ T cells derived from Irf4−/−(T) bone marrow retained a naive phenotype as long as they developed in the presence of WT cells (Fig. S3). These data indicate that the spontaneous activation of Irf4−/−(T) CD8+ T cells in intact Irf4−/−(T) mice is due to impaired peripheral tolerance in Irf4−/−(T) mice.
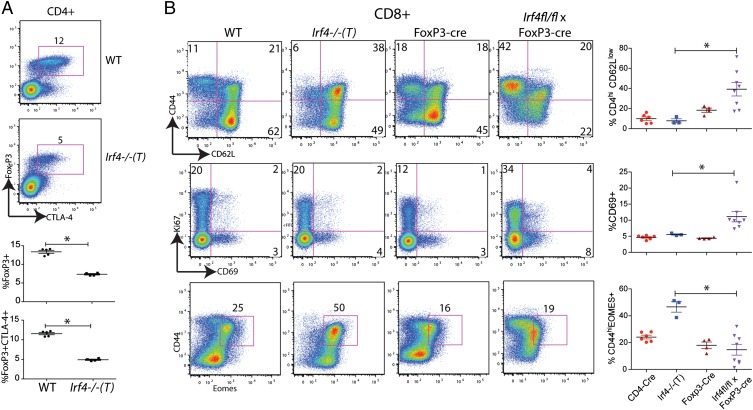
Based on these findings, we examined the phenotype of Foxp3+ T-regs in the Irf4−/−(T) mice. We observed a significant decrease in the proportion of CD4+Foxp3+ T-regs in the spleens of Irf4−/−(T) mice compared with WT mice (Fig. 3A). In addition, fewer of the Irf4−/−(T) FoxP3+CD4+ T cells expressed cytotoxic T lymphocyte-associated protein 4 (CTLA4) relative to WT FoxP3+ cells, suggesting a functional impairment in the Irf4−/−(T) T-regs. We also observed reduced expression of CTLA4 on Irf4−/−(T) CD4+Foxp3+ T-regs compared with WT T-regs. Collectively, these data support the conclusion that peripheral Irf4−/−(T) CD8+ T cells are activated due to a loss of peripheral tolerance resulting from FoxP3+ T-reg defects, as previously described (24).
Fig. 3.
Aberrant FoxP3+ regulatory T cells in Irf4−/−(T) mice. (A) CD4+ T cells from the spleens of WT and Irf4−/−(T) mice were analyzed for intracellular FoxP3 and CTLA-4. Numbers on the dot-plots represent the percentages of CD4+ T cells that are FoxP3+CTLA-4+. Below are compilations of the percentages of CD4+ T cells that are FoxP3+ or FoxP3+CTLA-4+. Data are representative of two independent experiments. Statistical significance was determined by the Mann–Whitney test. (B) Splenic CD8+ T cells from WT, Irf4−/−(T), FoxP3-cre control, and Irf4fl/fl × FoxP3-cre mice were analyzed for expression of CD44, CD62L, CD69, Ki67, and intracellular Eomes. Dot-plots display gated CD8+TCR-β+ cells; numbers indicate the percentages of cells in the indicated quadrants or gates. (Right) Compilations of data for the four groups of mice are shown. Statistical significance was determined by the Mann–Whitney test (*P = 0.02).
These data indicated that a component of the CD8+ T-cell phenotype observed in Irf4−/−(T) mice was not CD8+ T cell-intrinsic. To determine whether loss of IRF4 expression in the CD8+ T cells also contributed to their conversion to a memory cell phenotype, we compared Irf4−/−(T) CD8+ T cells with CD8+ T cells present in Irf4fl/fl × Foxp3-cre mice, with the latter being deficient for IRF4 only in FoxP3+ T-regs (24). This comparison revealed considerable differences between the peripheral CD8+ T cells present in the two lines of mice (Fig. 3B). Although the activated CD8+ T cells in the Irf4−/−(T) mice predominantly exhibited a memory phenotype (CD44hi CD62Lhi), activated IRF4-sufficient Irf4fl/fl × Foxp3-cre+ CD8+ T cells had a CD44hi CD62Llow phenotype associated with effector cells. Further, a greater proportion of CD8+ T cells from Irf4fl/fl × Foxp3-cre mice vs. those in Irf4−/−(T) mice showed evidence of recent activation, as indicated by CD69 up-regulation, although there was no statistically significant difference in Ki67 staining between any of the groups of mice. Finally, the transcription factor Eomes was up-regulated in a significantly higher fraction of Irf4−/−(T) CD8+ T cells compared with those in Irf4fl/fl × Foxp3-cre mice. These data indicate that both environmental factors, as well as a CD8+ T cell-intrinsic role for IRF4, are contributing to the phenotype of CD8+ T cells in Irf4−/−(T) mice.
