Abstract
Deregulation of microRNA (miRNA) expression can lead to cancer initiation and progression. However, limited information exists on the function of miRNAs in cancer maintenance. We examined these issues in the case of myeloproliferative diseases and neoplasms (MPN), a collection of hematopoietic neoplasms regarded as preleukemic, thereby representing early neoplastic states. We report here that microRNA-125a (miR-125a)–induced MPN display a complex manner of oncogene dependence. Following a gain-of-function genomics screen, we overexpressed candidate miR-125a in vivo, which led to phenotypes consistent with an atypical MPN characterized by leukocytosis, monocytosis, splenomegaly, and progressive anemia. The diseased MPN state could be recapitulated in a doxycycline-inducible mouse model. Upon doxycycline withdrawal, the primary MPN phenotypes rapidly resolved after the discontinuation of miR-125a overexpression. However, reinduction of miR-125a led to complex phenotypes, with some animals rapidly developing lethal anemia with extensive damages in the spleen. Forced expression of miR-125a resulted in elevated cellular tyrosine phosphorylation and hypersensitivity toward hematopoietic cytokines. Furthermore, we demonstrate that miR-125a targets multiple protein phosphatases. Our data demonstrate that miR-125a–induced MPN is addicted to its sustained overexpression, and highlight the complex nature of oncogenic miRNA dependence in an early neoplastic state.
Keywords: oncogene addiction, PTPN18, spleen fibrosis
The expression of microRNAs (miRNAs) is frequently deregulated in human cancers (1, 2). Alteration in miRNA gene expression or function can promote cancer initiation and progression in vivo, leading to the recognition that this class of small noncoding RNAs can function as both oncogenes and tumor suppressors (3–5). Much less is known, however, regarding the role of miRNAs in cancer maintenance. A special type of cancer maintenance is related to oncogene dependence/addiction, in which the maintenance of cancer phenotype is dependent on the sustained presence of one of the initial driving genetic lesions, even though cancer cells may have acquired additional mutations. Indeed, oncogene addiction has been demonstrated for a short list of protein-coding genes in vivo [e.g., c-myc, BCR/ABL, Her-2, and p53 (6–7)], many of which are clinically validated therapeutic targets (8, 9). In the case of miRNAs, withdrawal of ectopic miR-21 is known to cause regression in a mouse model of lymphoma (10), indicating that some established lymphomas may be dependent on miR-21 overexpression.
Interestingly, recent studies on p53 demonstrate that the phenotype of oncogene addiction can be dependent on the stage of cancer progression (11, 12). In mouse lung cancer models driven by the loss of p53 and mutant k-ras, genetic restoration of p53 leads to significant loss of tumor mass in more advanced tumor cells but not in the earlier, less aggressive ones. These studies suggest that early-stage neoplasms may provide a context for complex oncogene addition phenomena. However, dependence on oncogene behavior in early-stage neoplasms in vivo has not been much explored in other types of malignancies. Myeloproliferative diseases (also known as myeloproliferative neoplasms, MPNs) are a collection of hematopoietic diseases with common features (reviewed in refs. 13 and 14), such as overproduction of one or more myeloid/erythroid/platelet lineages and splenomegaly. In some patients, the MPN will progress to acute myeloid leukemia, leading to the notion that MPN is one of the preleukemic states of a full-blown myeloid leukemia.
In this study, we used genomics and genetics to explore the role of miRNAs in MPN and the dependence of the early neoplastic MPN state on an oncogenic miRNA. We report a barcode genetic screen with the use of a unique miRNA expression library, which led to the identification of miR-125 miRNAs in inducing hypersensitivity toward cytokine signaling in both cell line and primary bone marrow cells. Using a unique inducible model for miRNA expression, we show that miR-125a–induced MPN display a complex manner of dependence on the continued expression of miR-125a. Although the hematopoietic phenotypes of the initial miR-125a–induced MPN appear reversible, re-exposure to the same oncogenic miRNA causes significantly altered disease manifestation, with damages in multiple tissues indicating irreversible systemic changes. Molecularly, we identify several protein phosphatases as previously unrecognized targets of miR-125a and the associated elevation in cellular tyrosine phosphorylation. Our work demonstrates that oncogenic miRNA dependence is not limited to advanced disease states. Our work also implies that profound systemic changes could occur during an early neoplastic disease state and its remission, which then modifies the nature of the lesion upon re-encountering the same oncogene.
Results
Functional Genomics Screen Identified miR-125 Family miRNAs as Conferring Reduced Cytokine Dependence.
