Abstract
Listeria monocytogenes is a foodborne pathogen that crosses the intestinal barrier and disseminates within the host. Here, we report a unique comprehensive analysis of the impact of two Lactobacillus species, Lactobacillus paracasei CNCM I-3689 and Lactobacillus casei BL23, on L. monocytogenes and orally acquired listeriosis in a gnotobiotic humanized mouse model. We first assessed the effect of treatment with each Lactobacillus on L. monocytogenes counts in host tissues and showed that each decreases L. monocytogenes systemic dissemination in orally inoculated mice. A whole genome intestinal transcriptomic analysis revealed that each Lactobacillus changes expression of a specific subset of genes during infection, with IFN-stimulated genes (ISGs) being the most affected by both lactobacilli. We also examined microRNA (miR) expression and showed that three miRs (miR-192, miR-200b, and miR-215) are repressed during L. monocytogenes infection. Treatment with each Lactobacillus increased miR-192 expression, whereas only L. casei association increased miR-200b and miR-215 expression. Finally, we showed that treatment with each Lactobacillus significantly reshaped the L. monocytogenes transcriptome and up-regulated transcription of L. monocytogenes genes encoding enzymes allowing utilization of intestinal carbon and nitrogen sources in particular genes involved in propanediol and ethanolamine catabolism and cobalamin biosynthesis. Altogether, these data reveal that the modulation of L. monocytogenes infection by treatment with lactobacilli correlates with a decrease in host gene expression, in particular ISGs, miR regulation, and a dramatic reshaping of L. monocytogenes transcriptome.
Keywords: probiotics, virulence, vitamin B12
The intestinal tract is colonized by the largest microbial community of the human body. The intestinal microbiota plays a key role in host nutrition, immune cell homeostasis, and defense against pathogens (1). Analysis of microbiota composition and its effect on the host have blossomed in the last decade owing to the development of high throughput sequencing and the use of germ-free and gnotobiotic animals (2). Comparison between healthy individuals and patients with various clinical conditions has revealed significant differences in microbiota composition (3). These results strongly suggest that intervention aimed at changing or restoring microbiota composition may have an impact on health (4). The term “probiotics” defines live microorganisms that when administered in adequate amounts confer a health benefit on the host (5). Most current probiotics are lactic acid bacteria that belong to the normal intestinal microflora and exert numerous positive effects on human health (6). However, the mechanisms underlying these effects remain poorly understood.
Listeria monocytogenes is the etiological agent of listeriosis (7). To disseminate within the host, L. monocytogenes has to cross three barriers: the intestinal, blood–brain, and/or placental barriers. Crossing the intestinal barrier involves the interaction of L. monocytogenes internalin (InlA) with luminally accessible E-cadherin (Ecad) on enterocytes and goblet cells (8). This interaction is species specific and mouse Ecad, in contrast to human Ecad, does not interact with InlA (9). In a transgenic Fabpi–hEcad mouse line expressing human Ecad and in a knock-in E16P mouse line ubiquitously expressing a “humanized” Ecad, L. monocytogenes efficiently crosses the small intestine, cecum, and colon epithelial barriers (10, 11). The effects of the intestinal microbiota on L. monocytogenes infection have previously been assessed either in cell culture (12–14) or in vivo after L. monocytogenes i.p. or i.v. inoculation (15–18), two models that bypass the oral inoculation route. However, only limited information is available on the impact of lactobacilli on the pathophysiology of orally acquired listeriosis and on the mechanisms by which lactobacilli may be beneficial for the host in the context of an infection via the oral route (19, 20).
We took advantage of the knock-in E16P mouse line and of other recently developed tools such as mouse Affymetrix arrays containing miR probes and Listeria Affymetrix tiling arrays to perform a comprehensive analysis of the impact of two Lactobacillus species during orally acquired listeriosis. We analyzed the effect of Lactobacillus paracasei CNCM I-3689, a strain with in vitro antimicrobial and immunomodulatory properties (21) and Lactobacillus casei BL23, which mediates anti-inflammatory effects in a model of dextran sodium sulfate (DSS)-induced colitis in mice (22). We first established a germ-free colony of the E16P mouse line and showed that it develops systemic listeriosis upon oral challenge. We then showed that Lactobacillus administration decreases L. monocytogenes dissemination and down-regulates expression of immune genes normally induced upon L. monocytogenes infection, in particular IFN-stimulated genes (ISGs). We identified three miRs repressed upon L. monocytogenes infection in vivo and showed that their expression changes after treatment with the Lactobacillus. We also provide evidence that L. monocytogenes blocks gut IL-10 production induced by lactobacilli and enhances gut IL-22 production. Finally, we show that the two lactobacilli regulate L. monocytogenes genes and small RNA (sRNA) expression in particular genes involved in carbon and nitrogen utilization.
Results
Treatment with Lactobacillus Decreases L. monocytogenes Host Invasion.
We derived the E16P mouse line (10) as germ-free. We first established that the two lactobacilli L. paracasei CNCM I-3689 and L. casei BL23 (hereafter called L. paracasei and L. casei) persisted in the intestine after intragastric gavage. Six days after three consecutive gavages of 2 × 109 lactobacilli, the number of lactobacilli was identical in the intestinal (∼107/g for each lactobacilli) and cecal luminal contents of gnotobiotic E16P mice (∼108/g), demonstrating that both strains can colonize ileum and cecum (Fig. S1A). Persistence in the intestine was still observed after 3 wk (Fig. S1B).
