Abstract
Dispersion enables biofilm bacteria to transit from the biofilm to the planktonic growth state and to spawn novel communities in new locales. Although the chemotaxis protein BdlA plays a role in the dispersion of Pseudomonas aeruginosa biofilms in response to environmental cues, little is known about regulation of BdlA activity or how BdlA modulates the dispersion response. Here, we demonstrate that BdlA in its native form is inactive and is activated upon nonprocessive proteolysis at a ClpP-protease-like cleavage site located between the Per Arnt Sim (PAS) sensory domains PASa and PASb. Activation of BdlA to enable biofilm dispersion requires phosphorylation at tyrosine-238 as a signal, elevated c-di-GMP levels, the chaperone ClpD, and the protease ClpP. The resulting truncated BdlA polypeptide chains directly interact and are required for P. aeruginosa biofilms to disperse. Our results provide a basis for understanding the mechanism of biofilm dispersion that may be applicable to a large number of biofilm-forming pathogenic species. Insights into the mechanism of BdlA function have implications for the control of biofilm-related infections.
Keywords: protein interactions, dispersal, site-directed mutagenesis
The formation of bacterial biofilms, or surface-associated communities that are encased in a polymeric matrix, is a developmental process that is initiated with surface attachment by planktonic cells. In contrast, dispersion is the terminal stage of biofilm development, during which bacteria evacuate a mature biofilm and transition to a planktonic state. Biofilm dispersion can be induced by a variety of environmental cues, including changes in growth medium composition, pH, oxygen, and carbon concentrations, exposure to heavy metals and nitric oxide, exposure to the polysaccharide degrading enzyme dispersin B, and self-synthesized signaling molecules such as cis-2-decenoic acid (1–9). Dispersed cells are characterized by distinct gene expression and protein production patterns and increased susceptibility to antimicrobial agents compared with their sessile counterparts (2, 10–12). Moreover, although biofilms are considered the root cause of chronic and persistent infections (13), biofilm dispersion may be part of an inherent strategy of Pseudomonas aeruginosa to initiate a disseminating phenotype, causing acute and periodic infections.
Recent findings suggested that dispersion, not unlike surface attachment and initiation of biofilm formation, is a coordinated process coinciding with unique protein phosphorylation patterns and requiring specific regulatory events including phosphotransfer events (1–4, 14, 15). Regulation of biofilm dispersion has also been linked to the modulation of the intracellular signaling molecule cyclic di-GMP (c-di-GMP), high levels of which promote sessile growth, and low levels correlate with planktonic existence. Several proteins associated with dispersion have been shown to possess c-di-GMP–modulating activity (1–4, 14). These include the phosphodiesterases (PDEs) RbdA and DipA, which promote the return to free-swimming growth by reducing cellular c-di-GMP levels (15, 16). The notable exception is the P. aeruginosa chemotaxis transducer protein BdlA. BdlA was identified in a mutant screen with inactivation of bdlA rendering P. aeruginosa biofilms dispersion deficient in response to various environmental cues (4) and nitric oxide (14). The protein lacks the typical domains required for c-di-GMP modulation, but instead harbors a signal transduction/methyl-accepting chemotaxis (TarH/MCP) domain and two PAS domains (Fig. 1A) that are also found in RbdA and DipA. BdlA plays a key role in dispersion of P. aeruginosa biofilms, but little is known about the mechanism that modulates BdlA activity, in particular when considering that bdlA is expressed constitutively, with its expression elevated in dispersing cells (15).
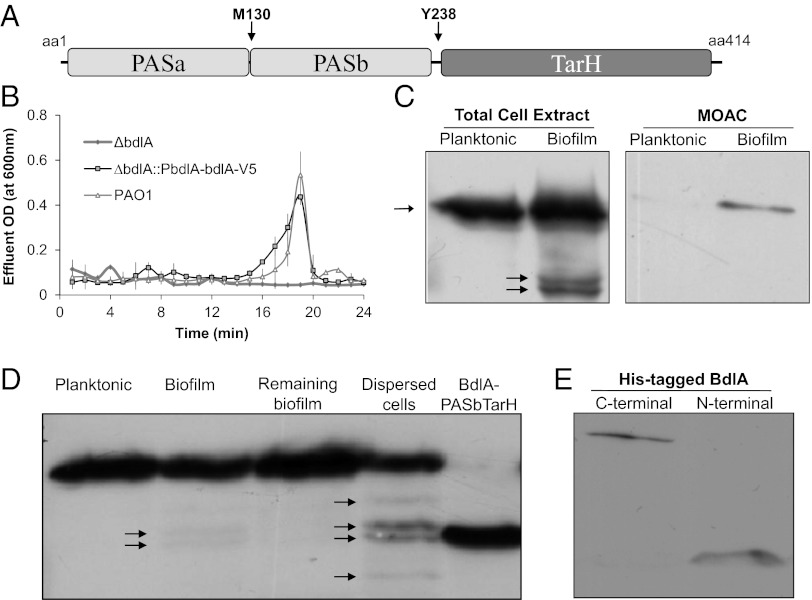
Fig. 1.