Both Cell-Intrinsic and Cell-Extrinsic Factors Contribute to the Presence of Innate/Memory CD8+ T Cells in Irf4−/−(T) Mice.
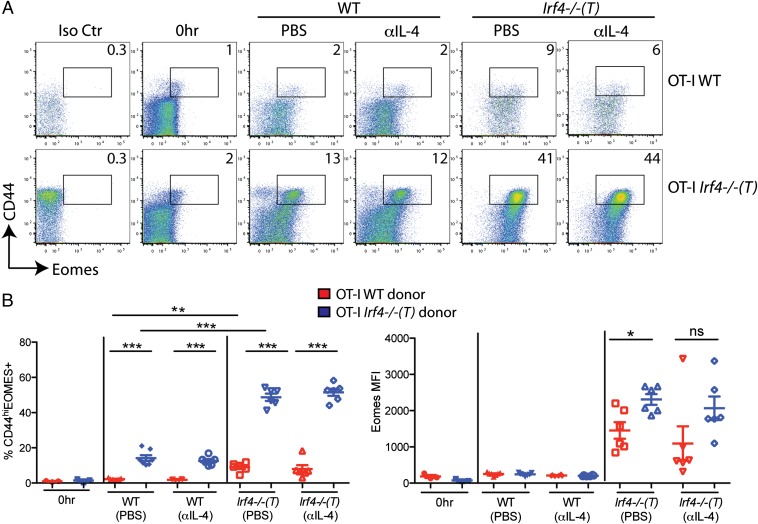
Irf4−/−(T) mice exhibit reduced peripheral tolerance and have increased numbers of innate/memory CD8+ T cells in their spleens. To determine the relative contribution of cell-intrinsic factors vs. factors contributed by the environment in Irf4−/−(T) mice (i.e., cell-extrinsic factors), we performed adoptive transfer experiments. In particular, we addressed a possible role for IL-4 in the environment of Irf4−/−(T) mice, because previous studies have indicated a key role for IL-4 in Eomes up-regulation during the development of innate CD8+ T cells in the thymus (7–9, 28, 29). For these experiments, a 1:1 mixture of naive CD8+ OT-I Rag1−/− and OT-I Rag1−/− Irf4−/−(T) (henceforth referred to as OT-I WT and OT-I Irf4−/−(T), respectively) splenocytes were transferred into WT or Irf4−/−(T) host mice. In addition, cohorts of mice received either PBS or 1 mg of anti–IL-4 neutralizing antibody 2–4 h before adoptive transfer. Donor cells were examined 44 h later for Eomes and CD44 expression.
As shown in Fig. 4, OT-I WT cells show little change when analyzed after 2 d in a WT host, whereas OT-1 Irf4−/−(T) CD8+ T cells had modestly increased proportions of CD44hi and Eomes+ cells in this environment. Neither group of cells in the WT hosts was affected by the presence of anti–IL-4 antibody. In contrast, following 2 d in Irf4−/−(T) hosts, both groups of donor cells showed increased expression of CD44 and Eomes, with a significantly enhanced effect on the OT-I Irf4−/−(T) cells. In the Irf4−/−(T) environment, IL-4 blocking caused a trend toward reduced Eomes up-regulation in both OT-I WT and OT-I Irf4−/−(T) cells, as assessed by the median fluorescence intensity (MFI) of Eomes staining on the entire cell population (Fig. 4B); however, these reductions were not statistically significant. These data also show that the majority of CD44lo cells, both OT-I WT and OT-I Irf4−/−(T) donor cells, have up-regulated Eomes in Irf4−/−(T) hosts. Because αIL-4 treatment did not abolish this effect, an alternative possibility is the presence of high levels of type I IFN in the OT-I Irf4−/−(T) mice; a strong type I IFN response has been shown to induce Eomes up-regulation specifically on CD44lo naive CD8+ T cells (30). Overall, these data indicate that the dramatic up-regulation of Eomes in Irf4−/−(T) CD8+ T cells is the result of both a cell-intrinsic defect caused by the absence of IRF4 plus an environmental factor present in the Irf4−/−(T) mice.
Fig. 4.