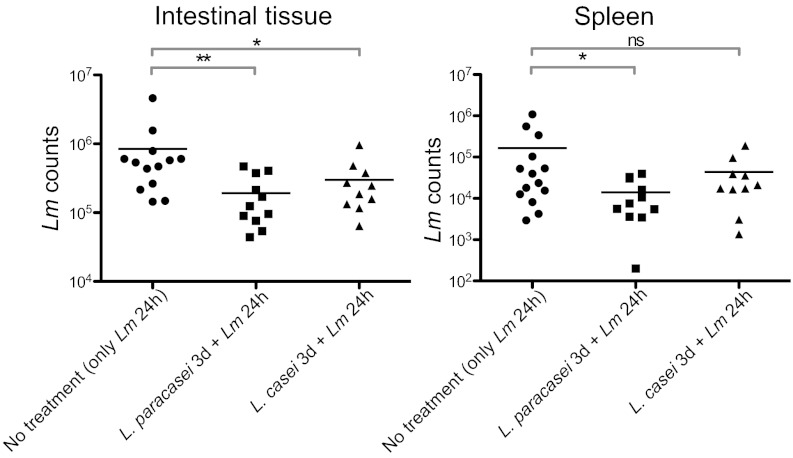
To determine whether Lactobacillus administration of germ-free E16P mice has an impact on L. monocytogenes translocation across the intestinal barrier and dissemination within the host, we evaluated L. monocytogenes counts in the ileal and cecal luminal contents, within the ileal and cecal tissue, and in the mesenteric lymph nodes, spleen, and liver. This showed that intragastric gavage with 2 × 109 L. paracasei and L. casei per mouse for 3 consecutive days prior to L. monocytogenes inoculation (5 × 109) significantly decreased L. monocytogenes counts in the intestinal tissue (Fig. 1), but neither in the cecum, mesenteric lymph nodes, and liver nor in the ileal and cecal luminal contents (Fig. S1C). Strikingly, L. paracasei also reduced L. monocytogenes dissemination to the spleen. We noticed that L. paracasei counts in the intestinal lumen, in contrast to those of L. casei, significantly decreased after L. monocytogenes infection (Fig. S1A), suggesting a competition between L. monocytogenes and L. paracasei in intestinal colonization.
Fig. 1.
Infection with L. monocytogenes (Lm) after Lactobacillus treatment. Lm counts in the intestinal tissue and in the spleen of gnotobiotic E16P mice that were monoassociated or not with the lactobacilli for 3 d and infected 3 d later with Lm for 24 h. Each dot represents one organ. Horizontal bars represent the mean for each condition. Statistical tests were performed using a Mann–Whitney test. Asterisks indicate a value considered statistically significant (*P < 0.05, **P < 0.01); NS, nonsignificant difference.
To demonstrate the relevance of our findings, we investigated whether lactobacilli are also able to limit L. monocytogenes colonization in conventional E16P mice and determined whether the two lactobacilli affect L. monocytogenes counts in mice: L. paracasei slightly but significantly decreased L. monocytogenes counts in the spleen, and L. casei significantly decreased L. monocytogenes counts in the spleen and the liver of conventional E16P mice (Fig. S1D).
Altogether these results establish that L. paracasei and L. casei can limit L. monocytogenes invasive infection. As L. paracasei and L. casei have similar inhibitory activity against L. monocytogenes in vitro (Fig. S2), their distinct effects probably rely on genomic differences and/or different interactions with the intestinal microbiota and the host.
Treatment with Lactobacillus Down-Regulates L. monocytogenes-Induced Immune Genes in the Intestine, in Particular IFN-Stimulated Genes.
We performed a whole genome transcriptional analysis of intestinal tissue in response to L. casei, L. paracasei, and L. monocytogenes, respectively, and compared it to that of germ-free E16P mice. We next analyzed whether treatment with each Lactobacillus affects host gene expression upon L. monocytogenes infection.
We found that expression of 87 and 71 genes significantly changed after 3 d of monoassociation with L. paracasei and L. casei, respectively (Fig. S3A). Only 18 genes were affected by both lactobacilli, suggesting that although the two species belong to the same genus, they do not behave identically. We next grouped these genes into canonical pathways using Ingenuity IPA software (IPA) (Fig. S4 A and B). Regulation of xenobiotic metabolism by cytochrome P450 and tryptophan metabolism were the only signaling pathways modulated by both lactobacilli. In conclusion, these results show that although L. paracasei and L. casei are taxonomically related, they each trigger a species-specific response.
L. monocytogenes alone was shown to affect the transcription of almost 1,000 genes 24 h postinfection: 493 genes were down-regulated and 494 were up-regulated in infected mice (Fig. S3B). Using IPA, we could show that these genes grouped in pathways involved in immune response, intracellular signaling, nuclear receptor signaling, and xenobiotic metabolism as well as amino acid, carbohydrate, and lipid metabolism (Fig. S4C). The most significantly induced genes (more than fivefold) were involved in immune responses, whereas genes related to host metabolism were highly repressed (more than fivefold). These results are in complete agreement with our initial study comparing the host response to L. monocytogenes, or to the related nonpathogenic noninvasive species L. innocua, or to Bacteroides thetaiotaomicron, a gut symbiont (23).
Strikingly, treatment with each Lactobacillus modulated expression of gene subsets during infection (Fig. S3C), with ISGs being the most affected by both lactobacilli (Fig. 2, Left). Among these 23 ISGs, 10 encode GTPases: 4 of the p47 immunity-related GTPase family, 4 guanylate-binding proteins of the p65 family (Gbps), and 2 immunity-related GTPases (M family). Other ISGs highly induced by L. monocytogenes and less induced in L. monocytogenes-infected mice treated with the two lactobacilli include Ifit1 and Ifit3 encoding IFN-induced proteins with tetratricopeptide repeats, Ifi205 IFN-activated gene, Oas2 2′–5′ oligoadenylate synthetase 2 and Mx2 encoding an IFN-induced GTP-binding protein. The differential expression of IfitI and Oas2 genes in the different conditions was confirmed by quantitative (q)RT-PCR (Fig. S5A). Some genes were modulated by only one of the two lactobacilli (Fig. S3 D and E): for instance, L. paracasei treatment affected expression of B3galt5 (UDP-Gal:β-GlcNAc β-1,3-galactosyltransferase), Car3 (carbonic anhydrase), Gstm3 (GST), Lrat (lecithin-retinol acyltransferase), and Nos2 (generation of reactive nitrogen species), whereas L. casei treatment affected expression of Chd7 (chromodomain helicase), Cubn (cubulin), and Lphn2 (latrophilin) (Fig. 2, Right). In conclusion, association with Lactobacillus, which alone only affects a small number of genes, modulates, in L. monocytogenes-infected mice, principally but not exclusively the expression of ISGs, which are among the most highly induced genes after L. monocytogenes infection.