Posttranslational modification and nonprocessive proteolysis of BdlA are growth-mode–dependent. (A) Domain structure of BdlA. MCP/TarH, homologue of Tar (Taxis toward aspartate and related amino acids) chemoreceptor domain; PAS, Per Arnt Sim sensory domain. Arrows indicate the position of the methionine-130 (M130) putative cleavage site and the tyrosine-238 (Y238) potential phosphorylation site. (B) cis-encoded (∆bdlA/pbdlA-bdlA-V5/His) C-terminally V5/His-tagged BdlA restores biofilm dispersion phenotype of ∆bdlA to wild-type levels. (C) Detection of cis-encoded V5/His-tagged BdlA in total cell extracts or MOAC-enriched phosphoproteomes of planktonic and biofilm P. aeruginosa cells. (D) BdlA processing is increased in dispersed cells, with the cleavage product being similar in size to BdlA lacking the PASa domain (BdlA-PASbTar). (E) The N-terminal BdlA-PASa remains detectable in cells as indicated using N- and C-terminally His-tagged BdlA proteins obtained from PAO1/pJN-His-bdlA and PAO1/pJN-bdlA-V5/His biofilms. Arrows indicate intact BdlA and its degradation products.
Here, we demonstrate that BdlA function is regulated via an unusual nonprocessive proteolytic cleavage, but still requires both truncated polypeptide chains (PASa, PASbTarH) to induce dispersion. Proteolysis was found to be biofilm specific, stimulated by increased c-di-GMP levels, and dependent on the protease ClpP, the chaperone ClpD, and BdlA phosphorylation at Y238.
Results
BdlA Is Phosphorylated in Growth-Mode–Specific Manner.
bdlA transcripts are detectable in P. aeruginosa regardless of the growth conditions (17), indicating that BdlA may be regulated posttranslationally. The activity of many chemotaxis transducer proteins is modulated via methylation/demethylation events. However, BdlA lacks orthodox methylation sites [(A/S)-X-X-E-(E/Q)-X-(A/T/S)-A-(A/S/T)] based on sequence alignments with the aerotaxis transducer Aer (4, 18). Moreover, nutrient-induced dispersion has been shown to be independent of the chemotaxis-specific methyltransferase B (CheB) (4), thus rendering methylation as a possible posttranslational modification less likely. Considering that dispersion by P. aeruginosa coincided with unique protein phosphorylation patterns and required phosphotransfer events with phosphatase inhibitors preventing dispersion (2, 4), we asked whether BdlA is differentially phosphorylated.
To ensure native BdlA levels and avoid overexpression/dosing effects, we generated a C-terminal V5/His-tagged BdlA construct under the control of the native bdlA promoter on the chromosome (PbdlA-bdlA-V5/His). The construct was capable of restoring the dispersion-deficient phenotype of ΔbdlA to wild-type levels (Fig. 1B) and was subsequently used to compare the detectable levels of BdlA in total cell extracts and the phosphoproteomes, enriched via metal-oxide affinity chromatography (MOAC), of P. aeruginosa planktonic and biofilm cells. Although V5/His-tagged BdlA was present in total cell extracts of both planktonic and biofilm cells, the construct was only detectable in the MOAC-enriched phosphoproteomes of biofilm, but not planktonic, cells (Fig. 1C). The findings indicated that BdlA is differentially modified because BdlA phosphorylation is biofilm specific. Liquid chromatography (LC)-MS/MS analysis of purified BdlA subsequently resulted in the identification of the tyrosine-238 (Y238) residue as a potential phosphorylation site. An additional posttranslational modification, an acetylation of lysine-226 (K226), was observed (Table S1).
In Vivo Degradation of PASa Domain of BdlA.
