Abstract
Depending on growth phase and culture conditions, cardiolipin (CL) makes up 5–15% of the phospholipids in Escherichia coli with the remainder being primarily phosphatidylethanolamine (PE) and phosphatidylglycerol (PG). In E. coli, the cls and ybhO genes (renamed clsA and clsB, respectively) each encode a CL synthase (Cls) that catalyzes the condensation of two PG molecules to form CL and glycerol. However, a ∆clsAB mutant still makes CL in the stationary phase, indicating the existence of additional Cls. We identified a third Cls encoded by ymdC (renamed clsC). ClsC has sequence homology with ClsA and ClsB, which all belong to the phospholipase D superfamily. The ∆clsABC mutant lacks detectible CL regardless of growth phase or growth conditions. CL can be restored to near wild-type levels in stationary phase in the triple mutant by expressing either clsA or clsB. Expression of clsC alone resulted in a low level of CL in the stationary phase, which increased to near wild-type levels by coexpression of its neighboring gene, ymdB. CL synthesis by all Cls is increased with increasing medium osmolarity during logarithmic growth and in stationary phase. However, only ClsA contributes detectible levels of CL at low osmolarity during logarithmic growth. Mutation of the putative catalytic motif of ClsC prevents CL formation. Unlike eukaryotic Cls (that use PG and CDP-diacylglycerol as substrates) or ClsA, the combined YmdB-ClsC used PE as the phosphatidyl donor to PG to form CL, which demonstrates a third and unique mode for CL synthesis.
Keywords: cardiolipin-deficient, bacteria, mass spectrometry
The anionic phospholipid cardiolipin (CL) (Fig. 1) is found primarily in energy-transducing membranes of most bacteria and almost exclusively in the mitochondria of eukaryotes, where it is essential for the optimal function of numerous enzymes involved in mitochondrial energy metabolism (1). The near-neutral pKa of one of its two phosphate groups (2) has been postulated to function as a proton trap during oxidative phosphorylation. Alterations in CL metabolism are associated with Barth syndrome (3, 4) caused by mutations in tafazzin, a transacylase involved in remodeling of the CL acyl chains (5, 6). Additionally, CL is involved in stabilization of higher order forms of the respiratory complexes (7, 8), promoting the ribbon-like assembly of adenosine 5'-triphosphate synthase dimers and maintaining the high curvature of the mitochondrial cristae (9).
Fig. 1.
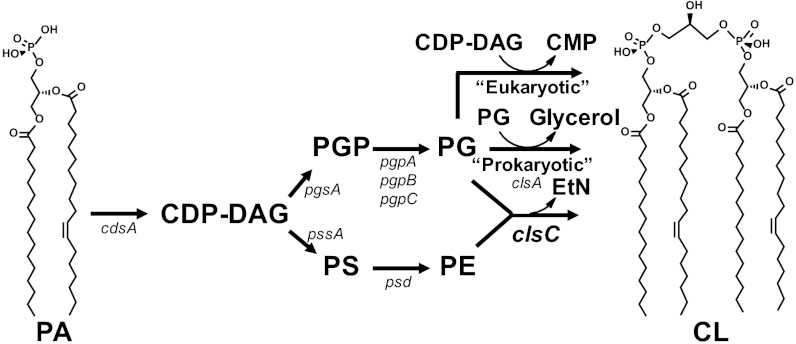
CL biosynthetic pathways. The structural genes encoding the enzymes responsible for each step are indicated. In E. coli, clsA has been shown to condense two PG molecules to form CL. The clsB gene product also catalyzes the formation of CL in E. coli, but its substrates have not been definitively established. In yeast (31), mammals (33), and actinobacteria (20), Cls (CRD1, hCLS1, and Sco1389 gene products, respectively) uses CDP-DAG and PG as substrates. The clsC gene product of E. coli catalyzes a third mode of CL synthesis, which is the subject of this report.
Mutations in the cls gene (10) (now annotated clsA) that encodes a cardiolipin synthase (denoted herein as ClsA) in Escherichia coli results in reduced CL production accompanied by sensitization of cells to low osmolarity (11) and reduced viability in long-term stationary phase (12). CL stabilizes the dimeric state of Sec-YEG protein channel complex (13), promotes polar localization of ProP (14), and activates respiratory complexes in E. coli (15). CL levels increase when cells enter stationary phase due to a 10-fold increase in ClsA activity (12). CL, a nonbilayer prone anionic lipid, is enriched at the cell poles and the cell division site (16), forming anionic lipid domains involved in the organization of molecular machines on the membrane surface responsible for initiation of DNA replication and cell division (17).