Fig. 2.
Transcriptomic analysis of the host response after Lactobacillus treatment and L. monocytogenes (Lm) infection. Effect of treatment with Lactobacillus on murine gene expression upon Lm infection. Mice were monoassociated or not monoassociated for 3 consecutive days (3 d) and infected 3 d later with Lm for 24 h. The heatmap presents a subset of host genes whose expression was significantly affected by the lactobacilli during Lm infection [false discovery rate, Benjamini and Hochberg approach (FDR-BH), P < 0.05]. Left, the three columns show values corresponding to the fold change (FC) of gene relative expression in Lm-infected mice (Lm), in mice treated with L. paracasei and infected by Lm (L. paracasei + Lm) and in mice treated with L. casei and infected by Lm (L. casei + Lm) compared with control mice (FC/control mice). Right, the two last columns show FC of gene relative expression in (L. paracasei + Lm) and (L. casei + Lm) mice compared with Lm-infected mice (FC/Lm-infected mice). White square in the heatmap indicates a gene whose expression was not significantly affected by the treatment with the lactobacilli. Asterisks indicate ISGs.
Treatment with Lactobacillus Modulates Expression of Several microRNAs During L. monocytogenes Infection.
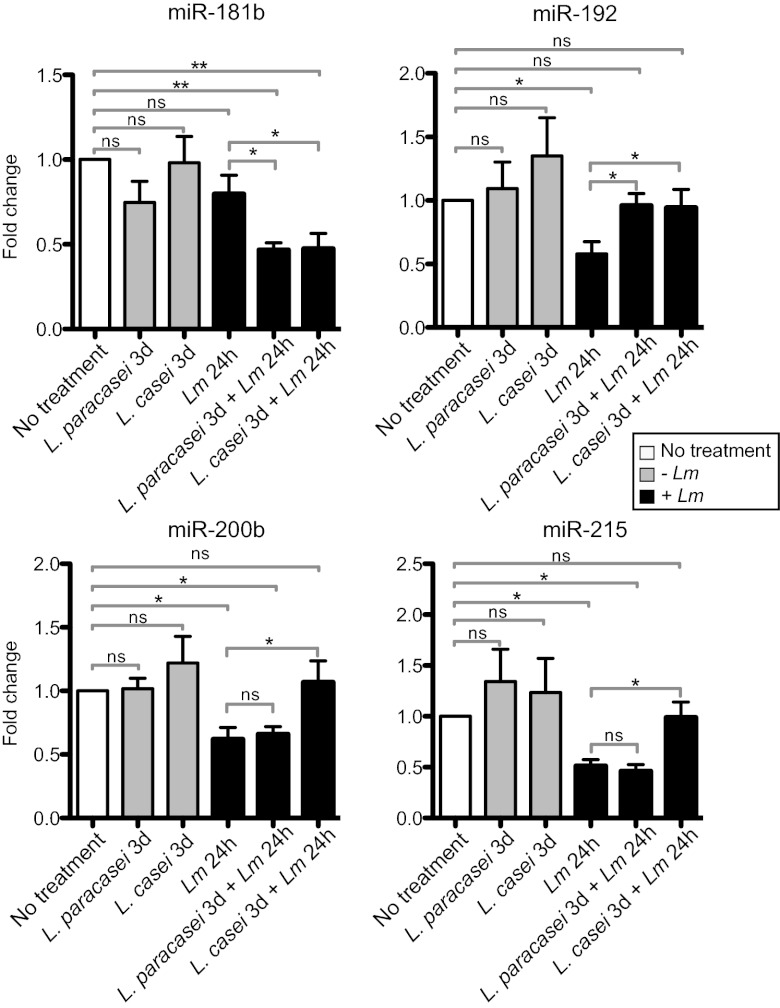
microRNAs (miRs) are endogenous small RNAs regulating gene expression by base pairing to messenger RNAs. miRs are involved in a variety of functions such as cellular growth, apoptosis, metabolism, immunity, and cancer (24). The role of miRs in microbiota–host interactions has only recently started to be investigated (25, 26). We used the Affymetrix Mouse Gene 1.0 ST arrays to investigate miR expression. Although these arrays have been designed primarily to analyze expression of protein-coding genes, 97 miR probe sets are also present on this array. Our array data combined with qRT-PCR analysis showed that in absence of treatment with each Lactobacillus, expression of miR-192, miR-200b, and miR-215 was repressed during L. monocytogenes infection and that treatment with each Lactobacillus increased miR-192 expression in infected mice (Fig. 3). Only association with L. casei led to higher expression of miR-200b and miR-215. In addition, we detected one miR, mirR-181b, whose expression was not affected during L. monocytogenes infection alone but decreased during infection after treatment with each Lactobacillus (Fig. 3). In conclusion, miRs are regulated by L. monocytogenes infection in vivo and treatment with each Lactobacillus, suggesting that these miRs in turn regulate host gene expression.
Fig. 3.
Detection of miRNA during infection. Relative expression of miR181b, miR-192, miR-200b, and miR-215 in mice monoassociated for 3 consecutive days (3 d) with L. paracasei or L. casei, or in mice that were monoassociated or not with the lactobacilli for 3 d and infected 3 d later with L. monocytogenes (Lm) for 24 h. Fold change is presented after standardization to the small nuclear RNA U6 and using control mice that have been normalized to 1 as reference. Data are represented as mean with SEM. Statistical tests were performed using a two-tailed Student t test. Asterisks indicate a value considered statistically significant (*P < 0.05, **P < 0.01); NS, nonsignificant difference.