Analysis of BdlA-V5/His production revealed an additional level of posttranslational processing: the protein appeared to be cleaved or degraded in a growth-mode–specific manner (Fig. 1C). Although only one band corresponding to intact BdlA was detectable in planktonic cells, additional bands corresponding to smaller protein species were present in biofilm cells, with abundance of these bands significantly increasing in dispersed cells (Fig. 1D). The cleavage products, however, were not detected in biofilm cells remaining following dispersion (Fig. 1D). The bands corresponding to intact BdlA and the dominating smaller protein bands were identified by LC-MS/MS analysis as BdlA. Comparison of molecular weights of the degradation products with intact BdlA indicated that the two dominating BdlA cleavage products were similar in size to BdlA-PASbTarH, a truncated BdlA lacking the N-terminally–located PAS domain, PASa (Fig. 1 A and D, Fig. S1). The finding suggested cleavage of approximately 120 amino acids of the N terminus of BdlA in biofilms and dispersed cells, but not in planktonic cells or biofilm cells that remained attached postdispersion (Fig. 1D). To determine whether the PASa domain is degraded or remains present in the cell following cleavage, BdlA was N-terminally His tagged. Subsequent immunoblot analysis indicated that the PASa domain of BdlA is cleaved, but not degraded (Fig. 1E), suggesting nonprocessive proteolysis of BdlA.
ClpD Is Required for BdlA-PAS Degradation and Dispersion.
The findings strongly suggested a specific mechanism of BdlA degradation to regulate BdlA levels and thus dispersion. We hypothesized that such specific cleavage would involve close interaction of a protease with BdlA. We therefore used coimmunoprecipitations using V5/His-tagged BdlA combined with 2D/PAGE to identify protein interaction partners. By doing so, we identified six proteins: a conserved hypothetical protein (PA1658), cystathionine gamma-lyase (PA0400), two proteins involved in LPS biosynthesis (WbpA, WbpB), oxygen-independent coproporphyrinogen oxidase HemN (PA1546), and a ClpA/B homolog PA0459.
ClpA and ClpB are chaperone proteins belonging to the Clp/Hsp100 ATPase family. Although both are involved in protein folding and stabilization, ClpA is also a regulatory component of the ClpAP protease that participates in regulatory degradation of protein aggregates (19). According to TIGRFAM conserved domain predictions, PA0459 harboring a chaperone_clpB domain may function in a manner similar to the ATP-dependent chaperone ClpB in protein folding and stabilization. Interaction between BdlA-V5/His and PA0459-HA was confirmed via pull-down assays (Fig. 2A, Fig. S2A). Due to its interaction with a protein central to the regulation of nutrient-induced dispersion, we named PA0459 ClpD. To determine whether ClpD is involved in BdlA processing under biofilm growth conditions, strains lacking and overexpressing clpD were analyzed for BdlA degradation. Overexpression of clpD resulted in significantly reduced abundance of intact BdlA, and lack of clpD expression significantly reduced PASa domain processing (Fig. 2B).
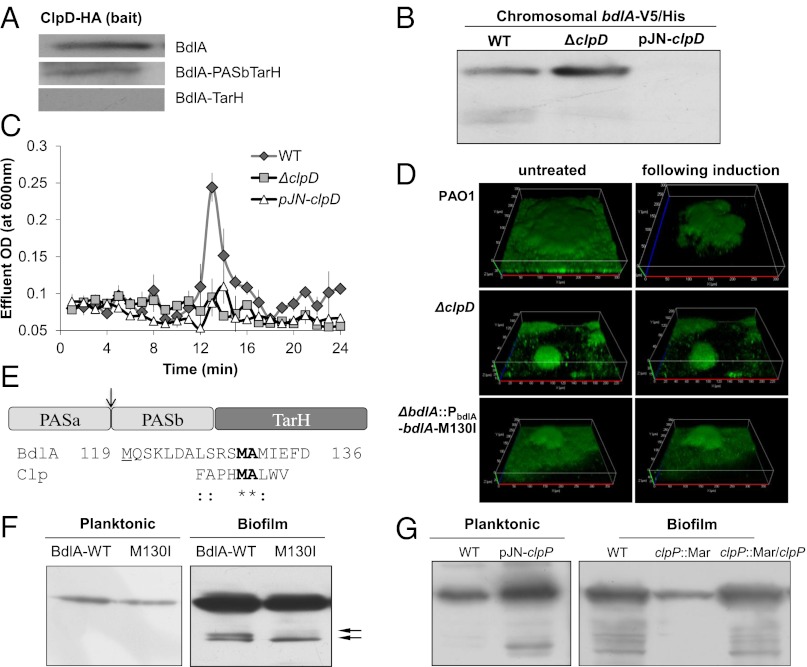
Fig. 2.