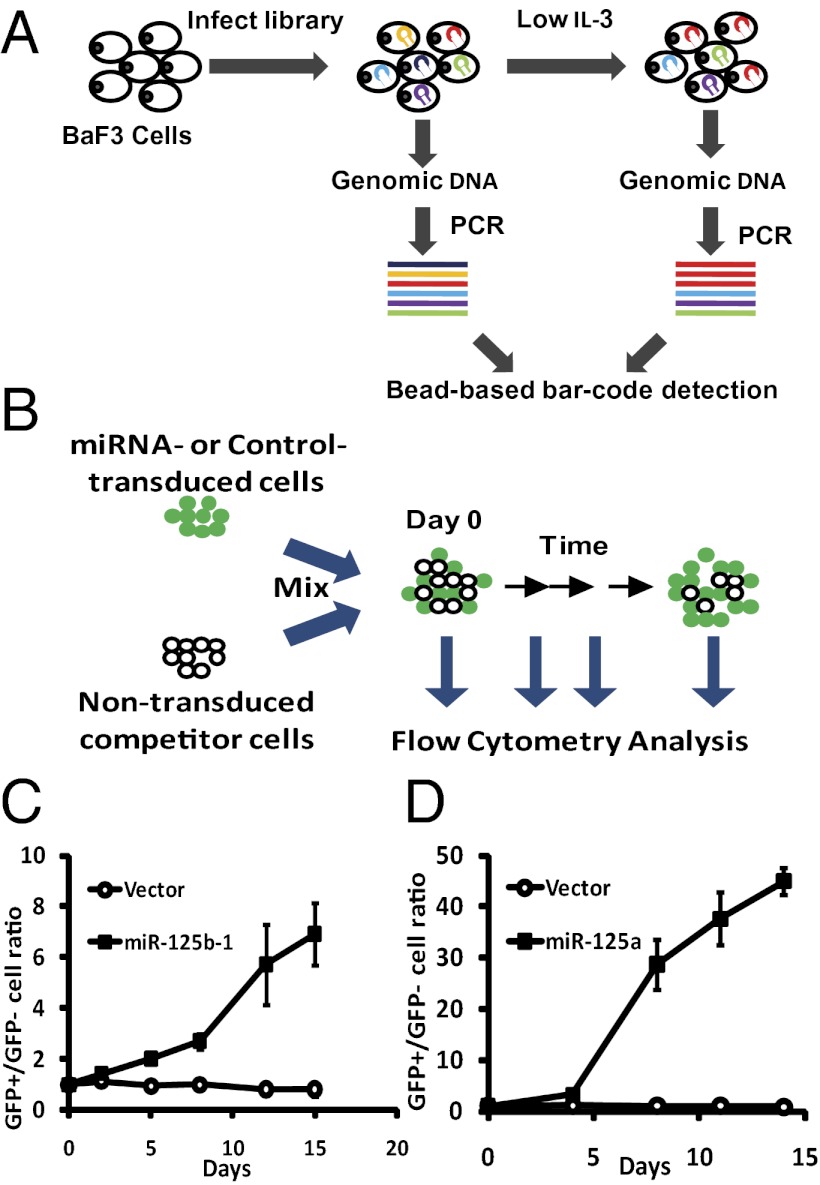
Hematopoietic cells from MPN patients often display hypersensitivity toward cytokines (15–17). To search for miRNAs that regulate cytokine sensitivity, we designed a barcode-based functional genomics screen in BaF3 cells, which strictly depends on IL-3 for survival and proliferation (18) (Fig. 1A). We constructed a retroviral miRNA expression library, by PCR-amplifying specific miRNA-encoding and flanking regions from human genomic DNA and inserting the amplified fragments into a retroviral vector. The resulted library contains 273 human miRNA expression constructs (see SI Methods and Dataset S1). The library vector supports overexpression in BaF3 cells (Fig. S1C). For the functional screen, we reasoned that BaF3 cells carrying a miRNA that confers reduced IL-3 dependence will have a growth advantage and be amplified under critically low IL-3 conditions. Consequently, this miRNA insert should be overrepresented in the cell pool after selection with low or no cytokine, compared with that before the selection. This overrepresentation should be detectable by quantifying the levels of construct-specific sequences (barcodes) in the genomic DNA of the transduced cell pool (Fig. 1A).
Fig. 1.
A functional screen identifies miR-125 family miRNAs conferring reduced cytokine dependence in BaF3 cells. (A) Schematic of the screen. BaF3 cells were infected with a pooled viral library containing 273 miRNAs. Genomic DNA was harvested before and after selection in low IL-3, amplified with construct-specific primers and detected on a bead-based platform. Colored hairpins and lines illustrate hypothetical miRNA integrations and amplicons. (B) A growth competition assay was used to validate candidate hits. BaF3 cells were infected with miR-125a, miR-125b or the control vector, which also expresses a GFP marker. Infected GFP+ cells were mixed with nontransduced cells. The percent GFP+ to percent GFP− ratios were measured by flow cytometry, following selection in low IL-3. Day 0 indicates the start of low IL-3 treatment. (C and D) Control vector, miR-125b (C) or miR-125a (D) constructs were used in the growth competition assay to assess their functions in BaF3 cells grown in low IL-3. Error bars represent SD.
We first asked whether there are miRNAs that can render complete IL-3 independence in BaF3 cells. We transduced BaF3 cells with the retroviral library, and then cultured them in complete absence of IL-3. Eight screen replicates from four independent viral transductions were performed (see Methods). Construct inserts were amplified with vector-specific biotinylated primers from the genomic DNA. PCR products were then hybridized to oligonucleotides coupled to a bead-based platform, which recognize mature miRNA sequences (1, 19) (Fig. 1A and Methods). However, no cells survived the zero IL-3 condition, suggesting that none of the miRNAs examined was capable of conferring complete IL-3 independence.
We next asked whether specific miRNAs can render reduced IL-3 dependence to allow preferential cell expansion under a critically low IL-3 concentration, which leads to slower proliferation as well as significant death. Screening in this condition identified miR-125b as one of the top candidate hits (Fig. S1 A and B and Dataset S1). MiR-125b belongs to the miR-125 family of miRNAs, which also includes miR-125a. Of note, the barcode for miR-125a was not detected above the detection threshold in the screen either before or after the low IL-3 selection, likely because of imperfect representation of constructs in the viral pool. Nevertheless, because of its sequence similarity with miR-125b, we also included miR-125a in the subsequent experiments. To validate the effect of miR-125a and miR-125b, we performed a growth competition assay (Fig. 1 B–D), which confirmed the role of these two miRNAs in inducing cytokine hypersensitivity in BaF3 cells. We used miR-125b-1 in all experiments because miR-125b-2 produces exactly the same mature product. Interestingly, the expression of miR-125a/b is highly enriched in hematopoietic stem cells (20–22). Translocation and overexpression have been reported in acute myeloid leukemia and MPN patients (23–25). Overexpression of miR-125a or miR-125b greatly enhances the hematopoietic stem cell pool and output, and can induce MPN in mice (21, 26). This result demonstrates the effectiveness of the screen to identify relevant MPN-inducing miRNAs, and suggest that miR-125 miRNAs may regulate cytokine signaling in primary cells.