High expression of Eomes by IRF4-deficient CD8+ T cells is dependent on both cell-intrinsic and cell-extrinsic factors in Irf4−/−(T) mice. Splenocytes from OT-I WT and OT-I Irf4−/−(T) mice were mixed 1:1 and adoptively transferred into WT or Irf4−/−(T) hosts and then injected i.p. with 1 mg of αIL-4 neutralizing antibody or PBS 2–4 h before adoptive transfer. CD8+ T cells were analyzed 44 h later with a viability stain, along with antibodies to CD8, Vα2, Vβ5, CD45.1, CD45.2, CD44, CD62L, and intracellular Eomes or an isotype control. (A) Dot-plots show Eomes vs. CD44 staining on gated live CD8+, Vα2+, and Vβ5+ cells distinguished by CD45.1 (WT) vs. CD45.2 [Irf4−/− (T)+] staining. Numbers indicate the percentages of CD44hiEomes+ cells. Iso Ctr, antibody control for Eomes staining; 0 h, cells analyzed directly ex vivo before adoptive transfer. (B) Compilations of data show percentages of CD44hiEomes+ cells (Left) and raw MFI of Eomes staining (Right) on live CD8+, Vα2+, and Vβ5+ cells distinguished by CD45.1 (WT) vs. CD45.2 [Irf4−/−(T)+] staining from three independent experiments with two to three host mice per group per experiment. Statistical significance was determined by a t test (*P = 0.01; **P = 0.001; ***P < 0.0001). ns, not significant.
IRF4 Is Required to Suppress Eomes Expression Following CD8+ T-Cell Activation.
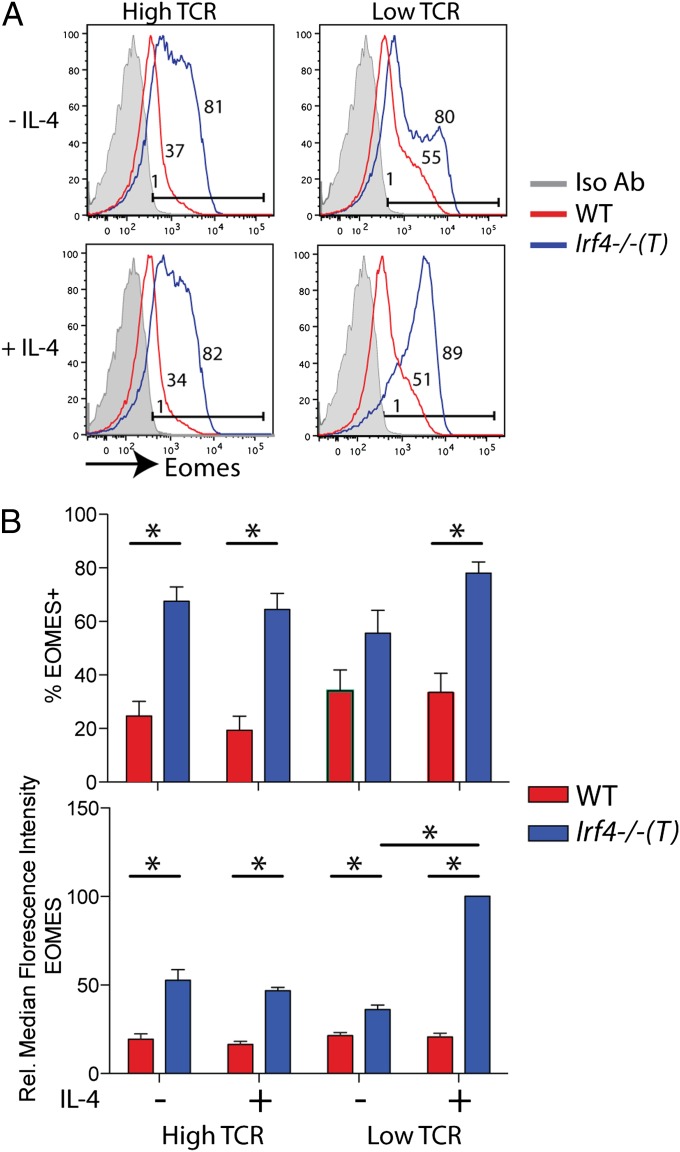
To address the cell-intrinsic role of IRF4 in CD8+ T cells directly, we performed in vitro stimulation assays using naive Irf4−/−(T) vs. WT CD8SP thymocytes. Cells were stimulated with high vs. low concentrations of CD3/CD28 antibodies, in the presence or absence of IL-4. We first determined that IL-4 had no impact on IRF4 expression in stimulated WT cells (Fig. S4A). As shown in Fig. 5, strong stimulation of Irf4−/−(T) CD8+ thymocytes led to significantly higher expression of Eomes compared with that seen following activation of WT CD8+ thymocytes (Fig. 5, high TCR). This increase included a greater percentage of cells expressing Eomes, as well as a higher expression of Eomes per T cell (Fig. 5B). Under these conditions, addition of exogenous IL-4 had no impact on Eomes expression in either WT or Irf4−/−(T) CD8+ thymocytes. When cells were stimulated with low concentrations of CD3/CD28 antibodies (Fig. 5, low TCR), there was a trend toward more Irf4−/−(T) CD8+ thymocytes up-regulating Eomes than WT cells, although this difference was not statistically significant; however, these conditions did lead to a significantly higher per cell expression of Eomes in the Irf4−/−(T) CD8+ thymocytes. Interestingly, in cells stimulated with low CD3/CD28 conditions, addition of exogenous IL-4 had no effect on WT cells but resulted in a dramatic increase in the per cell expression of Eomes in the Irf4−/−(T) CD8+ thymocytes (Fig. 5B). These data indicate that in WT CD8+ thymocytes, IRF4 normally suppresses Eomes up-regulation on TCR/CD28 stimulation.