Treatment with Lactobacillus Affects IFNγ Production in Spleen During Listeriosis.
Early production of IFNγ is a critical step of the immune response against L. monocytogenes (27). We detected IFNγ induction in the small intestine and spleen of L. monocytogenes-infected mice 24 h after infection (Fig. S6). A slight IFNγ production was detectable after 3 d of Lactobacillus monoassociation. Treatment with each Lactobacillus significantly reduced IFNγ production in the spleen of the L. monocytogenes-infected gnotobiotic mice but not in the small intestine (Fig. S6).
We also examined the production of IL-10, IL-22, and IL-2. IL-10 is an anti-inflammatory cytokine preventing excessive inflammation. IL-22 initiates an innate immune response against bacterial pathogens especially in epithelial cells such as enterocytes (28). IL-2, a pleiotropic cytokine, promotes the growth and differentiation of T cells and enhances the activity of natural killer cells (29). We showed that the level of IL-10 and IL-2 increased in the small intestine after 3 consecutive days of monoassociation with each Lactobacillus (Fig. S6). In contrast, IL-22 production was not detected in monoassociated mice. During L. monocytogenes infection without treatment with lactobacilli, the level of IL-22 increased, but not that of IL-10 and IL-2. Strikingly, L. monocytogenes infection down-regulated IL-10 and IL-2 production observed during monoassociation with L. paracasei and L. casei. However, L. casei treatment led to a higher IL-22 production in the small intestine of L. monocytogenes-infected mice (Fig. S6). Our data thus show that L. monocytogenes infection blocks IL-2 and IL-10 small intestinal production triggered after 3 d of monoassociation with the two lactobacilli, but promotes IL-22 production.
Treatment with Lactobacillus Reshapes the L. monocytogenes Protein-Coding Genes and sRNA Expression Program.
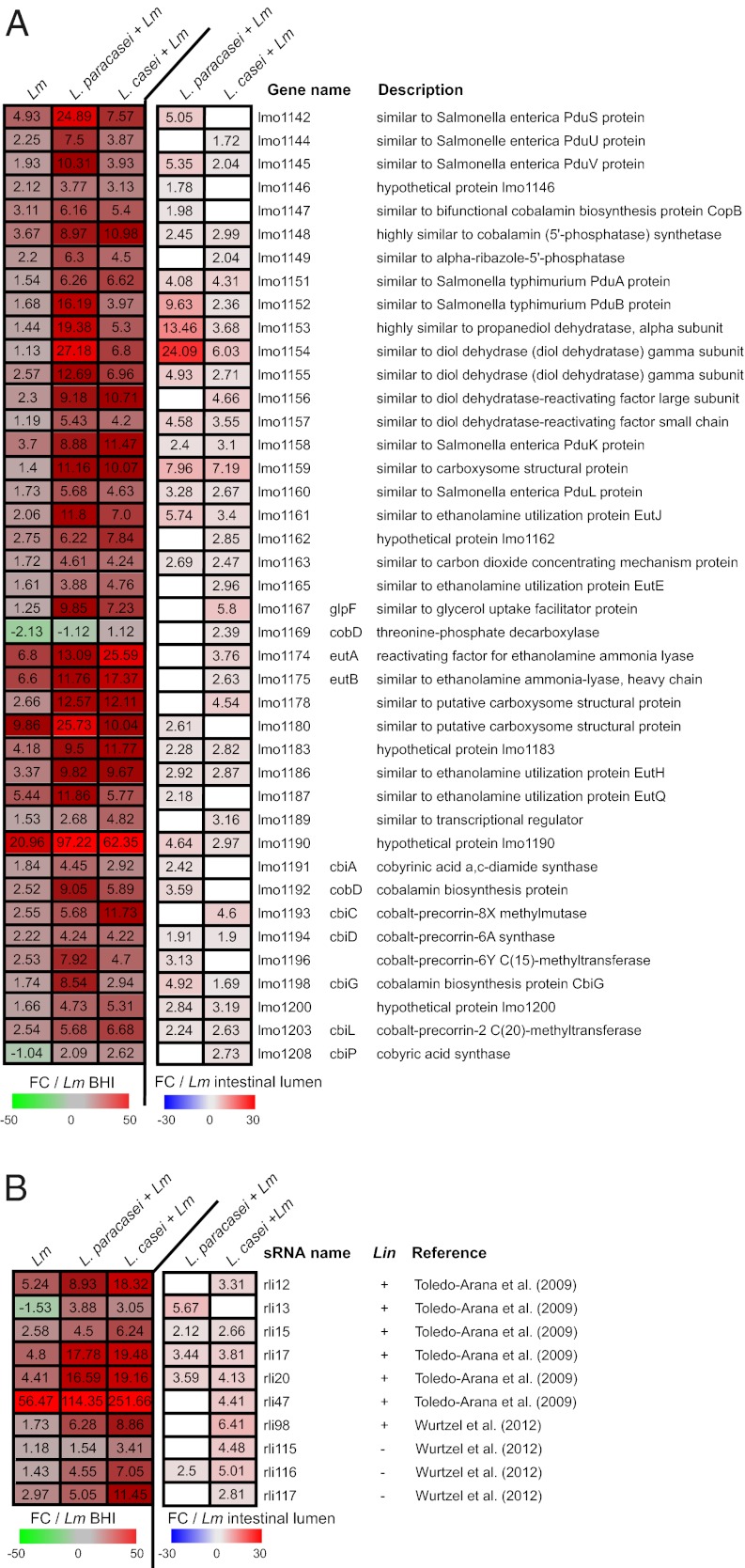
We used Listeria Affymetrix tiling arrays and L. monocytogenes RNAs retrieved from the ileo-cecal content of E16P gnotobiotic mice treated or not treated by the two lactobacilli to determine whether Lactobacillus treatment modulates L. monocytogenes genes and also sRNA expression. Indeed L. monocytogenes possesses a large repertoire of both cis- and trans-encoded RNAs but relatively little is known about their role in L. monocytogenes pathogenesis (30–33). We have shown previously an extensive transcriptional reshaping upon intestinal infection when comparing gene expression levels in bacteria either present in the ileo-cecal content of germ-free mice or grown in broth medium (31). Here, we compared L. monocytogenes gene expression in the ileo-cecal content of E16P germ-free mice during infection to that observed in broth medium: 520 L. monocytogenes protein-coding genes were significantly up-regulated (fold change, FC, from 2- to 130-fold) and 523 down-regulated (FC 2–102) (Fig. 4A and Fig. S7, first column); seven sRNAs were up-regulated (FC from 2- to 57-fold) and three sRNAs down-regulated (FC 2–10) (Fig. 4B and Fig. S8).