Constitutive Overexpression of miR-125a Leads to MPN.
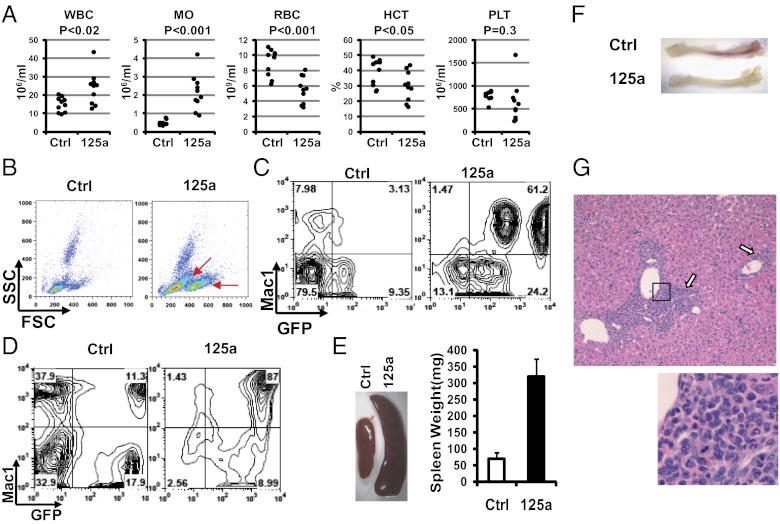
Although leukemia is induced by overexpression of miR-125b in several models (21, 24, 26–28), myeloproliferation could be the predominant effect of miR-125a overexpression (20, 29, 30). To address oncogene addiction in the context of MPN, we first assessed the effects of constitutive miR-125a overexpression in vivo to gauge the phenotypes of an inducible miR-125a model to be described later. Specifically, we transduced donor bone marrow cells with miR-125a or a control vector, each containing a GFP marker, and transplanted into lethally irradiated recipient animals. Although displaying prominent multilineage engraftment, miR-125a–recipient animals over time manifested multiple phenotypes indicative of an atypical MPN, including leukocytosis, monocytosis, splenomegaly, liver leukocyte infiltration, and progressive anemia (Fig. 2A and Fig. S2C). Leukocytosis in miR-125a recipients progressively worsened with time (Fig. S2A), which was the result of an increase in myeloid cells. Indeed, flow cytometry showed a significant elevation of the GFP+Mac1+ population in peripheral blood, which was accompanied by an increase in SSClowFSChi populations on scatter plots (Fig. 2 B and C). Consistent with this finding, the bone marrow of miR-125a recipients was dominated by myeloid cells (Fig. 2D and Figs. S3A and S4). The expanded myeloid population was composed of both monocytic and granulocytic lineages, with Ly6GhiLy6Clow and Ly6ChiLy6Glow cells expanded in the bone marrow (Fig. S4). The expansion of monocytic lineage is consistent with elevated monocyte counts in the peripheral blood (Fig. 2A).
Fig. 2.
Sustained miR-125a expression causes a MPN. (A) Complete blood counts of control vector (Ctrl) or miR-125a recipient mice at 4 mo posttransplantation. Each dot represents one animal. HCT, hematocrit; MO, monocytes; PLT, platelets; RBC. red blood cells; WBC, white blood cells. (B) Representative scatter plots of peripheral blood from recipient mice. SSC, side scatter; FSC, forward scatter. Red arrows indicate increases in SSClowFSChi populations. (C and D) Representative flow cytometry plots of peripheral blood (C) or bone marrow cells (D), detected with Mac1 and GFP. (E, Left) Representative spleens from control or miR-125a recipients. (Right) Measured spleen weight. n = 3 for Ctrl and n = 5 for miR-125a. P < 0.001. (F) Representative tibiae of miR-125a or Ctrl recipients. Note the pale bone color in miR-125a recipients. (G) Liver leukocyte infiltration in miR-125a recipient mice. A representative H&E-stained liver section (4× lens) is shown, with arrows indicating infiltrating hematopoietic cells. Images within the black box was magnified and shown below.
In addition to myeloid cell expansion, miR-125a recipients displayed several additional features analogous to MPNs seen in some human patients, including splenomegaly and liver leukocyte infiltration (Fig. 2 E and G). The enlarged spleens contained a higher percentage of myeloid cells accompanied with structural effacement (Fig. S3B). Over time, many of the miR-125a recipient mice developed anemia, as indicated by their decreased hematocrit and red blood cell count (Fig. 2A, Fig. S2C, and Dataset S2), and blood smears showed signs of stressed erythropoiesis (Fig S2B). Indeed, miR-125a recipients showed pale bone color (Fig. 2F) and reduced erythrocytes in their bone marrow (Fig. S3A). The presence of anemia is consistent with the notion that overproduction of myeloid cells in the bone marrow displaces normal erythropoiesis. Similar to human MPNs, mature myeloid cells were present in miR-125a recipients (Fig. S2B). Taken together, the data above showed that miR-125a can induce an atypical myeloproliferative neoplasm in recipient animals.
We observed similar myeloid expansion phenotypes with miR-125b recipients (Fig. S5). However, their phenotypes were always weaker compared with the miR-125a recipients in parallel experiments, possibly because of lower expression levels. We thus focused on miR-125a in the subsequent studies.
Inducible in Vivo Model of miR-125a Recapitulates the Myeloproliferative Phenotype.