Dispersion and BdlA processing is dependent on ClpD, ClpP, and a ClpP cleavage site. (A) Immunoblot analysis of pull-down assays demonstrating that complex formation between ClpD and BdlA depends primarily on PASa. (B) BdlA processing under biofilm growth conditions in strains lacking or overexpressing clpD. All strains express bdlA in cis. Deletion or overexpression of clpD renders P. aeruginosa dispersion deficient as indicated by measurements of effluent OD (C) and confocal microscopy (D). (D) Confocal images demonstrating that expression of the M130I variant of BdlA (∆bdlA/pbdlA-bdlA-M130I) does not rescue the dispersion-deficient phenotype of ∆bdlA biofilms. P. aeruginosa wild type was used as control. (E) Alignment of Clp cleavage consensus sequence and potential BdlA cleavage site. Possible Clp cleavage site in BdlA is located between PASa/b (see arrow) at position 130–131, which is 11 residues downstream of the methionine residue (position 119, residue is underlined) used for the generation of BdlA-PASbTarH. (F) Detection of the wild-type and M130I C-terminally V5/His-tagged variants of BdlA in planktonic and biofilm cells. (G) Inactivation of clpP impairs BdlA processing under biofilm growth conditions, and overexpression of clpP results in BdlA processing under planktonic growth conditions.
We next asked whether cleavage of BdlA by ClpD is a mechanism to modulate BdlA function in dispersion by testing biofilms lacking or overexpressing clpD for glutamate-induced dispersion. In accord with the role of BdlA in dispersion, the clpD overexpresser strain, which exhibits significantly reduced levels of BdlA (Fig. 2B), was unable to respond to dispersion cues (Fig. 2 C and D, Table S2). Biofilms of a strain inactivated in clpD, although containing higher levels of intact BdlA than those of the overexpresser or wild-type strains (Fig. 2B), did not disperse (Fig. 2 C and D). These findings suggested that not only the presence of BdlA, but also the regulated stage-specific cleavage of BdlA, is necessary for nutrient-induced dispersion.
Sequence-Specific Posttranslational Cleavage of BdlA Is Essential for Dispersion.
ClpA/B proteases recognize proteins in a sequence-specific manner (20), allowing the chaperones to interact exclusively with their target proteins. Pull-down assays demonstrated a sequence requirement for the ClpD–BdlA interaction (Fig. 2A). Although ClpD was found to interact with full-length BdlA or a truncated construct lacking the PASa domain (BdlA-PASbTarH), no ClpD interaction with a BdlA-TarH construct, which lacks both PAS domains, was observed (Fig. 2A, see Fig. S1 for overview of constructs). These findings suggested that the sequence recognized by ClpD is located within the PASb domain of BdlA or between the two PAS domains.
ClpP protease has been demonstrated in Escherichia coli to cleave a variety of proteins and peptides at a methionine-alanine (Met-Ala) peptide bond, with hydrolysis also occurring when Met was replaced by leucine (Leu) or tryptophane (Trp) (21). Analysis of the BdlA amino acid sequence revealed the presence of a Met–Ala pair at position 130–131, a region located between the two PAS domains that form a potential disordered coil. This pair is located 10 amino acids downstream from the start site of the BdlA-PASbTarH construct (Fig. 2E). No Trp–Ala pair was detected; however, three Leu–Ala pairs were located at the beginning of PASb at position 142–143, 149–150, and 159–160, which would result in truncated BdlA being smaller than BdlA-PASbTarH. In E. coli, various substitutions of the methionine residue of the Met–Ala pair have been shown to prevent substrate cleavage by ClpP including isoleucine substitution (21). Similarly, isoleucine substitution of BdlA-Met130 residue eliminated one of the two biofilm-specific BdlA cleavage products, as only the second cleavage product was detectable for the BdlA-M130I variant (Fig. 2F). Although it did not completely prevent BdlA processing, the substitution rendered BdlA nonfunctional with respect to dispersion, as the M130A variant was not capable of restoring nutrient-induced dispersion in ΔbdlA biofilms (Fig. 2D, Fig. S2B).