Fig. 4.
Listeria tiling analysis. (A) Effect of treatment with Lactobacillus on L. monocytogenes (Lm) gene expression during infection. Mice were monoassociated or not monoassociated for 3 consecutive days (3 d) and infected 3 d later with Lm for 24 h. Heatmaps present a subset of Lm genes (A) and all sRNAs (B) whose expression was significantly affected by the lactobacilli during Lm infection (t test P < 0.05). Left, the three columns show values corresponding to the fold change (FC) of gene (A) and sRNA (B) relative expression in ileal-cecal content from Lm-infected mice (Lm), in mice treated with L. paracasei and infected by Lm (L. paracasei + Lm) and in mice treated with L. casei and infected by Lm (L. casei + Lm) compared with Lm growing in broth medium (FC/Lm BHI). Right, the two last columns show FC of gene relative expression in (L. paracasei + Lm) and (L. casei + Lm) mice compared with Lm-infected mice (FC/Lm-intestinal lumen). White square in the heatmap indicates a gene whose expression was not significantly affected by treatment with the lactobacilli. Absence (−) or presence (+) of sRNAs in L. innocua (Lin) is indicated.
Treatment with each Lactobacillus affected the expression of the same 200 L. monocytogenes genes during infection. In addition, L. paracasei and L. casei also affected an additional specific set of 125 and 702 genes, respectively (Fig. S7). Remarkably, gene clusters involved in 1,2-propanediol and ethanolamine catabolism and in cobalamin (vitamin B12) biosynthesis were among the most significantly increased by the two lactobacilli (Fig. 4A and Fig. S5B).
Expression of several sRNAs, Rli15, Rli17, and that of Rli20 induced by L. monocytogenes infection, was higher after treatment with each Lactobacillus (Fig. 4B). We also found that expression of Rli12, Rli47, and Rli117, which was increased during infection, further increased after treatment with L. casei. On the contrary, we found examples like that of Rli116 whose expression was not affected by L. monocytogenes infection but increased after treatment with Lactobacillus (Fig. 4B and Fig. S5C).
Altogether our data show that treatment with each Lactobacillus decreases dissemination in the host, affects host gene expression, in particular ISGs, modulates miR regulation, and also triggers a significant reshaping in expression of L. monocytogenes protein-coding genes and sRNAs.
Discussion
We have performed a comprehensive study of the impact of two Lactobacillus species on orally acquired listeriosis. We show that both L. paracasei and L. casei are able to limit L. monocytogenes dissemination in vivo. We then show that lactobacilli affect the expression of a subset of host genes, in particular ISGs. Moreover, lactobacilli modulate the expression of three miRs, miR-192, miR-200b, and miR-215, repressed by L. monocytogenes infection. We also show that L. monocytogenes infection decreases gut IL-2 and IL-10 production triggered by lactobacilli and stimulates IL-22 production in the small intestine. Finally, treatment with each Lactobacillus significantly reshapes the expression program of L. monocytogenes protein-coding genes and of sRNAs. To our knowledge, this is a unique genome-wide transcriptomic analysis investigating the host and pathogen responses simultaneously and the expression of protein- and small noncoding RNA-encoding genes.
Our study highlights that the two lactobacilli behave slightly differently. It is increasingly recognized that chemicals produced by bacteria and their hosts control bacterial interactions and/or communication in the environment and in the host (1). Although lactic acid production by the two lactobacilli may contribute to the decrease in L. monocytogenes counts in organs, their distinct effects on L. monocytogenes infection probably mainly result from genomic differences and/or different interactions with the intestinal microbiota and the host.
Lactobacillus treatment affects the host response to L. monocytogenes infection. We had previously shown that this response is characterized by an up-regulation of genes involved in immune responses and a down-regulation of genes involved in lipid, amino acid, and energy metabolism (23). In agreement with this initial study, we show here that the host response to L. monocytogenes, is characterized by an induction of genes involved in immune response, intracellular signaling, nuclear receptor signaling, and xenobiotic metabolism as well as amino acid, carbohydrate, and lipid metabolism. More specifically, we show that treatment with Lactobacillus leads to a decrease of L. monocytogenes counts in host tissues and a down-regulated expression of ISGs at the intestinal level. Among L. monocytogenes-induced ISGs, genes encoding p47 GTPases have been shown to regulate innate immunity and inflammation (34) and their role in protecting L. monocytogenes infection has been recently highlighted (35). In contrast to type II IFN, which is beneficial to the host during listeriosis, type I IFN is detrimental (27, 36). More recently, type III IFN was shown to be produced upon L. monocytogenes infection (37, 38). It will thus be interesting to determine which type of IFN regulates the ISG subset affected by the Lactobacillus.