The pathway to CL biosynthesis in E. coli (Fig. 1) and almost all bacteria proceeds from phosphatidic acid (PA) to cytidine diphosphate-diacylglycerol (CDP-DAG) then via phosphatidylglycerophosphate (PGP) synthase to PGP, which is dephosphorylated to PG (18). The final step is a phosphatidyl transfer from one PG molecule to a second PG molecule to form CL and glycerol (19) catalyzed by a phospholipase d-type enzyme (ClsA) encoded by the clsA gene (10). CDP-DAG also provides the precursor to the major phospholipid phosphatidylethanolamine (PE, 70%). However, in eukaryotic cells, CDP-DAG is the phosphatidyl donor to PG to yield CL and cytidine monophosphate (CMP) (Fig. 1). A cls gene found in Streptomyces coelicolor and in most actinobacteria was recently shown to possess a CDP-DAG–dependent Cls (20). Based solely on informatics analysis, Trypanosoma brucei was suggested to encode a bacterial-type Cls (1). Thus, the terms prokaryotic- versus eukaryotic-type mechanism for CL synthesis may no longer be accurate.
Deletion of the clsA gene of E. coli resulted in no detectable CL in extracts of cells harvested during logarithmic growth in very low salt medium (1 g/L NaCl) (21, 22). However, stationary phase cells still accumulated CL, which strongly suggests the existence of another Cls. Two clsA paralog genes, ybhO and ymdC, exist in E. coli. The clsB (previously known as ybhO) gene product (ClsB) was shown to catalyze CL formation in vitro, but attempts to show in vivo activity were not successful (21). A double ∆clsAB mutant was constructed, but its lipid composition was not reported (23).
In this report, we demonstrate that clsC (previously annotated as ymdC) encodes a third and possibly last Cls. Deletion of all three genes (∆clsABC) resulted in a complete lack of CL. Each of the Cls contributes differently to the CL content under different growth conditions. Restoration of near–wild-type levels of CL production in the stationary phase by expression of clsC in the triple mutant requires the coexpression from the same operon of the preceding gene ymdB. Most significant is the demonstration that the mode of CL biosynthesis by the combined YmdB-ClsC follows neither the “eukaryotic” nor “prokaryotic” pathway but involves the transfer of a phosphatidyl moiety from PE to the terminal free hydroxyl of PG to form CL. Discovery of this third mode for CL biosynthesis has important implications for CL synthesis in many bacteria in which multiple cls genes have been identified mostly based on informatic analysis or incomplete substrate determination.
Results
Identification of Third cls Gene.
To determine the remaining source of CL, we focused on clsC (ymdC), whose gene product is homologous to clsA and clsB gene products (21). Each of the three proteins contains two HKD motifs characteristic of the phospholipase D superfamily (Fig. S1) (21). ClsC displays 23% identity and 39% similarity to ClsA and a 22% identity and 38% similarity to ClsB. However, sequence analysis did not predict any transmembrane helices in ClsC, unlike ClsA, which has two predicted transmembrane domains using the Phobius algorithm (24). The clsC gene is located at the 23rd min of the E. coli genome, whereas clsA and clsB are at the 28th min and17th min, respectively. See Tables S1 and S2 for a detailed description of strains and plasmids, respectively.