Fig. 5.
Naive Irf4−/−(T) CD8SP thymocytes up-regulate high levels of Eomes following TCR stimulation. (A) WT and Irf4−/−(T) CD8SP thymocytes were isolated and stimulated with 1 μg/mL CD3 plus 4 μg/mL CD28 (“high TCR”) or 0.1 μg/mL CD3 plus 0.4 μg/mL CD28 (“low TCR”) antibodies in the presence or absence of IL-4 (10 ng/mL) for 38 h. Cells were stained with antibodies to CD4, CD8, CD24, CD44, and intracellular Eomes or an isotype control. Histograms show Eomes expression relative to the isotype control (Iso Ab; gray-filled histograms) on gated CD8+CD24loCD44hi cells; numbers indicate the percentages of Eomes+ cells. (B) Compilations of data indicate percentages of Eomes+ cells (Upper) and relative (Rel.) MFIs of Eomes staining (Lower) for n = 4 experiments. Statistical significance was determined by the Mann–Whitney test (*P < 0.03).
Previous studies have indicated that IL-4 is sufficient to up-regulate Eomes in CD8+ T cells undergoing positive selection in the thymus (8, 28). To address whether IL-4 alone, in the absence of any TCR stimulation, is sufficient to induce Eomes expression in mature (CD24loTCR-βhi) CD8SP thymocytes, we cultured WT and Irf4−/−(T) cells with IL-4 in the presence vs. the absence of low concentrations of αCD3/CD28. IL-7 was added to all cultures to promote thymocyte survival. As shown in Fig. S4B, in the absence of overt TCR/CD28 stimulation, IL-4 + IL-7 alone led to a very minimal increase in Eomes expression in both WT and Irf4−/−(T) mature CD8SP thymocytes compared with cells analyzed directly ex vivo. When combined with the low strength of TCR/CD28 signaling, IL-4 + IL-7 promoted a high level of Eomes expression in Irf4−/−(T) mature CD8SP thymocytes, with only an incremental effect on the WT cells. These latter conditions also induced Tbet up-regulation in both populations of thymocytes, with only a modest difference between WT and Irf4−/−(T) cells (Fig. S4C). These data indicate that a combination of weak or impaired TCR signaling plus IL-4 is required for robust Eomes up-regulation in CD8SP thymocytes.
ITK Inhibition Synergizes with IL-4 to Promote Eomes Up-Regulation.
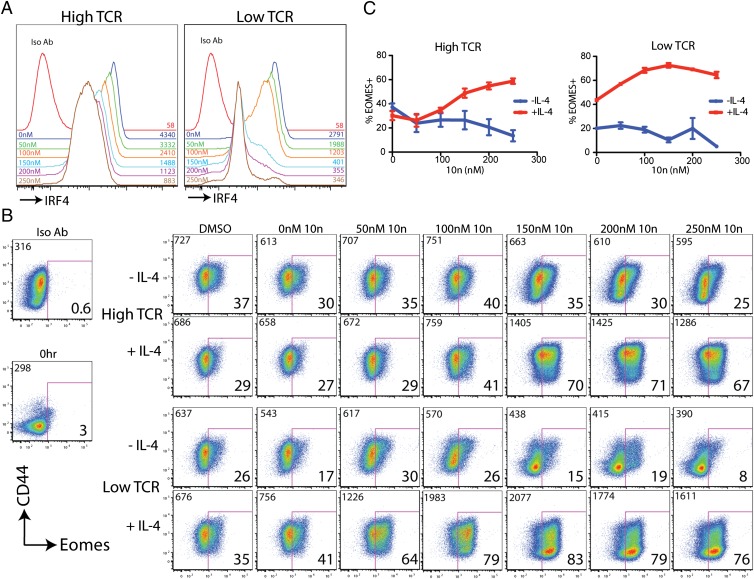
The data presented thus far suggest that ITK activation downstream of the TCR is required for maximal IRF4 up-regulation, and thus for suppression of Eomes expression following CD8+ T-cell activation. To test this prediction, we stimulated naive OT-I WT CD8+ T cells with high or low concentrations of CD3/CD28 antibodies in the presence of a small-molecule inhibitor of ITK, 10n (31). As shown in Fig. 6, ITK inhibition reduced the levels of IRF4 expressed in each activated T cell, as indicated by the dose-dependent decrease in the MFI of IRF4 staining. The ability of 10n to inhibit IRF4 up-regulation was seen in both high TCR and low TCR stimulation conditions. Analysis of Eomes expression in these cells revealed that high TCR stimulation led to a modest up-regulation of Eomes, which was not enhanced by ITK inhibition. However, when exogenous IL-4 was added to these cultures, we observed substantial Eomes up-regulation in the presence of high concentrations of 10n. This effect was further enhanced following low TCR stimulation, where IL-4 in combination with 10n led to robust Eomes expression.