To determine the protease responsible for BdlA cleavage, we tested mutants inactivated in clpP and PA0451 (a ClpP homolog encoded in close proximity to clpD). BdlA degradation was observed in PA0451 mutant biofilms (Fig. S2C). In contrast, no BdlA degradation was observed in ∆clpP biofilms and overexpression of clpP resulted in observable degradation of BdlA under planktonic growth conditions indicating that ClpP was responsible for the nonprocessive cleavage of BdlA (Fig. 2G).
Both BdlA Cleavage Products Are Required for Dispersion.
The correlation between BdlA cleavage and nutrient-induced dispersion, together with the observation that BdlA processing is elevated in dispersed cells, suggested a role for BdlA cleavage in regulating the dispersion response. To determine whether PASa cleavage is a mechanism by which the activity of BdlA is modulated, ΔbdlA mutant biofilms complemented with intact or truncated BdlA, which were soluble and produced at levels comparable to the wild-type protein, were tested for dispersion in response to glutamate. Although ΔbdlA/bdlA biofilms dispersed, ΔbdlA biofilms complemented with BdlA lacking the PASa (BdlA-PASbTarH), PASab (BdlA-TarH), or TarH (BdlA-PASab) domains did not disperse (Fig. 3A, Fig. S1). The observations indicated that truncated BdlA peptides are inactive, and that the presence of a single specific BdlA cleavage product is not sufficient for dispersion.
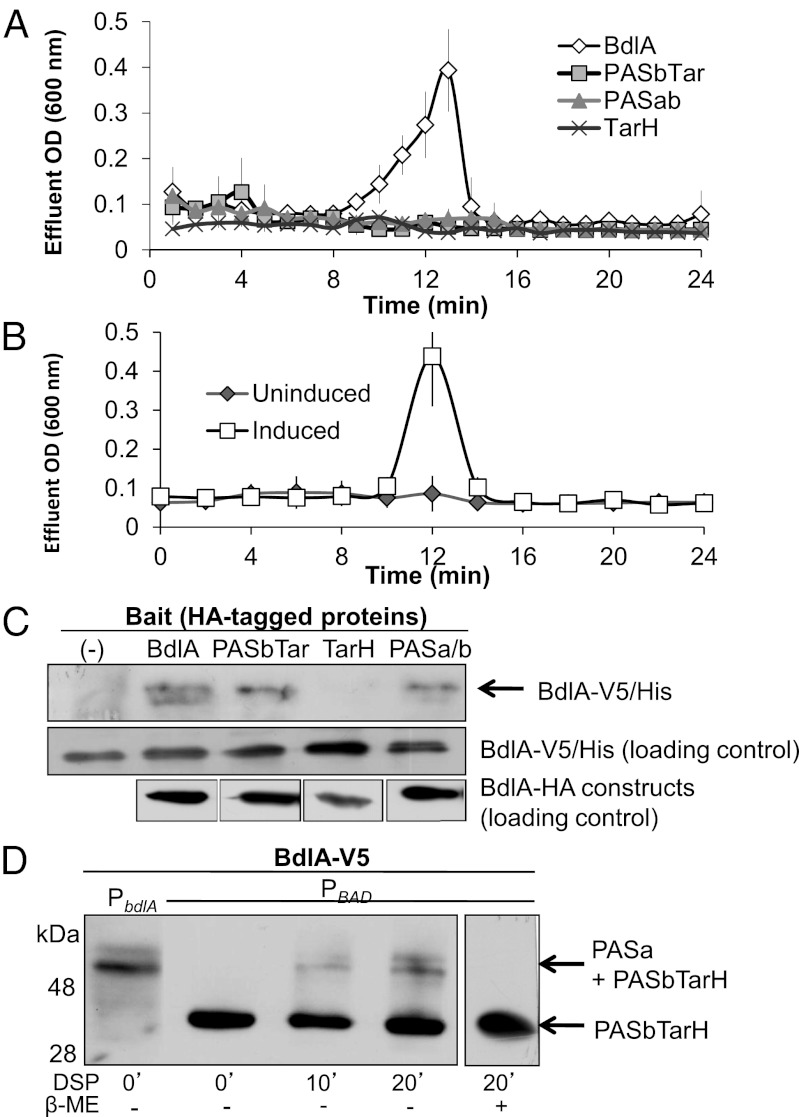
Fig. 3.