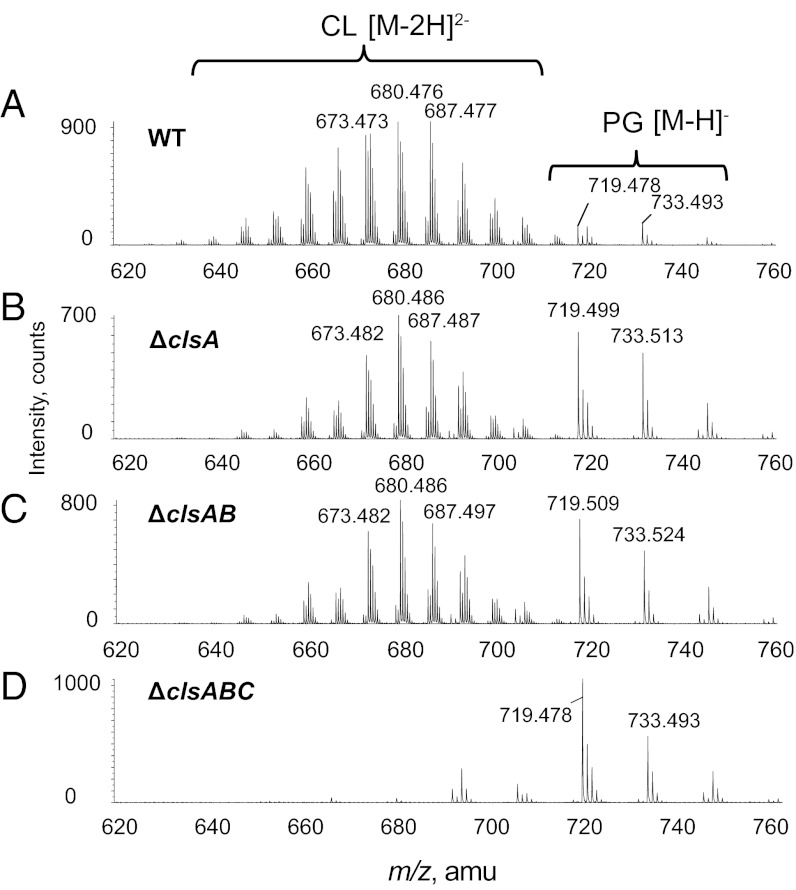
The total lipids extracted from cells grown to stationary growth phase were analyzed by normal phase liquid chromatography–tandem mass spectrometry (LC/MS/MS), which allowed the separation and identification of PG, CL (eluting at 13–14 min), monolyso CL, PE, PA, and bislyso CL (Fig. S2). Unless otherwise noted, cells were grown at 37°C in rich LB medium containing 10 g/L NaCl. The corresponding mass spectra acquired during the CL time window (Fig. 2A) showed major CL species as the doubly charged [M-2H]2− ions (m/z 630–710). The ion at m/z 680.476 matches the doubly charged CL with two 16:0 fatty acids, one 16:1 fatty acid and one 17:1 cyclopropane-containing fatty acid. The major PG species were detected as the singly charged [M-H]− ion at m/z 719.479 for PG (16:0/16:1) and at m/z 733.493 for PG (16:0/17:1). Compared with the wild type (WT) (W3110 denotes isogenic parent strain) (Fig. 2A), the ∆clsA (Fig. 2B) and ∆clsAB mutants (Fig. 2C) still accumulated CL in the stationary phase with an increase in the PG species contrary to previous results for the latter (21). The ∆clsABC mutant displayed a complete disappearance of CL, monolyso CL, and bislyso CL while maintaining elevated PG (Fig. 2D and Fig. S2). The complete lack of CL substantiates clsC functioning as a Cls.
Fig. 2.
Triple deletions of three Cls genes results in complete depletion of CL. Strains of E. coli were created lacking all various cls genes. Cells were cultured and grown into stationary phase. Their lipid extracts were examined by LC/MS/MS. Accumulation of CL was found in the WT (A), ∆clsA (B), and ∆clsAB (C) strains. Only ∆clsABC (D) showed complete disappearance of CL, consistent with ClsC being a third Cls. Experiments with [32P]PO4 labeling confirmed this conclusion (Fig. S3).
TLC analysis (Fig. S3 A and B) of stationary phase cells grown in 5 g/L NaCl and labeled with 5 μCi/mL of [32P]PO4 revealed the same defect found with mass spectrometry. WT cells contained the three major phospholipids, PE, PG, and CL, at the expected levels, and the triple deletion mutant showed no detectible CL. The area corresponding to CL (Fig. S3B) registered a background level signal (X1 = 0.02%); the faint spot (X2 = 0.11%) indicates an above background signal. PE level was unchanged, and PG increased by the amount of CL loss.
The triple mutant, like the clsA single deletion mutant (12), showed reduced viability during long-term incubation in the stationary phase, but no added effects were found when grown in LB medium at 37°C.
Expression of CL Synthases.
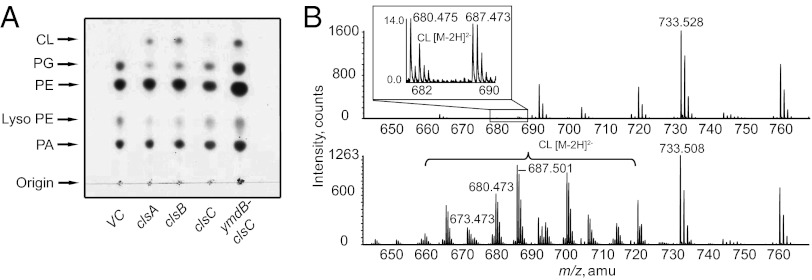
The three cls genes with ribosome-binding sites were cloned into the arabinose inducible pBAD30 vector (25) and individually transformed into the ∆clsABC mutant. The transformants were grown to stationary (A600 of ∼2.0) phase in medium with ampicillin and 0.2% arabinose for lipid analysis. By charring of the TLC plate, CL accumulation was seen with the overexpression of ClsA or ClsB (Fig. 3A). Previous work could only confirm in vitro activity of ClsB (21). The production of CL from the overexpression of ClsC was not be detected. However, MS analysis revealed small but measurable amounts of CL (Fig. 3B, Upper, Inset), confirming low in vivo ClsC activity.