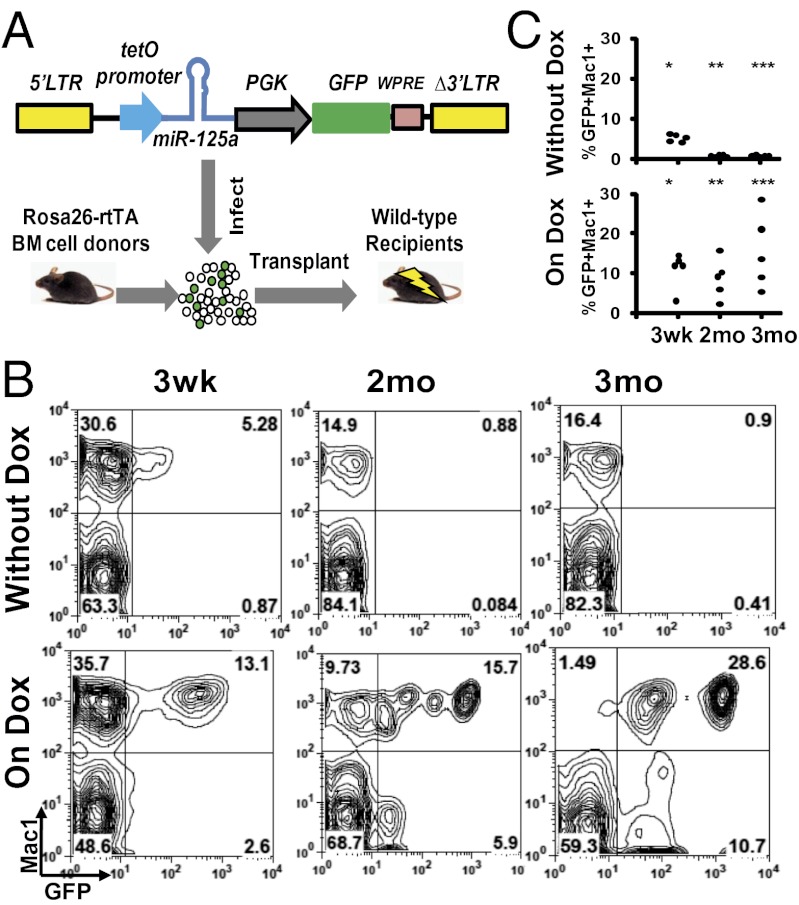
To address whether miR-125a–induced MPN is dependent on this miRNA, we established an inducible expression system for miR-125a in vivo. We engineered a lentiviral vector (hereafter referred to as “i125a”), with the expression of miR-125a under the control of a tetracycline-inducible promoter, and with a constitutively expressed GFP marker to label the transduced cells (Fig. 3A). Bone marrow cells from Rosa26-rtTA mice (31) were transduced with the i125a vector and transplanted into lethally irradiated wild-type recipients to permit inducible miR-125a expression specifically in the hematopoietic compartment.
Fig. 3.
An in vivo model of inducible miR-125a expression. (A) Schematic of the inducible system. Bone marrow cells from Rosa26-rtTA mice were transduced with a lentivirus encoding miR-125a under the tet-O promoter, and transplanted into irradiated wild-type animals to achieve inducible miR-125a expression specifically in the hematopoietic compartment. (B and C) i125a recipient mice transplanted with a low dose of transduced HSPCs were maintained with or without Dox water. Peripheral blood cells were analyzed by flow cytometry at indicated time points with Mac1 and GFP. Representative flow cytometry plots (B) and the GFP+Mac1+ percentages (C) are shown. *P < 0.05, **P < 0.03, ***P < 0.03.
To evaluate the performance of this inducible system, we tested whether induction of miR-125a leads to an expansion of transduced hematopoietic cells, particularly the myeloid cells. To maximize the readout sensitivity, we first transplanted a limited number of transduced hematopoietic stem and progenitor cells (HSPCs). Recipient mice receiving i125a-transduced cells were randomly divided into two groups, one of which was put on doxycylcine water (on Dox), whereas the other group was maintained on regular water (without Dox). We confirmed that the expression level of miR-125a could be modulated by doxycycline in recipient mice (Fig. S6A). Mice that were maintained without Dox had lower levels of GFP+ cells in their peripheral blood, which became more pronounced with time. By 2 mo, mice without Dox had significantly lower levels of engraftment and lower percentages of Mac1+GFP+ cells (< 2%) (Fig. 3 B and C). In contrast, 100% (five of five) of mice on Dox showed higher levels of GFP+ myeloid cells at 3 wk posttransplantation, and lasted until at least 3 mo posttransplantation (Fig. 3 B and C). These data indicate that inducible expression of miR-125a can lead to myeloid expansion in a Dox-dependent manner. However, no overt abnormality in differential blood parameters was observed at this input cell dosage.