Fig. 6.
Inhibition of ITK synergizes with IL-4 to up-regulate high levels of Eomes following TCR stimulation. Naive peripheral CD8+ T cells were isolated from OT-I WT mice and were stimulated with 1 μg/mL CD3 plus 4 μg/mL CD28 (“high TCR”) or 0.1 μg/mL CD3 plus 0.4 μg/mL CD28 (“low TCR”) antibodies in the presence or absence of IL-4 (10 ng/mL) for 31 h. Cultures were supplemented with the small-molecule ITK inhibitor, 10n, at the indicated concentrations or with DMSO alone at the highest concentration used. Cells were stained with a viability dye, and antibodies to CD8, CD69, CD44, intracellular IRF4, Eomes, or isotype controls. (A) Histograms show IRF4 expression relative to the isotype control (Iso Ab). The concentrations of 10n (Left) and the MFI of IRF4 staining (Right) are indicated in each histogram. All data are from cells stimulated in high TCR (Left) or Low TCR (Right) in the absence of exogenous IL-4. (B) Dot-plots show Eomes vs. CD44 staining in cells stimulated with high TCR (Upper two rows) or low TCR (Lower two rows) and supplemented with IL-4 or 10n as indicated. Numbers at the upper left of each dot-plot indicate the MFI of Eomes staining. Numbers at the lower right of each dot-plot indicate the percentage of Eomes+ cells. Data are representative of four experiments. (C) Compilations of data indicate percentages of Eomes+ cells for each condition.
The ability of IL-4 to enhance Eomes up-regulation was not due to an unanticipated role for IL-4 in suppressing IRF4 expression, because OT-I WT cells stimulated with “high” or “low” TCR stimulation each expressed slightly higher amounts of IRF4 in the presence vs. the absence of IL-4 (Fig. S5A). We also ruled out effects of 10n, or the lack of IRF4, on IL-4 signaling, based on analysis of STAT6 phosphorylation in response to IL-4 stimulation (Fig. S5). Altogether, these findings demonstrate that impaired CD3/CD28 signaling, either via inhibition of ITK or by eliminating IRF4, synergizes with IL-4 to promote high expression of Eomes in naive CD8+ T cells. Furthermore, these data show a striking similarity to the altered development of innate-like CD8+ T cells observed in the thymus of Itk−/− mice.
Discussion
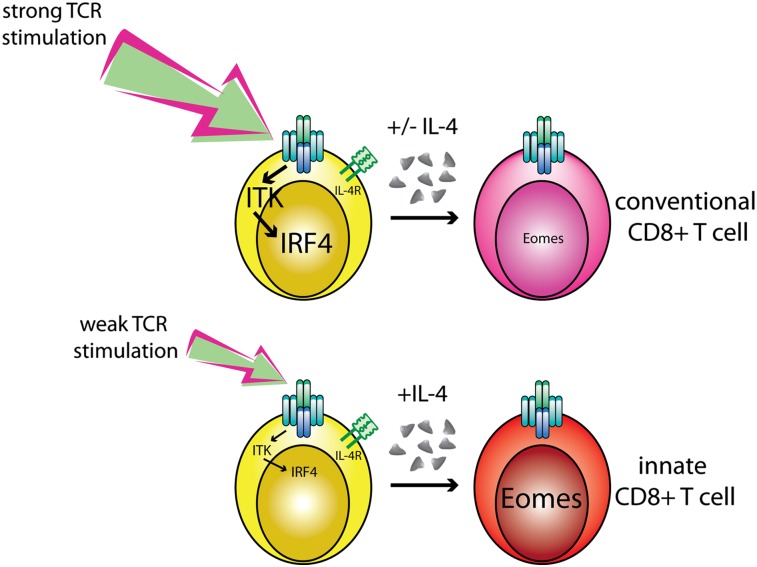
This study has identified the transcriptional regulator IRF4 as a key downstream mediator that converts differences in TCR signal strength into distinct programs of gene expression. We show that robust up-regulation of IRF4 expression in T cells requires the Tec family tyrosine kinase, ITK, and that, in turn, IRF4 suppresses the up-regulation of Eomes following TCR stimulation. These data suggest a model to account for the enhanced development of innate CD8+ T cells seen in Itk−/− mice. During T-cell development in the thymus, IRF4 is transiently up-regulated during the maturation of DP into CD4SP and CD8SP thymocytes, and in the absence of ITK, the up-regulation of IRF4 in CD8SP cells is impaired. Together with the excess IL-4 present in the Itk−/− environment, a consequence of alterations in other T-cell lineages, this low TCR signaling that drives CD8SP maturation leads to high expression of Eomes, and thus to conversion of conventional naive cells into innate CD8+ T cells. In turn, high levels of Eomes promote expression of CD122, IFN-γ, CXCR3, and other genes associated with a CD8+ memory T-cell phenotype (8, 32, 33).