Fig. 3.
ClsC needs YmdB to fully complement CL synthesis. Cls were expressed from plasmid pBAD30 in ∆clsABC strain BKT12, induced with 0.2% arabinose, and grown to stationary phase. (A) One-dimensional TLC developed with solvent 4 followed by charring revealed clsA (pBAD30-A) and clsB (pBAD30-B) restored CL production, whereas clsC (pBAD30-C) could only produce a trace amount of CL; no CL was detected in the vector control (VC). Coexpression from the same promoter (pBAD30-YC) of both ymdB and its neighbor clsC restored CL levels to higher levels than clsC by itself. (B) Lipid extracts from strains carrying plasmid pBAD-C or pBAD-YC were analyzed by LC/MS/MS. Vector control showed no CL production, whereas clsC expression alone only produced trace amounts of CL, with its doubly charged ion peaks being magnified in the boxed inset. Expression of ymdB-clsC resulted in high levels of CL production (Lower).
The clsC gene is separated by one base pair in the same operon from the preceding gene, ymdB, which encodes a protein containing a macro domain with a predicted adenosine diphosphate (ADP) ribose-binding potential (26). The entire two-gene operon was cloned into pBAD30. Co-overexpression of both proteins in the ∆clsABC mutant resulted in a CL level comparable to that with overexpression of clsA or clsB individually (Fig. 3 A and B, Lower). Further investigation as to the relationship between clsC and ymdB was carried out using LC/MS/MS lipid analysis. An alternative triple knockout BKT21 (∆clsAB, ΔymdB::KanR) still contained CL, albeit at a greatly reduced level similar to the triple mutant complemented with a plasmid copy of clsC. However, complementation of BKT21 with a plasmid harboring only the ymdB gene did not increase CL levels over the vector control. Additionally, strain BKT22 (∆clsABC, ΔymdB::KanR) still contained low levels of CL when clsC and ymdB were simultaneously expressed independently from different but compatible plasmids. However, coexpressing the cls and ymdB genes from the same operon (Fig. 3) fully complemented BKT22 with a high level of CL.
CL Levels in Single and Double Deletions in the cls Genes.
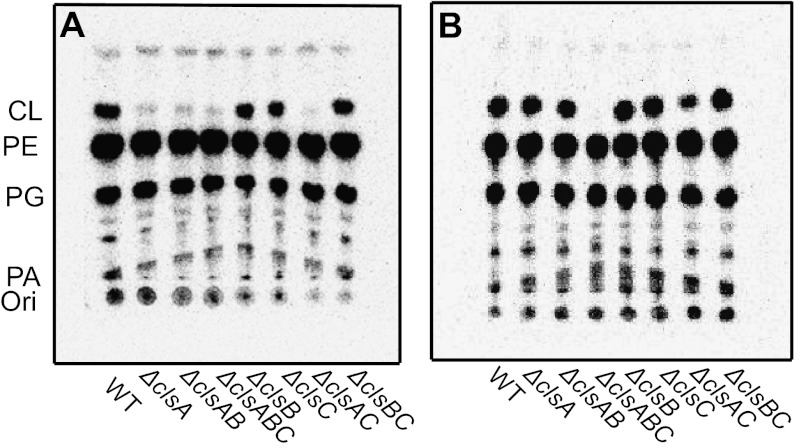
All combinations of WT, single, double, and triple chromosomal deletions of cls were grown to both midlog phase (M) or stationary phase (S) in the presence or absence of 5 μCi/mL of [32P]PO4 in LB medium containing 5 g/L or 10 g/L NaCl, respectively. TLC analysis of radiolabeled lipids from cells in logarithmic growth (Fig. 4A and Table S2, M rows) showed high levels of CL (∼5%) only in strains expressing clsA. The strains lacking clsA, including the triple cls null strain, displayed a trace spot (0.3–0.6%) with mobility similar to CL. This trace spot detected in ∆clsABC was scraped and extracted using Bligh-Dyer solvents. LC/MS/MS analysis indicated the only major lipid species to be undecaprenyl-phosphate (C55-P) (m/z 845.663) and decaprenyl-phosphate (C50-P) (m/z 777.607). No spot corresponding to CL was detected when 2D TLC was used to analysis midlog phase cells lacking clsA or the ∆clsABC mutant in stationary phase (Fig. S4). The TLC analysis of radiolabeled lipids extracted from cells grown to stationary phase (Fig. 4B and Table S2, S rows) revealed that all mutants with the exception of the cls triple deletion mutant produced high levels of CL (∼11% except for ∆clsAC at 2.2%). Therefore, in logarithmic growth phase (in 5 g/L of NaCl), ClsA contributes all of the CL while ClsB and ClsC contribute no detectible CL. All Cls contribute CL in stationary phase.