We also show in this study that treatment with both lactobacilli modulates expression of genes of the cytochrome P450 family (Cyp) and L. paracasei treatment affects expression of three glutathione S-transferases. Enterocytes have the ability to metabolize drugs or xenobiotics by numerous pathways involving phase I and II reactions (39). In phase I, enzymes such as Cyp P450 oxidases modify xenobiotics that are conjugated to polar compounds in phase II reactions catalyzed by enzymes such as GST before being processed and transported out from the cells. Gene expression in the Cyp P450 family is regulated by NF-κB and nuclear receptors such as CAR, PXR, RXR, PPAR, FXR, and LXR, which mediate gene regulation through chromatin remodeling and histone modifications (40, 41). Importantly, metabolism of xenobiotics by the Cyp P450 and LXR, PXR, and RXR signaling pathways were among the most significantly regulated pathways 24 h postinfection with L. monocytogenes: e.g., 19 Cyp genes from the P450 family and more than 30 genes encoding solute carrier proteins regulating transport of many substances. Altogether these data suggest that the so-called responses to xenobiotics mediated by the Cyp P450 and nuclear receptors could be involved in the control of host invasion by microbes. The Cyp P450 system consists of a large subfamily of enzymes that are genetically polymorphic. Because it has been proposed that this polymorphism may affect the capacity of individual patients to absorb and metabolize drugs (42), Cyp P450 polymorphism may also be involved in differences in susceptibility to infectious diseases.
The field of miRs and bacterial infections is still in its infancy. Two recent papers have reported that L. monocytogenes promotes changes in the level of several miRs in epithelial cells and in macrophages in vitro (43, 44). It has also been shown that miR-29 regulates immune responses to L. monocytogenes by targeting IFNγ mRNA in vivo (45). Here, we show that treatment with Lactobacillus leads to a higher expression of miR-200b, miR-215, and miR-192, which are normally repressed during L. monocytogenes infection and to a lower expression of miR-181b, which does not vary during infection. The miR-200 family has been reported to induce epithelial differentiation and suppress epithelial–mesenchymal transition by inhibiting translation of zinc finger E-box–binding homeobox ZEB1 and ZEB2, two transcriptional repressors of E-cadherin, in several types of cancers (46). Strikingly, using the Target Scan Mouse 6.0 software, we identified mRNAs predicted to be targeted by miR-200b, whose expression in our transcriptomic data anticorrelates with that of miR-200b: Nlr5, Slf4, Ly6c, Ifi203, Gm12250, and Gbp6 encoding, respectively, the NOD-like receptor NLR5, a member of the Schlafen family involved in cell growth and T-cell development, the lymphocyte antigen 6, an IFN-induced protein, and the IrgB10 and Gbp6 GTPases, respectively. Interestingly, miR-192 which has the same “seed region” as miR-215 is also predicted to target Nlr5, Gbp6, and Slf5 mRNAs as well as the GTPases encoded by the genes Irgm2 and Tgtp1. Increase in expression of these miRs by one or two of the lactobacilli may be responsible for the lower expression of the corresponding genes after treatment with the Lactobacillus. It will be important to decipher among the hundreds of genes predicted to be targeted by these miRs (Fig. S9) those involved in lactobacilli-mediated effects.
We also demonstrate here that treatment with Lactobacillus reshapes L. monocytogenes transcriptome. Both protein-coding and sRNAs genes were affected. Work in progress addresses the mode of action of these sRNAs and their targets. We previously showed a role of the stress-responsive alternative σ factor, Sigma B (σB) in the expression of the invasion proteins, InlA and InlB, and other surface proteins (31). Here, we found that treatment with lactobacilli increases expression of such σB-regulated genes, illustrating the possible stress encountered by L. monocytogenes in the intestinal lumen in the presence of the two lactobacilli. More importantly, we identified several gene clusters encoding enzymes allowing L. monocytogenes to catabolize intestinal carbon and nitrogen sources such as 1,2-propanediol and ethanolamine and to synthesize cobalamin (vitamin B12), a cofactor for enzymes involved in ethanolamine and 1,2-propanediol degradation whose expression was increased by the presence of each Lactobacillus. 1,2-Propanediol is produced by the fermentation of plant sugars rhamnose and fucose (47). Ethanolamine is a compound mainly derived from phosphatidyl ethanolamine in epithelial cell membranes that only certain bacteria can use as a source of carbon and/or nitrogen (48). Many reports have shown that L. monocytogenes, Clostridium perfringens, Enteroccocus faecalis, and Salmonella enterica possess a highly similar organization of the ethanolamine gene cluster, which is absent in most other bacteria, suggesting that ethanolamine utilization contributes to virulence (49, 50). Interestingly, it has recently been shown that intestinal inflammation allows Salmonella to use ethanolamine and compete with microbiota (51). It is probably also the case for L. monocytogenes. In L. monocytogenes, a lack of ethanolamine ammonia lyase leads to an attenuated proliferation in epithelial cells (52) and all genes involved in ethanolamine catabolism are up-regulated in the intestine of infected gnotobiotic mice (31). However, our results suggest that in gnotobiotic mice, increasing the expression of these gene clusters does not confer a selective advantage to L. monocytogenes because treatment with both lactobacilli decreases L. monocytogenes translocation and dissemination in the host. It is possible that in the absence of lactobacilli, L. monocytogenes uses various carbon and energy sources including ethanolamine and 1,2-propanediol. After treatment with lactobacilli, a competition between L. monocytogenes and the lactobacilli is likely taking place for carbon and nitrogen availability forcing L. monocytogenes to use mostly ethanolamine not utilizable by lactobacilli (53), suggesting that the competition between L. monocytogenes and the lactobacilli for carbon and nitrogen availability is a critical step for a successful invasion.
To our knowledge, our study is a unique comprehensive approach that analyzes simultaneously in the host and the pathogen the molecular events occurring after treatment with two lactobacilli species. It paves the way for a thorough and systematic analysis of the key components mediating the protecting effect of lactobacilli.