The precise mechanism by which IRF4 suppresses Eomes expression is not yet known. IRF4 can function either as a transcriptional repressor or as an activator, depending on the cellular context and its binding partners (34). One simple scenario is that IRF4, together with a corepressor, binds to the Eomes locus and represses transcription of the gene. To date, our efforts to detect IRF4 binding to consensus binding sites in the region surrounding the Eomes promoter by ChIP assays have been unsuccessful. It may be that IRF4 binds to a distal region of the Eomes locus not yet investigated in our assays; alternatively, the regulation of Eomes expression by IRF4 may be indirect, mediated by other factors that are the direct targets of IRF4.
In addition to ITK, several other proteins have been identified as playing a role in conventional vs. innate T-cell development in the thymus. These include SLP76, Id3, CBP, and Klf2 (6–10). Mice carrying genetic alterations in each of these genes share the general properties of enhanced innate CD8+ T-cell development accompanied by excess IL-4 production. In the case of SLP-76 Y145F knock-in mice, the connection to the pathway described herein is clear, because Y145 of SLP-76 is required for ITK recruitment to the linker for activation of T cells (LAT)/SLP-76 adapter complex following TCR stimulation (35, 36); thus, our data predict that T cells from SLP-76 Y145F knock-in mice would also be impaired in IRF4 up-regulation following TCR stimulation. In the case of KLF2, CBP, and Id3, potential links to a pathway involving IRF4 are less apparent. CBP, the coactivator “Creb-binding protein,” may be required for IRF4 transcriptional activation downstream of the TCR and ITK, as it is for other ITK-regulated genes, such as Egr2, Egr3, and IL-2 (10). Id3, an antagonist of E-protein transcriptional activators, and the transcription factor Klf2 are highly expressed in naive T cells, and are normally down-regulated following CD8+ T-cell activation and differentiation (9, 37, 38); for these two factors, genetic deficiencies in T cells may lead to spontaneous up-regulation of CD8+ effector genes, even in the absence of robust TCR stimulation. However, it also remains possible that the Klf2 and Id3 deficiencies promote innate CD8+ T-cell development by a completely CD8+ nonintrinsic mechanism, acting solely on the CD4+ NKT-like population that overproduces IL-4 in these mice.
Peripheral CD8+ T cells in Irf4−/−(T) mice spontaneously convert to a memory T-cell phenotype, expressing high levels of CD44, CD62L, CXCR3, CD122, and Eomes. We show that there are two components to this process. First, Irf4−/−(T) mice lack functional Foxp3+ regulatory T cells, as has been described for mice bearing a specific deletion of Irf4 only in FoxP3+ T cells (24). As a result, the majority of peripheral T cells in Irf4−/−(T) mice become activated. The outcome of this activation is then determined by whether the conventional T cells are IRF4-deficient or -sufficient. In the Irf4fl/fl × FoxP3-cre mice, where conventional T cells can express IRF4, T-cell activation proceeds down a pathway resembling effector cell differentiation. A greater proportion of the CD8+ T cells in these mice express CD69 and down-regulate CD62L compared with those in the Irf4−/−(T) mice, which lack IRF4. In contrast, a higher percentage of Irf4−/−(T) CD8+ T cells express Eomes than in the Irf4fl/fl × FoxP3-cre mice. These data indicate that the memory/innate cell phenotype of Irf4−/−(T) CD8+ T cells is due to a combination of both cell-extrinsic and cell-intrinsic factors.
These in vivo data, together with our findings from in vitro stimulations of Irf4−/−(T) CD8SP thymocytes, indicate that IRF4 up-regulation following CD8+ T-cell activation functions to suppress Eomes expression. One interesting possibility is that this pathway functions early in the response to infection to promote robust effector cell generation; following this early response, as antigen is cleared and TCR signaling subsides, IRF4 expression would wane, allowing the up-regulation of Eomes and the generation of memory T cells. It is also possible that within a heterogeneous population of T cells responding to an infection, those with lower TCR affinity or with exposure to limiting amounts of antigen would be induced directly into a memory lineage based on weak IRF4 up-regulation failing to suppress Eomes expression.