Fig. 4.
Growth phase dependence of CL synthesis in cls mutants grown in 5 g/L NaCL. WT or the indicated cls deletion strains were grown in LB medium containing 5 g/L of NaCl and [32P]PO4 to either midlog phase (A, OD600 of 0.5–0.7) or to stationary phase (B, overnight culture). Phospholipids were extracted and separated by one-dimensional TLC using solvent 1 followed by imaging as described in the Materials and Methods. Results of quantification of radiolabeled spots are shown in Table S2. The trace signal at the position of CL in A was not CL, as evidenced by lack of a CL species when the same samples were analyzed by 2D TLC (Fig. S4). Only background signal at the position of CL was detected for the ∆clsABC strain in B.
For cells grown in 10 g/L of NaCl, LC/MS/MS was used for semiquantification of CL levels relative to PE by dividing the chromatographic peak areas of the most abundant CL (66:2) by the chromatographic peak area of the most abundant PE (16:0/16:1). Essentially the same results (Fig. S5 A and B) were found, except CL was detected in the ∆clsA and ∆clsAB cells growing logarithmic phase. The effect of growth media composition is investigated in greater detail later.
Mutation of HKD Motifs of ClsC.
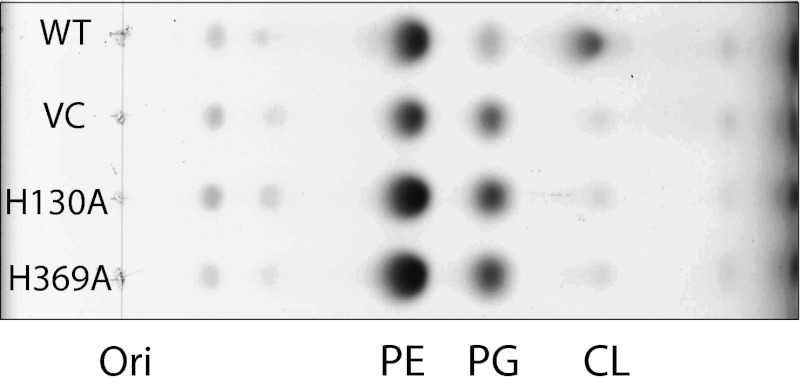
To further substantiate that ClsC is a Cls, two HKD motifs characteristic of the phospholipase D superfamily were individually changed to H130A or H369A (Fig. S1) and coexpressed in tandem with ymdB from plasmids introduced into the triple mutant. Lipids extracted from cells (OD600 ∼0.5) and analyzed by one-dimensional TLC with charring (Fig. 5) and LC/MS (Fig. S6) showed no detectible CL, further confirming ClsC to be a Cls.
Fig. 5.
Mutations in the HKD motifs of ClsC block CL synthesis. The WT without a plasmid or ∆clsABC strain carrying vector control (VC), pBAD-YC H130A, or pBAD-YC H369A (indicating mutation in ClsC HKD motif) was grown to stationary phase in the presence of 0.2% arabinose. TLC analysis of lipid extracts using solvent 4 and charring showed no increase in the CL spot over the vector control for ClsC, in which either HKD motif was mutated but PG was elevated.
Increase in CL as a Function of Medium Osmolarity.
As shown in Fig. 4A and Fig. S7, midlog cells lacking ∆clsA and ∆clsAB mutants contain low levels of CL when grown in 10 g/L but not 5 g/L NaCl. The proportion of CL is increased in E. coli grown in medium with high osmolarity adjusted with either an ionic or a nonionic osmoticum, demonstrating that the range of osmolarity rather than the chemical nature of the osmoticum is important for up-regulation of CL biosynthesis (27). All mutant combinations were radiolabeled in medium containing 5 g/L NaCl and 0.6 M mannitol (28) and analyzed for lipid composition by 2D TLC at midlog phase (Table S2, row M*); WT cells and all mutant combinations except ∆clsABC showed increased CL when grown in high (M*) versus low (M) osmolarity medium. In high osmolarity medium, mutants expressing only clsA, clsC, or clsB showed 12%, 2.6%, or 0.3% CL, respectively. Single deletions in these genes contained 2.2%, 15%, or 12% CL, respectively, versus 16% for WT cells. Therefore, only ClsA makes detectible CL in low-osmolarity medium during logarithmic growth and contributes the majority of CL in high-osmolarity medium.
Previously Undescribed Mechanism for ClsC.