Restoration of the ∆bdlA dispersion phenotype to wild-type levels requires both truncated polypeptides of BdlA. (A) BdlA requires all conserved domains for functions, as constructs lacking any of the domains do not rescue the ∆bdlA dispersion-deficient phenotype. (B) Dispersion is restored in ∆bdlA upon coexpression of bdlA-PASa and bdlA-PASbTar from two separate vectors (∆bdlA/pJN-PASa/pMJT-PASbTar). Uninduced, no arabinose was added to growth medium; induced, arabinose was present. (C) BdlA oligomerization as determined using pull-downs depends on the presence of the PAS domains. A pull-down control (-) was performed in the absence of an HA-tagged bait protein. (D) PASbTarH interacts with PASa in PAO1/pJN-bdlA-V5 (PBAD) as indicated using in vivo cross-linking and probing with anti-V5 antibodies. PAO1::PbdlA-bdlA-V5 (PbdlA) was used as control. Cross-linking with dithiobis(succinimidyl propionate) (DSP) was reversed using β-mercaptoethanol (β-ME).
Considering that PASa is cleaved but not degraded (Fig. 1 D and E), we determined whether separately encoded truncated PASa and PASb-TarH domains would restore dispersion by ΔbdlA biofilms. As shown in Fig. 3B, the dispersion-deficient phenotype of ΔbdlA biofilms was restored to wild-type levels when ΔbdlA was complemented with separately encoded PASa and PASb-TarH constructs. This finding strongly indicated that BdlA is active upon cleavage of the PASa domain only when both truncated parts of BdlA (PASa and PASb-TarH) are present at the same time.
To determine whether the two truncated BdlA fragments are capable of interacting, pull-down assays using V5/His- and HA-tagged intact and truncated BdlA proteins were used. BdlA interacted with intact BdlA and truncated BdlA-PASbTarH and BdlA-PASab constructs (Fig. 3C). No interaction was observed for BdlA-TarH, lacking both PAS domains, indicating that BdlA oligomerizes via its PAS domains (Fig. 3C). Complex formation via the two PAS domains was confirmed by in vivo cross-linking using the C-terminally–tagged BdlA, which demonstrated that the BdlA-PASa domain interacted with BdlA-PASbTarH (Fig. 3D). Likewise, PASa–PASa complex formation was observed when N-terminally His-tagged BdlA was used (Fig. S3). Together, these findings suggested that following BdlA processing, the resulting cleavage products likely interact to form a complex that is required for the induction of dispersion.
Phosphorylation at Y238 Is a Signal for BdlA Nonprocessive Proteolysis.
To determine how the processing of BdlA is regulated, we first determined clpD levels by RT-PCR. No difference in clpD mRNA abundance, ClpD protein levels, or interaction with BdlA under biofilm and planktonic growth conditions was observed, indicating that cues other than ClpD availability regulate BdlA cleavage. As posttranslational modifications have been reported to impair or activate proteolytic cleavage (19) and because BdlA cleavage correlated with phosphorylation (Fig. 1C), we asked whether phosphorylation at Y238 is essential for BdlA cleavage. Alanine substitution of Y238 eliminated BdlA cleavage (Fig. 4A). Moreover, a BdlA-Y238A variant, which was soluble and produced at levels comparable to the wild-type protein (Fig. 4A), failed to rescue the dispersion-deficient phenotype of the bdlA mutant (Fig. 4 B and C and Table S2). The findings indicate that the Y238 residue is essential for BdlA function in triggering dispersion, probably by Y238 phosphorylation acting as a signal for BdlA processing.
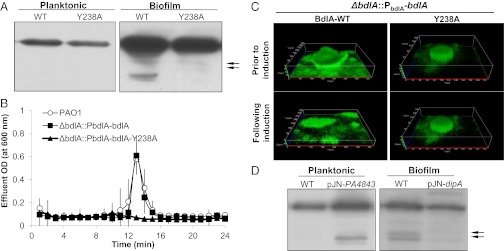
Fig. 4.
BdlA-Y238 phosphorylation and elevated c-di-GMP levels are required for BdlA cleavage. (A) Detection of the wild-type and Y238A C-terminally V5/His-tagged variants of BdlA in planktonic and biofilm PAO1 cells reveals that Y238 is required for BdlA cleavage. (B and C) Expression of wild-type (∆bdlA/pbdlA-bdlA), but not of the Y238A variant (∆bdlA/pbdlA-bdlA-Y238A), restores dispersion of ∆bdlA biofilms to wild-type levels. (D) Overexpression of cyclase PA4843 correlates with BdlA cleavage under planktonic growth conditions and dipA overexpression impairs BdlA processing under biofilm growth conditions.