Evidence from mixed bone marrow chimeras indicates that an excess of klf2-deficient, Itk−/−, or id3−/− T cells overproducing IL-4 can promote the conversion of WT CD8+ T cells into the innate lineage (7–9). These data indicate that reduced or impaired TCR signaling within the developing CD8SP thymocyte is not necessarily required for Eomes up-regulation and the expression of effector/memory T-cell genes. However, the efficiency of this conversion is reduced compared with that seen with intact Itk−/− or conditional klf2−/− mice, as was particularly apparent when OT-1 TCR transgenic CD8+ T cells were tracked as the WT population (8). These data suggest that signaling differences intrinsic to the CD8+ cells contribute to this phenotype. It is likely that among a heterogeneous population of developing CD8SP thymocytes, there are differences in the strength of TCR signaling, which, in turn, may lead to varying levels of IRF4 up-regulation. As we demonstrate, CD8SP thymocytes that are unable to express IRF4 have a dramatically increased tendency to up-regulate Eomes, particularly in the presence of high levels of IL-4. Further, impaired TCR signaling via pharmacological inhibition of ITK in naive CD8+ T cells leads to robust Eomes expression, in conjunction with IL-4. Together, these data demonstrate the synergism of low TCR signal strength and IL-4 in promoting the innate/memory pathway of differentiation.
In response to infection, innate CD8+ T cells appear to provide rapid effector functions that limit the spread of the pathogen. This has been most clearly demonstrated in a Listeria monocytogenes system, in which innate CD8+ T cells produce high levels of IFN-γ as bystander T cells, and thereby reduce bacterial titers at the peak of the infection (39). Bystander CD8+ T cells have also been shown to up-regulate Eomes and to acquire effector functions during virus infections in a response that was dependent on weak TCR interactions and cytokines, in this case, type I IFNs; this latter response was particularly striking because it occurred in >70% of the bystander T cells (30). These studies demonstrate that the appropriate combination of low TCR signaling plus selected cytokines, such as IL-4 or type I IFNs, can promote the rapid acquisition of CD8+ T-cell effector functions via the up-regulation of Eomes in a sizeable population of naive T cells. Thus, this innate CD8+ T-cell response may provide a protective advantage similar to that of true innate cells, such as natural killer cells.
Methods
Mice.
Mice were bred and housed in specific pathogen-free conditions at the University of Massachusetts Medical School (UMMS) in accordance with institutional animal care and use committe guidelines. WT C57BL/6 mice were purchased from the Jackson Laboratories. OT-I TCR transgenic Rag1−/− mice were purchased from Taconic. CD4-Cre mice were a gift from Joonsoo Kang (UMMS). Itk−/− mice (40) were developed and bred at UMMS. Irf4fl/fl mice have been described previously (13). CD4-Cre transgenic (C57BL/6) or C57BL/6 mice were used as WT controls.
Antibodies and Staining.
FoxP3-FITC, IRF4-Allophycocyanin (APC), CD44-efluor 450, CD127-phycoerythrin (PE) Cy7, CD62L-APC eFluor 780, CD62L-APC eFluor 700, TCR-β–APC eFluor 780, Tbet-PeCy7, Vα2-peridinin chlorophyll protein (PerCP) eFluor 710, CD45.1 APC eFluor 780, Eomes-Alexa Fluor 488, Eomes-Alexa Fluor 647, and Eomes-PerCP eFluor 710 antibodies were purchased from eBioscience. Ki-67-FITC, CD69-PE, CD24 human serum albumin-FITC, CD4-V500, CTLA4-PE, CD124-PE, phospho-STAT6–APC, CD45.2-V500, and TCR-β–Alexa Fluor 700 antibodies were purchased from BD Pharmingen. CD8-PE-Texas Red (PETR) and Live/Dead Violet were purchased from Invitrogen. Intracellular staining for IRF4 was performed as previously described (14) or was performed using the manufacturer’s protocol. CD1d tetramer was obtained from the National Institutes of Health Tetramer Core Facility. To compare spleens from Irf4−/−(T) and Irf4fl/fl × FoxP3-cre mice, splenocytes were harvested and RBCs were lysed at UMMS and Memorial Sloan–Kettering Cancer Center, respectively; stored at 4 °C overnight; and stained the following day. Cells retained >90% viability. Samples were analyzed on an LSRII flow cytometer (Becton Dickinson), and data were further analyzed using FlowJo (Tree Star).
Cell Cultures.
To assay for cytokine production, cells were stimulated with 10 ng/mL phorbol 12-myristate 13-acetone and 2 μg/mL ionomycin (Sigma–Aldrich) for 4 h in the presence of 0.7 μg/mL Golgi Stop and 1 μg/mL Golgi Plug (Becton Dickinson). For longer term cultures (2–3 d), wells were coated with AffiniPure Goat anti-Armenian hamster IgG (H + L) (1 μg/mL; Jackson ImmunoResearch), followed by anti-CD3 and anti-CD28 (high TCR, 1 μg/mL + 4 μg/mL; low TCR, 0.1 μg/mL + 0.4 μg/mL) (Ebioscience). IL-7 and IL-4 were purchased from R&D Systems and used at a concentration of 10 ng/mL. Peripheral T cells were isolated from splenocytes by depleting B220+ cells using magnetic affinity cell sorting (MACS; Miltenyi) beads, and they were cultured at a concentration of 1 × 106 cells/mL. To isolate naive CD8SP thymocytes, CD4+ cells were depleted using MACS beads and then cultured at 1 × 106 cells/mL. Cells were then harvested, stained, and analyzed by flow cytometry. The ITK inhibitor, 10n (31), was synthesized at the National Institute of Health’s Chemical Genomics Center and dissolved in DMSO at 100 mM, before dilution into cell culture media at the indicated working concentrations.