To investigate the mode of CL synthesis by the coexpressed YmdB-ClsC, we constructed an E. coli mutant that cannot make PG (UE54, ∆pgsA) and also lacked all four genes (∆clsABC, ∆ymdB::KanR) associated with CL synthesis (BKT29) (Fig. 1). Plasmids expressing ClsA or YmdB-ClsC (as an operon) were transformed into BKT29. These strains were grown to stationary phase with 0.05% arabinose, and the CL synthetic capability of the membrane fraction was determined using synthetic phospholipids (5 μM) with unnatural fatty acid composition as substrates.
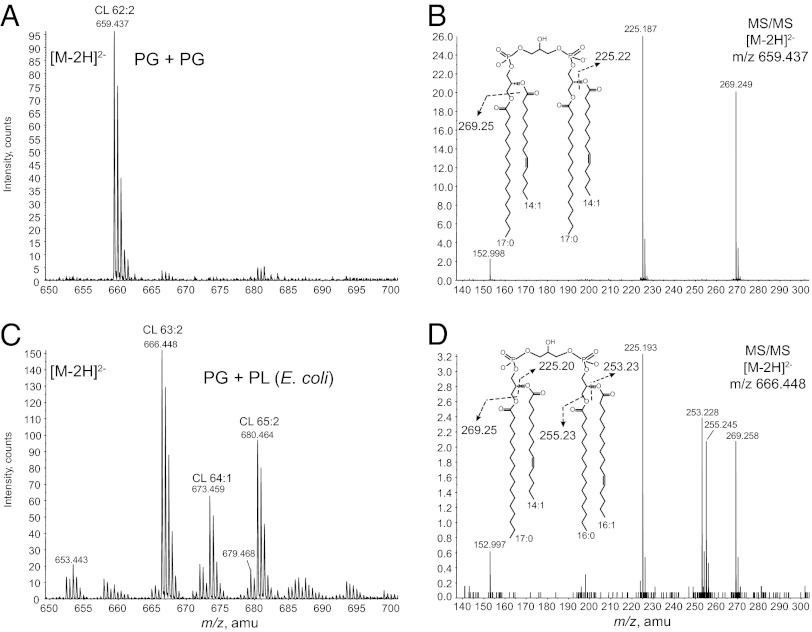
Extracts of cells expressing ClsA produced a major CL molecule, detected at m/z 659.437 for the [M-2H]2– ion, whose molecular weight is consistent with the condensation of two synthetic PG (17:0/14:1) molecules (measured mass:1320.926 Da; expected mass: 1320.910 Da) (Fig. 6A). In addition, collision-induced dissociation MS/MS of the m/z 659.437 ion produced major ions at m/z 225.187 and m/z 269.249, corresponding to the carboxylic anions of C14:1 fatty acid and C17:0 fatty acid, respectively (Fig. 6B). In contrast, cell extracts expressing YmdB-ClsC produced a heterogeneous distribution of CL species, with the major peaks being detected at m/z 666.448 for CL (63:2), m/z 673.459 for CL (64:1), and m/z 680.464 for CL (65:2) (Fig. 6C). MS/MS analysis showed that the major CL species fatty acids were derived from synthetic PG and those of endogenous lipids in E. coli. For example, MS/MS spectrum (Fig. 6D) of the major peak at m/z 666.448, revealed the fatty acyl composition of this CL species to be m/z 269.258 (17:0) and m/z 225.193 (14:1) ions from the synthetic PG (17:0/14:1) as well as m/z 255.245 (16:0) and m/z 253.228 (16:1) ions corresponding to the major fatty acids in E. coli phospholipids. Addition of synthetic CDP-DAG (17:0/18:1) along with synthetic PG to the reaction mixture did not change the MS/MS pattern of CL made by ClsA or YmdB-ClsC over that with synthetic PG alone, which ruled out the use of CDP-DAG as the endogenous substrate.
Fig. 6.
YmdB-ClsC differs from ClsA in their substrates. Cell membranes derived from strain BTK29 (∆pgsA, ∆clsABC, ∆ymdB) expressing clsA or ymdB-clsC from plasmid pBAD30 were used with a synthetic PG (17:0/14:1) as substrate to synthesize CL. (A) Membranes from cells with overexpressed ClsA primarily produced a single species of CL consistent with the condensation of two PG molecules. (B) MS/MS spectrum of the [M-2H]2– ion at m/z 659.4 of the CL made by ClsA contained only the carboxylic anions of fatty acids derived from the synthetic PG (17:0/14:1). (C) YmdB-ClsC produced a heterogeneous distribution of CL species. (D) MS/MS spectrum of the [M-2H]2– ion at m/z 666.448 of the most abundant CL formed by YmdB-ClsC contained carboxylic anions of fatty acids derived from both the endogenous sources (16:0/16:1) and those from the synthetic PG (17:0/14:1).