Materials and Methods
Additional information on the strains and reagents and on the experimental protocols is provided in SI Materials and Methods.
Bacterial Strains.
L. monocytogenes EGDe strain was grown in brain heart infusion (BHI) medium (Difco) at 37 °C. L. paracasei CNCM I-3689 and L. casei BL23 strains were grown in de Man, Rogosa, and Sharpe (MRS) medium (Oxoid) at 37 °C.
Animals and Generation of a Germ-Free Mouse Line.
All experiments involving mice were handled in accordance with the Pasteur Institute guidelines for animal welfare. We derived the knock-in E16P mouse line (10) as germ-free mice (Taconic).
Monoassociation and Treatment with Lactobacillus.
For each Lactobacillus, mice were inoculated orally with 2 × 109 bacteria diluted in 200 μL of PBS. Mice (n ≥ 3 per group) were either (i) monoassociated with each Lactobacillus for 3 consecutive days (3 d) (or for 24 h when required) and killed 6 d after the first inoculation or (ii) monoassociated with the lactobacilli for 3 consecutive days and infected 3 d later with L. monocytogenes for 24 h.
Infection.
Mice (n ≥ 3 per condition) were infected orally with 5 × 109 bacteria diluted in 200 μL of PBS supplemented with 300 μL of CaCO3 (50 mg/mL) for 24 h. Serial dilutions of the inoculum were plated to control the number of L. monocytogenes inoculated in mice.
Bacterial Counts.
The spleen was directly disrupted in PBS. Intestinal fragments were incubated 2 h in DMEM containing 100 μg/mL gentamicin and disrupted in PBS. Serial dilutions were plated on BHI plates and incubated for 2 d at 37 °C.
Antimicrobial Activity.
The antimicrobial activity was tested as previously described (21).
Mouse Gene Chip Analysis.
RNAs from the ileal tissue were extracted and purified using classical TRIzol/chloroform protocol. Labeled cDNA was synthesized from 200 ng total RNAs using NuGEN Applause WT-Amp Plus ST systems (NuGEN Technologies). Labeled samples were hybridized to Affymetrix MoGene 1.0 ST GeneChips and scanned with an Affymetrix GeneChip Scanner 3000, generating cell intensity files for each array. Gene-level expression values were derived from the CEL file probe-level hybridization intensities using the model-based robust multichip average algorithm (54). Canonical pathways have been identified using the Ingenuity IPA application using a P value calculated by Fisher's exact test, right tailed.
Listeria Tiling Array Analysis.
Total bacterial RNAs from the ileo-cecal content were extracted and treated as previously described (31); extracted RNAs from each mouse (n = 3 for each condition) were treated separately. Total RNAs (200 ng) were amplified using the MessageAmpII-Bacteria kit (Ambion) before fragmentation using the 5× fragmentation buffer. A total of 7.5 μg of amplified and fragmented RNA was used per chip. Sample preparation for each chip was then processed following the Affymetrix GeneChip Expression Analysis Technical Manual (P/N 702232 Rev. 2) as previously described (31).
Supplementary Material
Acknowledgments
We thank J. R. Mellin, N. Sesto, and A. Tessier for fruitful discussions and the Animalerie Axénique de Micalis (ANAXEM) platform at Institut National de la Recherche Agronomique (INRA) and the Pasteur animal facilities. This work received financial support from Danone Research Project 704033, European Research Council (ERC) Advanced Grant 233348, ERC Starting Grant 261157, Programme Transversal de Recherche Pasteur-INRA PTR-288, Institut Pasteur, Institut National de la Santé et de la Recherche Médicale, Ville de Paris, Fondation pour la Recherche Médicale, and the Banque Nationale de Paris-Paribas Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212809109/-/DCSupplemental.
References
- 1.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.Gordon JI, Klaenhammer TR. A rendezvous with our microbes. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4513–4515. doi: 10.1073/pnas.1101958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez-Bello MG, Blaser MJ. Do you have a probiotic in your future? Microbes Infect. 2008;10:1072–1076. doi: 10.1016/j.micinf.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrogosz WJ, Peacock TJ, Hassan HM. Evolution of the probiotic concept from conception to validation and acceptance in medical science. Adv Appl Microbiol. 2010;72:1–41. doi: 10.1016/S0065-2164(10)72001-3. [DOI] [PubMed] [Google Scholar]
- 6.Masood MI, Qadir MI, Shirazi JH, Khan IU. Beneficial effects of lactic acid bacteria on human beings. Crit Rev Microbiol. 2011;37:91–98. doi: 10.3109/1040841X.2010.536522. [DOI] [PubMed] [Google Scholar]
- 7.Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikitas G, et al. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J Exp Med. 2011;208:2263–2277. doi: 10.1084/jem.20110560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lecuit M, et al. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 1999;18:3956–3963. doi: 10.1093/emboj/18.14.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Disson O, et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 11.Lecuit M, et al. A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 12.Coconnier MH, et al. Inhibition of adhesion of enteroinvasive pathogens to human intestinal Caco-2 cells by Lactobacillus acidophilus strain LB decreases bacterial invasion. FEMS Microbiol Lett. 1993;110:299–305. doi: 10.1111/j.1574-6968.1993.tb06339.x. [DOI] [PubMed] [Google Scholar]
- 13.Coconnier MH, Liévin V, Bernet-Camard MF, Hudault S, Servin AL. Antibacterial effect of the adhering human Lactobacillus acidophilus strain LB. Antimicrob Agents Chemother. 1997;41:1046–1052. doi: 10.1128/aac.41.5.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corr SC, Gahan CG, Hill C. Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunol Med Microbiol. 2007;50:380–388. doi: 10.1111/j.1574-695X.2007.00264.x. [DOI] [PubMed] [Google Scholar]
- 15.de Waard R, Garssen J, Vos JG, Claassen E. Modulation of delayed-type hypersensitivity and acquired cellular resistance by orally administered viable indigenous lactobacilli in Listeria monocytogenes infected Wistar rats. Lett Appl Microbiol. 2002;35:256–260. doi: 10.1046/j.1472-765x.2002.01171.x. [DOI] [PubMed] [Google Scholar]
- 16.dos Santos LM, et al. Monoassociation with probiotic Lactobacillus delbrueckii UFV-H2b20 stimulates the immune system and protects germfree mice against Listeria monocytogenes infection. Med Microbiol Immunol (Berl) 2011;200:29–38. doi: 10.1007/s00430-010-0170-1. [DOI] [PubMed] [Google Scholar]
- 17.Kim YG, et al. Probiotic Lactobacillus casei activates innate immunity via NF-kappaB and p38 MAP kinase signaling pathways. Microbes Infect. 2006;8:994–1005. doi: 10.1016/j.micinf.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Sato K. Enhancement of host resistance against Listeria infection by Lactobacillus casei: role of macrophages. Infect Immun. 1984;44:445–451. doi: 10.1128/iai.44.2.445-451.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bambirra FH, et al. Protective effect of Lactobacillus sakei 2a against experimental challenge with Listeria monocytogenes in gnotobiotic mice. Lett Appl Microbiol. 2007;45:663–667. doi: 10.1111/j.1472-765X.2007.02250.x. [DOI] [PubMed] [Google Scholar]
- 20.Corr SC, et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patent Application Publication 2011. Novel strain of Lactobacillus paracasei subspecies paracasei having antimicrobial and immunomodulary properties. US 2011/0150852 A1.