Adoptive Transfers.
WT, Irf4−/−(T), and Itk−/− hosts were injected i.p. with 1 mg of αIL-4 neutralizing antibody (clone 11B11; BioXCell) or an equal volume of PBS 2–4 h before adoptive transfer. Splenocytes were harvested from OT-I WT and OT-I Irf4−/−(T) mice, and equal numbers of cells from each population were mixed and transferred into WT or Irf4−/−(T) hosts via tail vein injections. After 44 h, splenic CD8+ T cells in the recipients were analyzed by flow cytometry.
Supplementary Material
Acknowledgments
We thank Regina Whitehead, Catherine Yin, and Sharlene Hubbard for technical assistance and Dr. Alexander Rudensky, John Evans III, and Dr. Iivari Kleino for the contribution of reagents. This work was supported by National Institutes of Health Grants AI084987 and AI083505.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 16420 (volume 109, number 41).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205742109/-/DCSupplemental.
References
- 1.Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 3.Atherly LO, et al. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Broussard C, et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. and correction (2006) 25:849. [DOI] [PubMed] [Google Scholar]
- 5.Dubois S, Waldmann TA, Müller JR. ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc Natl Acad Sci USA. 2006;103:12075–12080. doi: 10.1073/pnas.0605212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan MS, et al. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinreich MA, et al. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuyama T, et al. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol. 2009;29:3894–3904. doi: 10.1128/MCB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama T, et al. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 13.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 14.Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Mittrücker HW, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. [PubMed] [Google Scholar]
- 16.Staudt V, et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Chung Y, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohoff M, et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci USA. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brüstle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 20.Tominaga N, et al. Development of Th1 and not Th2 immune responses in mice lacking IFN-regulatory factor-4. Int Immunol. 2003;15:1–10. doi: 10.1093/intimm/dxg001. [DOI] [PubMed] [Google Scholar]
- 21.Ahyi A-NN, Chang H-C, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immunol. 2009;183:1598–1606. doi: 10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honma K, et al. Interferon regulatory factor 4 differentially regulates the production of Th2 cytokines in naive vs. effector/memory CD4+ T cells. Proc Natl Acad Sci USA. 2008;105:15890–15895. doi: 10.1073/pnas.0803171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber M, et al. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc Natl Acad Sci USA. 2008;105:20846–20851. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 27.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 28.Min HS, et al. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. J Immunol. 2011;186:5749–5757. doi: 10.4049/jimmunol.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon SM, et al. Requirements for eomesodermin and promyelocytic leukemia zinc finger in the development of innate-like CD8+ T cells. J Immunol. 2011;186:4573–4578. doi: 10.4049/jimmunol.1100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall HD, Prince AL, Berg LJ, Welsh RM. IFN-alpha beta and self-MHC divert CD8 T cells into a distinct differentiation pathway characterized by rapid acquisition of effector functions. J Immunol. 2010;185:1419–1428. doi: 10.4049/jimmunol.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riether D, et al. 5-Aminomethylbenzimidazoles as potent ITK antagonists. Bioorg Med Chem Lett. 2009;19:1588–1591. doi: 10.1016/j.bmcl.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. Cutting Edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol. 2006;177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- 33.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohoff M, Mak TW. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat Rev Immunol. 2005;5:125–135. doi: 10.1038/nri1552. [DOI] [PubMed] [Google Scholar]
- 35.Bunnell SC, et al. Biochemical interactions integrating Itk with the T cell receptor-initiated signaling cascade. J Biol Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 36.Su YW, et al. Interaction of SLP adaptors with the SH2 domain of Tec family kinases. Eur J Immunol. 1999;29:3702–3711. doi: 10.1002/(SICI)1521-4141(199911)29:11<3702::AID-IMMU3702>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 37.Ji Y, et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12:1230–1237. doi: 10.1038/ni.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang CY, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhanji S, Chow MT, Teh H-S. Self-antigen maintains the innate antibacterial function of self-specific CD8 T cells in vivo. J Immunol. 2006;177:138–146. doi: 10.4049/jimmunol.177.1.138. [DOI] [PubMed] [Google Scholar]
- 40.Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]