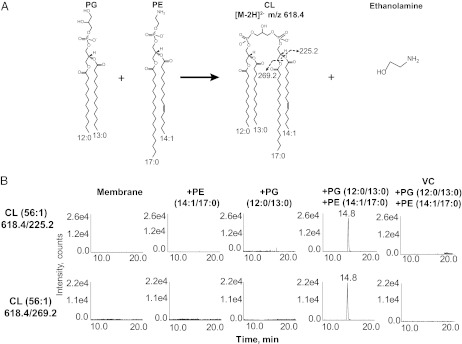
PE and PA were the two major phospholipid species that might serve as phosphatidyl donors to PG. The same in vitro assay system was used with the addition of synthetic PG (12:0/13:0) and either synthetic PE or PA with 14:1/17:0 acyl chains in both cases (Fig. 7A shows the expected PE-derived CL). To determine whether phosphatidyl moieties from PE or PA were incorporated into CL without the interference of possible endogenous lipids, we used the multiple reaction monitoring (MRM) method performed on a triple quadrupole instrument in which the first (Q1) and last (Q3) mass analyzers were used as mass filters to ensure that only the expected CL species are detected. If ClsC uses the aforementioned substrates, the expected CL would have a doubly charged [M-2H]2– ion at m/z 618.4 (Fig. 7A), which is selected by the first mass filter. After the MS/MS collision cell, the last mass filter should isolate the m/z 225.2 (14:1) and 269.2 (17:0) ions, corresponding to the exogenous acyl chains derived from the synthetic substrates. With exogenous PA, PG, PE alone, or PG with PA, the expected CL species (14.8 min) with the respective fatty acyl compositions were not found (Fig. 7B). However, the addition of exogenous PG and PE together in the assay mixture created a 500-fold increase in signal at 14.8 min when selecting for the expected mass and fatty acyl composition of CL (Fig. 7A) made from PE and PG (Fig. 7B). The absence of signal with other combinations of exogenous phospholipids indicates that CL synthesis was dependent on ClsC with PG and PE as cosubstrates.
Fig. 7.
ClsC uses PG and PE as substrates. BKT29 (∆pgsA, ∆clsABC)–derived membranes expressing ymdB-ClsC (from plasmid pBAD-YC) were reacted with synthetic PG (12:0/13:0) and either synthetic PE (17:0/14:1) or synthetic PA (17:0/14:1). The CL product was detected by normal phase LC coupled with the multiple reaction monitoring (MRM) protocol performed on a 4000 Q-Trap mass spectrometer. (A) Proposed scheme of synthesis of CL from PG (12:0/13:0) and PE (17:0/14:1). The CL product was expected to be detected as the doubly charged [M-2H]2– at m/z 618.4. The MS/MS product ions should include m/z 225.2 and m/z 269.2, corresponding to the carboxylic anions of the 14:1 and 17:0 fatty acids (derived from the synthetic PE), respectively. (B) LC-MRM detection of the CL product (14.8 min) formed from the synthetic PG (12:0/13:0) and synthetic PE (17:0/14:1). The ion pairs of 618.4/225.2 and 618.4/269.2 indicate the MRM transitions from the CL precursor ion ([M-2H]2– at m/z 618.4) to its two product ions at m/z 225.2 and 269.2, respectively. No predicted CL species were detected with the other combinations of substrates or with membranes derived from BKT29 transformed with the plasmid vector (VC).
Discussion
E. coli has three paralogs of Cls dispersed over the genome. Each contains two phospholipase D HKD motifs. The first mutation in a Cls gene, clsA, of E. coli was reported in 1978 (10), and a null gene was later made by kanamycin-resistant cassette replacement (29). The next Cls gene, clsB, was reported in 2000 (21). Although clsB was shown to catalyze CL formation in vitro, its in vivo activity could not be demonstrated. The attempt to demonstrate in vivo activity was based on the incorrect assumption that CL production by ClsB could rescue the temperature-sensitive phenotype of a pssA and clsA double mutant using growth medium containing 5 g/L NaCL, which we demonstrated does not support the synthesis of CL by ClsB during the logarithmic growth. In this study, we demonstrated in vivo CL synthesis by expression of clsB in the absence of clsAC in cells grown to stationary phase. The low amounts of CL detected in ∆clsA strains was postulated to be a minor product of the pssA gene product (phosphatidylserine synthase) because the latter also contains the HKD phospholipase D motif (29). However, our demonstration of a third gene, clsC, encoding ClsC and the absence of CL in a ∆clsABC mutant exclude this possibility. CL levels as a function of cls genes, growth phase of cells, and osmolarity of the growth medium fully explain many of the anomalies (including the above) with respect to CL synthesis. It is clear that growth conditions and genetic backgrounds must be carefully controlled when studying the role of CL in cell function.