DipA and Increased c-di-GMP Levels Contribute to the Nonprocessive Proteolysis of BdlA.
Regulation of biofilm dispersion has been linked to the modulation of c-di-GMP, requiring the PDE DipA for reduction of c-di-GMP levels upon induction of dispersion and for the maintenance of biofilm c-di-GMP levels (17). We therefore asked whether DipA and c-di-GMP levels play a role in BdlA processing. Overexpression of dipA in PAO1::PbdlA-bdlA-V5/His impaired BdlA cleavage in biofilms (Fig. 4D). Conversely, we found that BdlA processing could be induced in planktonic cells via overexpression of the cyclase PA4843 (Fig. 4D), suggesting a strong correlation between BdlA activation and c-di-GMP levels.
Discussion
BdlA is essential for dispersion of the pathogen P. aeruginosa in response to a variety of environmental cues. The findings that BdlA processing is essential for function and is induced in dispersed but not planktonic cells indicate the presence of a sophisticated mechanism to control and regulate the last stage of biofilm development by P. aeruginosa. Although posttranslational modification of proteins is common in bacteria, it is usually limited to the modification of amino acids including phosphorylation or glycosylation or the cleavage of N-terminally–located signal peptides or domains. The posttranslational modification of BdlA involved the cleavage of BdlA for activation, and still required both truncated polypeptide chains (PASa, PASbTarH) to induce dispersion. In addition to changes in the primary structure of BdlA, our findings indicated changes in the tertiary/quaternary structure as truncated BdlA polypeptide chains oligomerized, probably via the PAS domains. It is unclear whether the truncated BdlA polypeptides are required to interact to enable dispersion, but the ability of PAS domains to oligomerize has been demonstrated previously to convert various input stimuli into signals that propagate to downstream components by modifying protein–protein interactions (22, 23). To our knowledge, such extensive posttranslational modification involving cleavage combined with changes in the protein structure have not been described for bacterial proteins. However, several eukaryotic proteins and peptide hormones have been shown to be cleaved and subsequently rearranged for activation. The best-known example is the peptide hormone insulin, which is cut twice to remove an internal peptide (24, 25). The posttranslational modification of BdlA is reminiscent of the posttranslational modification of peptide hormones.
In a similar way to insulin, BdlA appears to be cleaved in a nonprocessive manner. We propose that upon transition to surface-associated growth, a subpopulation of the biofilm cells exhibits cleavage of BdlA, in response to a phosphorylation signal at the Y238 residue, thus readying the cells for a dispersion response in the event that changing environmental conditions are encountered. A proposed model of BdlA activity regulation is summarized in Fig. S4. However, although the cleavage products of BdlA were detectable in biofilm cells, with levels of these peptides increasing in dispersed cells, BdlA cleavage was not complete and no cleavage products were detectable in biofilms remaining attached following dispersion. The findings indicate heterogeneity within the biofilm population with only a subpopulation of cells (harboring cleaved BdlA) capable of responding via dispersion to changes in environmental conditions. Such heterogeneity has been previously observed for native dispersion triggered by the signaling molecule cis-2-decenoic acid, with cells within larger clusters preferentially undergoing native dispersion responses (8).
Based on the BdlA site-specific cleavage and mutant analyses, the protease likely responsible for the nonprocessive cleavage of BdlA is ClpP. It is also likely that BdlA cleavage is dependent on the ClpB-type protein ClpD. Considering that ClpB-type chaperones are known to be involved in protein folding and formation and stabilization of multiprotein complexes (19), it is also likely that interaction of ClpD with BdlA is independent of proteolysis but instead indicates a role of ClpD before and following restructuring of BdlA, with the nonprocessive proteolysis occurring in response to phosphorylation and elevated c-di-GMP levels (Fig. 4, Fig. S4). While ClpB-type chaperones are involved in the formation and dissociation of multiprotein complexes, PAS domains are known to be involved in complex or heterooligomer formation. Three PAS-domain–containing proteins (BdlA, DipA, and RbdA) have been identified to be required for P. aeruginosa biofilm dispersion (4, 16, 17). BdlA and DipA were recently demonstrated to interact, with the interaction resulting in increased DipA levels, increased PDE activity, and reduced biofilm biomass accumulation (26). Although dispersion has been correlated with decreased c-di-GMP levels (17), we demonstrate here a requirement for increased c-di-GMP levels to enable BdlA processing. Based on these recent findings, one may speculate that BdlA is part of a multiprotein dispersion signaling network that links dispersion cues not only to c-di-GMP modulation by fine-tuning DipA activity but also to the modulation of BdlA itself. We thus put forward a model (Fig. S5) in which BdlA processing, in addition to ClpP, ClpD, and phosphorylation of Y238, also requires elevated c-di-GMP levels. Active BdlA in turn correlates with DipA activation, decreasing c-di-GMP levels, induction of phenotypes associated with dispersion, and ultimately, BdlA inactivation upon return to planktonic growth (Fig. S5).