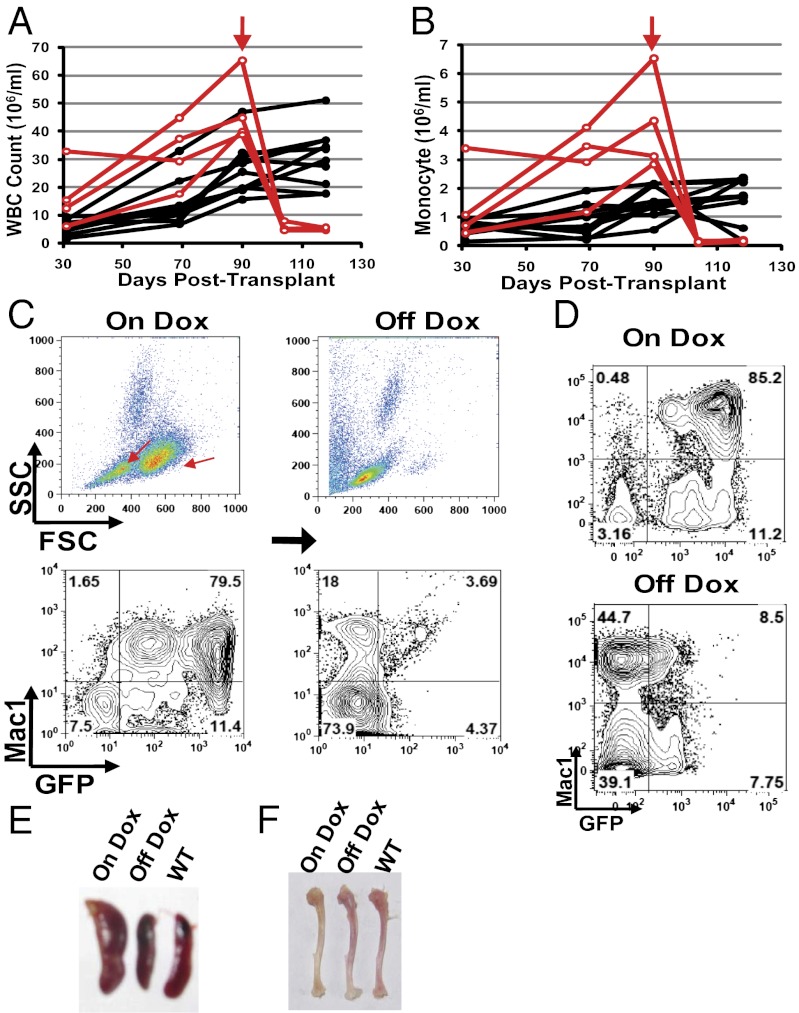
We then modeled the MPN phenotype by transplanting a larger number of transduced HSPCs. Indeed, induction of miR-125a recapitulated the phenotypes associated with the atypical MPN observed with the constitutive miR-125a model. The mice developed leukocytosis and monocytosis, which worsened with prolonged Dox induction (Fig. 4 A and B, and Fig. S6B). In contrast, mice treated with Dox alone did not exhibit such phenotypes (Dataset S3). Significant increases in GFP+Mac1+ cells occurred in both peripheral blood and bone marrow, leading to GFP+ cells dominating the Mac1+ myeloid population (Fig. 4 C and D), with associated expansion of the characteristic SSClowFSChi populations (Fig. 4C). Furthermore, the mice developed splenomegaly (Fig. 4E). We only observed progressive anemia in a small number of i125a mice on Dox (Fig. S7A). Pale bone color was nonetheless present even though the red cell parameters remained within the normal range for most animals (Fig. 4F and Fig. S7A). This observation is consistent with the notion that the anemic phenotype is a consequence of myeloid expansion disrupting normal erythropoiesis, which may exhibit different severity depending on the extent of myeloproliferation. Taken together, the data above show that the inducible i125a model recapitulates the MPN phenotypes of the constitutive model.
Fig. 4.
Dependence on miR-125a expression in the MPN model. (A and B) i125a recipient mice were maintained on doxycyline (on Dox) after transplantation and their hematopoietic parameters were monitored over time. Each line represents one animal. Dox was switched off (off Dox) for a subgroup of these animals (red lines), at the time point indicated by the vertical red arrows. Peripheral blood WBC (A) and monocyte (B) counts are shown. P < 0.01 for WBC and P < 0.02 for monocytes, comparing before and after 2 wk of switching off Dox. n = 4 for off Dox and n = 10 for on Dox. A representative cohort is shown. (C and D) Representative flow cytometry analysis of peripheral blood (C) and bone marrow cells (D). The same animal on Dox and off Dox is shown in C. Red arrows in C indicate the characteristic expanded populations on scatter plot. (E and F) Spleens and tibiae of mice that are wild-type, or i125a recipients maintained on Dox, or switched off Dox.
miR-125a–Induced Primary MPN Phenotypes Are Dependent on Its Sustained Overexpression.
Having established an in vivo inducible model of miR-125a, we then asked whether the MPN is dependent on miR-125a overexpression. i125a mice that were kept on Dox were monitored for their peripheral blood parameters. Four mice on Dox with high white blood cell counts at 3 mo posttransplant were switched off doxycycline (off Dox), whereas the rest of the cohort was maintained on Dox induction. Within 2 wk after switching off Dox, the peripheral white blood cell counts and monocyte counts returned to normal range (Fig. 4 A and B, and Fig. S8A), whereas mice kept on Dox continued to have high white cell and monocyte counts (Fig. 4 A and B). The reduction in white cell count in the off Dox mice was accompanied by a strong reduction in GFP+ percentage in both peripheral blood and bone marrow (Fig. 4 C and D), indicating that the decrease of i125a-GFP cells was underlying the normalization of blood parameters. Further examination showed that the spleen size of off Dox mice returned back to normal size, indistinguishable from that of wild-type animals (Fig. 4E). These data indicate that the overproduction of myeloid cells was corrected after stopping inducible miR-125a expression.
We next examined whether erythropoiesis was restored after switching off Dox. Indeed, the bone color of the off Dox mice returned to a level of redness similar to that seen with wild-type animals (Fig. 4F), indicating more effective bone marrow erythropoiesis. This finding prompted us to examine the few mice that did develop significant anemia with Dox induction. Invariably, significant rebound of red cell parameters was observed after switching off Dox (Fig. S7B). Even in mice that did not show red cell parameters below the anemic cutoff, discontinuation of Dox administration led to improved red blood cell parameters in three out of four animals (Fig. S7C). Taken together, the data above show remarkable correction of the hematopoietic phenotypes of MPN after switching off miR-125a induction.
Repeated miR-125a Overexpression Results in Complex Outcomes.