- 22.Rochat T, et al. Anti-inflammatory effects of Lactobacillus casei BL23 producing or not a manganese-dependant catalase on DSS-induced colitis in mice. Microb Cell Fact. 2007;6:22. doi: 10.1186/1475-2859-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecuit M, Sonnenburg JL, Cossart P, Gordon JI. Functional genomic studies of the intestinal response to a foodborne enteropathogen in a humanized gnotobiotic mouse model. J Biol Chem. 2007;282:15065–15072. doi: 10.1074/jbc.M610926200. [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalmasso G, et al. Microbiota modulate host gene expression via microRNAs. PLoS ONE. 2011;6:e19293. doi: 10.1371/journal.pone.0019293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue X, et al. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J Immunol. 2011;187:5879–5886. doi: 10.4049/jimmunol.1100535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stavru F, Archambaud C, Cossart P. Cell biology and immunology of Listeria monocytogenes infections: Novel insights. Immunol Rev. 2011;240:160–184. doi: 10.1111/j.1600-065X.2010.00993.x. [DOI] [PubMed] [Google Scholar]
- 28.Rutz S, Ouyang W. Regulation of interleukin-10 and interleukin-22 expression in T helper cells. Curr Opin Immunol. 2011;23:605–612. doi: 10.1016/j.coi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: New insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 32.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Wurtzel O, et al. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacMicking JD. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 2004;25:601–609. doi: 10.1016/j.it.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Kim BH, et al. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 36.Stockinger S, Decker T. Novel functions of type I interferons revealed by infection studies with Listeria monocytogenes. Immunobiology. 2008;213:889–897. doi: 10.1016/j.imbio.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 37.Bierne H, et al. Activation of type III interferon genes by pathogenic bacteria in infected epithelial cells and mouse placenta. PLoS ONE. 2012;7:e39080. doi: 10.1371/journal.pone.0039080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebreton A, et al. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science. 2011;331:1319–1321. doi: 10.1126/science.1200120. [DOI] [PubMed] [Google Scholar]
- 39.Omiecinski CJ, Vanden Heuvel JP, Perdew GH, Peters JM. Xenobiotic metabolism, disposition, and regulation by receptors: From biochemical phenomenon to predictors of major toxicities. Toxicol Sci. 2011;120(Suppl 1):S49–S75. doi: 10.1093/toxsci/kfq338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishimoto M, et al. Nuclear receptor mediated gene regulation through chromatin remodeling and histone modifications. Endocr J. 2006;53:157–172. doi: 10.1507/endocrj.53.157. [DOI] [PubMed] [Google Scholar]
- 41.Zordoky BN, El-Kadi AO. Role of NF-kappaB in the regulation of cytochrome P450 enzymes. Curr Drug Metab. 2009;10:164–178. doi: 10.2174/138920009787522151. [DOI] [PubMed] [Google Scholar]
- 42.van Schaik RH. CYP450 pharmacogenetics for personalizing cancer therapy. Drug Resist Updat. 2008;11:77–98. doi: 10.1016/j.drup.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Izar B, Mannala GK, Mraheil MA, Chakraborty T, Hain T. microRNA response to Listeria monocytogenes infection in epithelial cells. Int J Mol Sci. 2012;13:1173–1185. doi: 10.3390/ijms13011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnitger AK, et al. Listeria monocytogenes infection in macrophages induces vacuolar-dependent host miRNA response. PLoS ONE. 2011;6:e27435. doi: 10.1371/journal.pone.0027435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma F, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 46.Kurashige J, et al. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg Oncol. 2012;19(Suppl 3):656–664. doi: 10.1245/s10434-012-2217-6. [DOI] [PubMed] [Google Scholar]
- 47.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsoy O, Ravcheev D, Mushegian A. Comparative genomics of ethanolamine utilization. J Bacteriol. 2009;191:7157–7164. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchrieser C, Rusniok C, Kunst F, Cossart P, Glaser P. Listeria Consortium Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: Clues for evolution and pathogenicity. FEMS Immunol Med Microbiol. 2003;35:207–213. doi: 10.1016/S0928-8244(02)00448-0. [DOI] [PubMed] [Google Scholar]
- 50.Garsin DA. Ethanolamine utilization in bacterial pathogens: Roles and regulation. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiennimitr P, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joseph B, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamada N, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.