Future research will determine whether BdlA is directly involved in dispersion signal propagation and modulation of c-di-GMP levels via protein–protein interactions. However, the mechanism described here to enable dispersion and thus, the transition from the surface associated to planktonic state may be applicable to a large number of pathogenic species, as BLAST and precomputed BLAST link (BLINK) analysis revealed the presence of BdlA orthologs in the genomes of numerous pathogenic strains including Pseudomonas syringae, Yersinia sp., Helicobacter sp., and Pantoea sp. These pathogens are capable of forming biofilms with the motile/sessile transitions recently implicated as switches in pathogenicity phenotypes. Thus, biofilm dispersion may be a contributor to the migration of bacteria to locations remote from an initial site of infection with cells dispersed from biofilms causing acute and periodic infections.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains and plasmids are described in Table S3; oligonucleotides are described in Table S4. Strain construction and site-directed mutagenesis is described in SI Materials and Methods. P. aeruginosa was grown at 37 °C in Luria–Bertani (LB) or Vogel-Bonner Minimal Media (VBMM) as described in SI Materials and Methods.
Biofilm Growth and Biofilm Dispersion.
Biofilms were grown in tube reactors in VBMM and dispersion experiments carried out as described in SI Materials and Methods using glutamate as inducer.
Protein Identification.
The identity of BdlA and its degradation product were determined following tryptic digest using an LC-MS/MS-based approaches as previously described (10).
Purification of Phosphorylated Proteins.
Phosphorylated proteins were purified using MOAC as previously described (10).
Immunoblot Analysis and Pull-downs.
Immunoblots were performed using whole-cell lysates with equal amounts of total protein used in each lane. Antibodies and detection methods are described in SI Materials and Methods. Pull-downs were carried out using protein A/G agarose beads (Cell Signaling) with anti-HA, anti-V5, or anti-His antibodies as previously described (27, 28).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant 1R01 A107525701A2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207832109/-/DCSupplemental.
References
- 1.Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol. 2005;7:894–906. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- 2.Sauer K, et al. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thormann KM, Saville RM, Shukla S, Spormann AM. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol. 2005;187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barraud N, et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Applegate DH, Bryers JD. Effects of carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol Bioeng. 1991;37:17–25. doi: 10.1002/bit.260370105. [DOI] [PubMed] [Google Scholar]
- 7.James GA, Korber DR, Caldwell DE, Costerton JW. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J Bacteriol. 1995;177:907–915. doi: 10.1128/jb.177.4.907-915.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies DG, Marques CNH. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2004;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Southey-Pillig CJ, Davies DG, Sauer K. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 14.Barraud N, et al. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basu Roy A, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194(11):2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An S, Wu J, Zhang LH. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol. 2010;76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy AB, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bibikov SI, Miller AC, Gosink KK, Parkinson JS. Methylation-independent aerotaxis mediated by the Escherichia coli Aer protein. J Bacteriol. 2004;186:3730–3737. doi: 10.1128/JB.186.12.3730-3737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenal U, Hengge-Aronis R. Regulation by proteolysis in bacterial cells. Curr Opin Microbiol. 2003;6:163–172. doi: 10.1016/s1369-5274(03)00029-8. [DOI] [PubMed] [Google Scholar]
- 20.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 21.Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- 22.Card PB, Erbel PJA, Gardner KH. Structural basis of ARNT PAS-B dimerization: Use of a common beta-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 23.Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duckworth WC, Stentz FB, Heinemann M, Kitabchi AE. Initial site of insulin cleavage by insulin protease. Proc Natl Acad Sci USA. 1979;76:635–639. doi: 10.1073/pnas.76.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White MF, Kahn CR. The insulin signaling system. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 26.Petrova OE, Sauer K. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol. 2012 doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrova OE, Schurr JR, Schurr MJ, Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.