To model reoccurrence of oncogenic lesion following a period of remission, we assessed the effects of miR-125a re-exposure in our inducible model. Four mice (from two cohorts) that had their initial MPN corrected after withdrawing Dox treatment were administered Dox for a second time. Instead of a uniform manifestation of the same MPN seen with the first Dox induction, the repeated Dox treatment led to unexpected complex disease outcomes. Only in one of four mice was a similar MPN induced with the second exposure (Dataset S4, mouse #4). Surprisingly however, three of four mice quickly became moribund, at which point eight of nine mice that remained on Dox all of the time were still alive (Dataset S4) (P = 0.05 Fisher’s exact test). This rapid lethality occurred within 5–7 wk upon reinitiation of miR-125a overexpression. Myeloproliferation and splenomegaly were absent or only mild (Dataset S4). Detailed hematopoietic and pathological examination showed that lethal anemia was present in mice (two of four) (Dataset S4) with severe spleen damage. Stromal collapse and fibrosis is prominent in these spleens (Figs. S9A and S10). Importantly, splenic lesions of this nature was not present in recipient animals that received control vector-transduced cells, or remained on continuous Dox induction (Figs. S9A and S10), or in the constitutive 125a model (Fig. S3B), strongly suggesting that it was a consequence of off-Dox treatment. Notably, spleen was also the major site of extramedullary hematopoiesis with constitutive miR-125a overexpression (Fig. 2E and Fig. S3B), with the enlarged spleen supporting red cell production to compensate for ineffective bone marrow erythropoiesis. The severe spleen damage and relatively normal spleen size in these mice provide a plausible explanation for the development of lethal anemia. Finally, rapid lethality (one of four mice) was seen with extreme hematopoietic infiltration into the lung (Fig. S9B). The data above demonstrate that re-exposure to miR-125a is associated with complex outcomes, with some animals developing systemic changes that alter the overall disease manifestation.
miR-125a Potentiates Cytokine Sensitivity, Targets Protein Phosphatases, and Elevates Cellular Protein Phosphorylation.
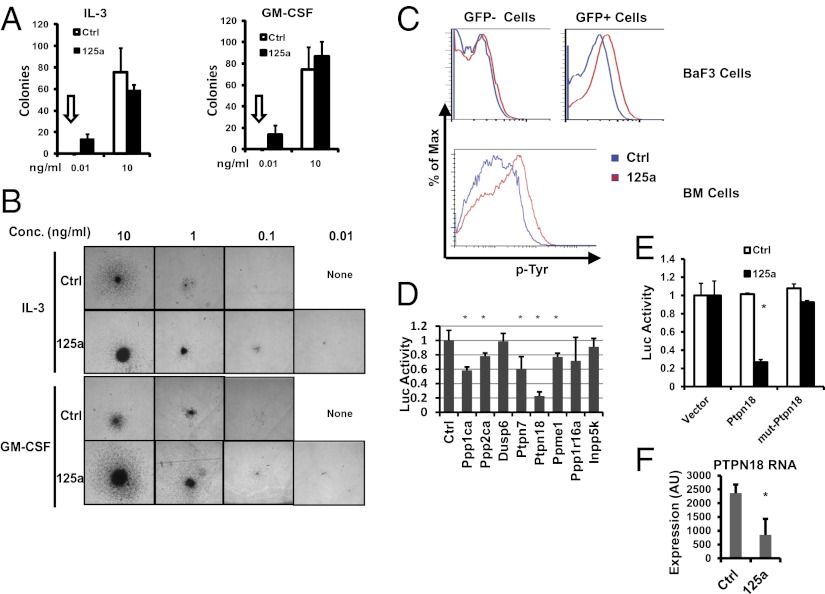
To shed light on the mechanism of miR-125a, we extended on the finding that miR-125a induces IL-3 hypersensitivity in BaF3 cells and asked whether cytokine hypersensitivity could also be seen with bone marrow cells. To this end, we performed myeloid colony formation assays with bone marrow cells from miR-125a recipients or control vector recipients, using different concentrations of IL-3 or GM-CSF. At the highest dose of either cytokine, similar numbers of colonies were formed by miR-125a and control bone marrow cells, indicating that forced expression of miR-125a did not increase the frequency of clonogenic progenitors at permissive growth conditions with abundant supportive signaling (Fig. 5A). At a low concentration (0.01 ng/mL) of IL-3 or GM-CSF however, control bone marrow cells completely lost the ability to produce colonies, whereas miR-125a bone marrow still gave rise to small but robust colonies (Fig. 5 A and B). Even in the complete absence of IL-3 or GM-CSF, a small number of colonies were still present in miR-125a cultures (Fig. S8B). These data indicate that miR-125a potentiates cytokine sensitivity in primary cells.
Fig. 5.
miR-125a regulates cytokine hypersensitivity and targets multiple protein phosphatases. (A and B) miR-125a-recipient bone marrow cells show cytokine hypersensitivity. Bone marrow cells from control vector (Ctrl) or miR-125a recipient animals were plated for colony formation assay with indicated concentrations of IL-3 or GM-CSF. Colonies were scored and photographed on day 10. (A) Colony numbers per well. n = 4 for each condition. Arrows indicate that zero colonies were observed for Ctrl bone marrow cells at the corresponding concentration. (B) Representative colony morphology at indicated concentrations (conc) of IL-3 or GM-CSF. (C) Cellular phospho-tyrosine (p-Tyr) levels increased in miR-125a-expressing cells. BaF3 cells were transduced with a control construct or miR-125a construct, with GFP labeling transduced cells. Cells were cultured under low IL-3 and low serum condition, and p-Tyr levels were measured with flow cytometry, gating on both GFP+ (transduced) and GFP− (nontransduced) fractions. Bone marrow cells from control-vector or miR-125a recipients were analyzed with p-Tyr. Because of the minimal levels of GFP− cells in miR-125a recipients, only data on GFP+ cells are shown. All flow cytometry plots were representative of two or more experiments. (D) The 3′ UTR luciferase reporters of indicated genes were analyzed with miR-125a. Normalized luciferase activities are shown, with a value of 1 indicating no effect on the 3′ UTR. n = 6 for control, PTPN7, PTPN18, and PPP1R16A; n = 3 for others. *P < 0.05. (E) Wild-type PTPN18 3′ UTR reporter, and a mutant reporter with mutation in a putative miR-125a site, were assayed with control or miR-125a construct. Normalized luciferase activities are shown. The activity of the UTR-less reporter in the presence of control and miR-125a construct is set to 1. n = 3. *P < 0.05. (F) Erythroid myeloid lymphoid cells transduced with control or miR-125a construct were measured for PTPN18 mRNA expression using microarray. n = 3. *P < 0.05. For all figures, error bars represent SD.
Regulation of signaling downstream of cytokines involves protein phosphorylation and de-phosphorylation in multiple signaling cascades (reviewed in ref. 32). We thus examined whether phosphatases, including tyrosine phosphatases, may be targeted by miR-125a. TargetScan predicts multiple phosphatases as potential miR-125a targets. To validate these predictions, we cloned the 3′ UTRs of eight phosphatases or related genes into luciferase reporters. miR-125a significantly down-regulated the 3′ UTR reporters of multiple protein phosphatases, including protein tyrosine phosphatases PTPN18 and PTPN7 (Fig. 5D). miR-125a also targeted serine/threonine phosphatases PPP1CA and PPP2CA (Fig. 5D), similar to miR-125b (33). We next used PTPN18 as an example for further characterization, which is a gene with enriched expression in HSPCs (34–36). Mutation of a putative miR-125 binding site in PTPN18 3′ UTR abolished the targeting by miR-125a (Fig. 5E), demonstrating a direct targeting effect. Although we were not able to assess the endogenous protein level because of the lack of a reliable antibody, we did observe a down-regulation of the endogenous PTPN18 mRNA with miR-125a overexpression (Fig. 5F). These data demonstrate that miR-125a targets protein phosphatases and suggest its modulation of cellular protein phosphorylation. This is indeed true, because BaF3 cells and primary bone marrow cells expressing miR-125a showed higher protein tyrosine phosphorylation than corresponding controls (Fig. 5C). These findings illustrate the protein phosphatases as a class of common targets by classic hematopoietic oncogenes, such as BCR-ABL (37), and a noncoding small RNA miR-125a.
Discussion
In this study, we examined the role of miR-125a in an in vivo model of MPN and report the complex nature of dependence on an oncogenic miRNA. Unlike the previously reported case of oncogene addiction to miR-21, which resulted in lymphoma formation in mice (10), overexpression of miR-125a resulted in a neoplasm with excessive expansion of the myeloid lineage, representing an early neoplastic stage. Contrasting the lack of oncogene addiction when p53 is restored in early-stage lung adenocarcinomas (18, 19), switching off exogenous miR-125a led to rapid resolution of MPN phenotypes and normalization of hematopoietic parameters. These data demonstrate that MPN induced by miR-125a is dependent on its continued overexpression. However, a repeat of miR-125a overexpression led to significant altered phenotypes, with some mice showing severe damages in spleen and lung. These data show the complexity of oncogenic miRNA dependence and suggest that irreversible systemic damages caused by oncogene inhibition could be an important modifier for host response to a subsequent wave of oncogenic events.
We demonstrated and validated the utility of a unique barcode functional genomics screen system that identified miR-125 miRNAs as conferring reduced cytokine dependence. Although we have not validated other candidates, we believe the list of candidates can be interesting miRNAs to explore in the future. The miRNA expression library constructed in this study is one of the few libraries available for the broader miRNA community. Unlike previous efforts that used microarrays for detecting barcode changes (38, 39), we used a unique bead-based platform for barcode detection. Compared with microarray or deep sequencing, the bead-based detection platform is much more cost-effective, and is flexible enough to incorporate additional barcode detectors (1). This platform provides the feasibility to detect large numbers of samples at low cost, a feature that can be particularly useful when considering detailed time-course experiments, with many replicates or experimental conditions. We further envision that this system can be applied to in vivo functional screens, which likely will require multiple cohorts of animals over multiple time points.
In this study, we also show that miR-125a induces cytokine hypersensitivity both in cell line and in bone marrow cells. This hypersensitivity is reflected by increased global tyrosine phosphorylation in cells. We demonstrate that miR-125a targets multiple protein phosphatases, providing a possible mechanistic explanation for the effects on protein phosphorylation and cytokine sensitivity. Strikingly, mice defective in several phosphatases display MPN-like phenotypes (40–44). In addition, severe lung pathology is present in mice deficient in tyrosine phosphatase PTPN6 (44) and SH2-containing inositol-5-phosphatase (SHIP) (41). Furthermore, potentiation of colony growth at low cytokine concentrations and an increase in colony sizes were documented for mice null of SHIP (41). These observations indicate that disruption of phosphatase activity can lead to MPN, and suggest incomplete inhibition of multiple phosphatases by miR-125a may lead to cooperative outcomes in MPN formation. Other targets of miR-125 have been previously characterized by us and other groups (reviewed in ref. 45) and are still growing in numbers. With a long list of over 800 predicted targets (TargetScan), it is highly likely that cooperation among multiple targets, including protein phosphatases, is responsible for the overall MPN phenotypic manifestation.
Methods
miRNA Functional Screen.
The library of miRNA expression vectors was pooled and used to generate pooled viral mixture for infection. Genomic DNA from infected BaF3 cells was harvested before and after low IL-3 selection, and was PCR-amplified using biotinylated primers only present in the vector. PCR products were hybridized to colored beads (Luminex) coupled with antisense sequences to mature miRNAs. See SI Methods for more details.
Bone Marrow Transplantation.
Lethal irradiation was delivered at 9 Gy. HSPC viral transduction and transplantation was performed as described before (20). Unless otherwise specified, all mice used were wild-type C57Bl6/J purchased from the Jackson Laboratory. For experiments with the inducible i125a, Rosa26-rtTA mice were used as donors. Dox was supplied in drinking water at 1 mg/mL. See SI Methods for more details.
Additional Methods.
See SI Methods for information on constructs and plasmids, cell culture, animal maintenance and histology, quantitative PCR, flow cytometry, colony formation assay, and statistics. See Dataset S5 for primers and enzymes.
Supplementary Material
Acknowledgments
We thank Rita Schlanger, Hao Zhang, Judy Wang, and Sharon Lin for their technical assistance. This work was supported in part by National Institutes of Health Grant R01CA149109 (to J.L.) and American Cancer Society Institutional Research Grant 58-012-51 (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213196109/-/DCSupplemental.
References
- 1.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 2.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res. 2011;157:216–225. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Xue W, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 8.Sharma SV, Settleman J. Oncogene addiction: Setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080, discussion 3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 10.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 11.Feldser DM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junttila MR, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468:567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine RL, Gilliland DG. Myeloproliferative disorders. Blood. 2008;112:2190–2198. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orazi A, Germing U. The myelodysplastic/myeloproliferative neoplasms: Myeloproliferative diseases with dysplastic features. Leukemia. 2008;22:1308–1319. doi: 10.1038/leu.2008.119. [DOI] [PubMed] [Google Scholar]
- 15.Dai CH, Krantz SB, Means RT, Jr, Horn ST, Gilbert HS. Polycythemia vera blood burst-forming units-erythroid are hypersensitive to interleukin-3. J Clin Invest. 1991;87:391–396. doi: 10.1172/JCI115009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi S, et al. Circulating megakaryocyte progenitors in myeloproliferative disorders are hypersensitive to interleukin-3. Br J Haematol. 1993;83:539–544. doi: 10.1111/j.1365-2141.1993.tb04688.x. [DOI] [PubMed] [Google Scholar]
- 17.Prchal JF, Axelrad AA. Letter: Bone-marrow responses in polycythemia vera. N Engl J Med. 1974;290:1382. doi: 10.1056/nejm197406132902419. [DOI] [PubMed] [Google Scholar]
- 18.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo S, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell RM, et al. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatley ME, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bousquet M, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enomoto Y, et al. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia. 2011;25:1849–1856. doi: 10.1038/leu.2011.166. [DOI] [PubMed] [Google Scholar]
- 25.Marcucci G, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhuri AA, et al. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci USA. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ooi AG, et al. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo S, Scadden DT. A microRNA regulating adult hematopoietic stem cells. Cell Cycle. 2010;9:3637–3638. doi: 10.4161/cc.9.18.13174. [DOI] [PubMed] [Google Scholar]
- 30.Gerrits A, et al. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2012;119:377–387. doi: 10.1182/blood-2011-01-331686. [DOI] [PubMed] [Google Scholar]
- 31.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Bezbradica JS, Medzhitov R. Integration of cytokine and heterologous receptor signaling pathways. Nat Immunol. 2009;10:333–339. doi: 10.1038/ni.1713. [DOI] [PubMed] [Google Scholar]
- 33.Le MT, et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 2011;7:e1002242. doi: 10.1371/journal.pgen.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng J, Daimaru L, Fennie C, Lasky LA. A novel protein tyrosine phosphatase expressed in lin(lo)CD34(hi)Sca(hi) hematopoietic progenitor cells. Blood. 1996;88:1156–1167. [PubMed] [Google Scholar]
- 35.Dosil M, Leibman N, Lemischka IR. Cloning and characterization of fetal liver phosphatase 1, a nuclear protein tyrosine phosphatase isolated from hematopoietic stem cells. Blood. 1996;88:4510–4525. [PubMed] [Google Scholar]
- 36.Huang K, Sommers CL, Grinberg A, Kozak CA, Love PE. Cloning and characterization of PTP-K1, a novel nonreceptor protein tyrosine phosphatase highly expressed in bone marrow. Oncogene. 1996;13:1567–1573. [PubMed] [Google Scholar]
- 37.Sattler M, et al. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. J Biol Chem. 2000;275:24273–24278. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- 38.Izumiya M, Okamoto K, Tsuchiya N, Nakagama H. Functional screening using a microRNA virus library and microarrays: A new high-throughput assay to identify tumor-suppressive microRNAs. Carcinogenesis. 2010;31:1354–1359. doi: 10.1093/carcin/bgq112. [DOI] [PubMed] [Google Scholar]
- 39.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 40.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helgason CD, et al. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shultz LD, et al. Mutations at the murine motheaten locus are within the hematopoietic cell protein-tyrosine phosphatase (Hcph) gene. Cell. 1993;73:1445–1454. doi: 10.1016/0092-8674(93)90369-2. [DOI] [PubMed] [Google Scholar]
- 43.Tsui HW, Siminovitch KA, de Souza L, Tsui FW. Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat Genet. 1993;4:124–129. doi: 10.1038/ng0693-124. [DOI] [PubMed] [Google Scholar]
- 44.Ward JM. Pulmonary pathology of the motheaten mouse. Vet Pathol. 1978;15:170–178. doi: 10.1177/030098587801500203. [DOI] [PubMed] [Google Scholar]
- 45.Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;31:1354–1359. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.