The most remarkable finding was that ClsC, dependent on the HKD phospholipase D motifs, synthesized CL by a different mode using PE as the phosphatidyl donor to PG rather than CDP-DAG (“eukaryotic” mode) or a second PG (“bacterial” mode). Existence of this mode of synthesis challenges the simple informatics approach to classifying a Cls from all organisms as discussed for other bacteria below. A recent report from T. brucei suggested the presence of a bacterial-type Cls (30) based solely on informatics analysis without determination of in vitro synthase activity, so it is unclear which “prokaryotic” mode is used. All other eukaryotes, as exemplified by Saccharomyces cerivisiae (CRD1) (31) and human (CLS1) (32) genes, encode a Cls that uses CDP-DAG and PG as substrates (33, 34). S. cerevisiae ∆crd1 strains lack CL but have not been analyzed under a wide spectrum of growth conditions to determine if other modes of CL synthesis may exist. Whether somatic cells or plant species contain alternative modes of CL synthesis remains an open question.
In many bacteria, either single or multiple genes are present that encode proteins with Cls activity and/or share homology with the E. coli cls genes. In only a few cases has PG independent of CDP-DAG been identified as a substrate and in no case has other phospholipid substrates been ruled out. In fact, previous in vitro determination of the substrates used by E. coli ClsB only established PG as a substrate and did not rule out involvement of membrane-derived PE as a second substrate (21). We suspect that ClsB may use additional substrates and PG. The set of three homologous genes encoding Cls only exists in a few specific prokarya: Salmonella, Escherichia, and Shigella in Eneterobactera; Pseudomonia in Gammaproteobacteria; and Burkholderia and Bordetalla in Betaproteobacteria. Three cls paralogs (clsA [formerly known as ywnE], ywjE, and ywiE) have been identified in Bacillus subtilis, but only ywnE has been shown to encode a Cls with a dominant role in CL synthesis (35). Interestingly, a triply null mutant of Bacillus subtilis still contains CL as determined by MS. Two genes (cls1 and cls2) encoding a Cls were identified in Staphylococcus aureus. These two genes together appear to account for nearly all CL formation, because deletion of both genes showed essentially no detectable CL accumulation in the stationary phase of growth (36). Methanomicrobia of Archea maintains a single copy clsA homolog with conserved functional motifs. Although rare in eukarya, strong homologs of bacterial Cls exist in Caebirgabditis japonica and Ciona intestinalis, which most likely use a CDP-DAG independent mode of CL synthesis.
ClsC is the proposed catalytic component of a possible two-enzyme complex containing YmdB, which is also a remarkable finding for a Cls. ClsC contains the repetition of the HKD motif found in the two other Cls. Like ClsB, ClsC contains no predicted transmembrane helices; however, both activities were found in the membrane fraction, suggesting they are membrane-associated proteins. Like ClsA, ClsC overexpression is toxic to cells and can only be maintained by low induction. The role of YmdB in enhancing ClsC activity and necessity for transcription from the same polycistronic operon is unknown. The ymdB-clsC operon is present in a group of bacteria that are closely related to E. coli, such as Salmonella, Shigella, Cronobacter, Citrobacter, and Enterobacter. YmdB belongs in the macro family of proteins, which is characterized by an ADP-binding domain (37). In vitro studies show YmdB can deacetylate the sirtuin product of O-acetyl-ADP-ribose and reform ADP ribose in vitro (37). The macro domain usually precedes the sirtuin gene as a neighbor. In this case, ymdB does not precede a sirtuin gene because ClsC was shown to be involved in CL synthesis. YmdB has also been shown to be an RNase III inhibitor by protein binding (38).
Overall, we have shown that ClsC is a third and possibly last Cls of E. coli. Triple deletions of clsA, clsB, and clsC resulted in the complete depletion of CL in E. coli cells. The availability of the full complement of cls genes will facilitate studies of the regulation of CL biosynthesis and the physiologic importance of CL in E. coli and other bacteria. Most significant is the finding that YmdB-ClsC, an apparent unusual two-component complex, uses PG and PE in an unexpected mode of CL biosynthesis. This finding should stimulate more careful characterization of the multiple homologous cls genes in other bacteria and eukaryotic cells. Why E. coli has two modes for the synthesis of CL remains an intriguing question.
Materials and Methods
Mutants with multiple chromosomal deletions of cls genes were derived from the Keio single deletion collection. Lipid extraction was accomplished through an acidic Bligh-Dyer method. Total lipids were analyzed by either TLC or normal phase LC/MS using a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Foster City, CA). The LC-MRM analysis for assaying the ClsC activity was carried out using a 4000 Q-Trap hybrid triple quadrupole linear ion-trap mass spectrometer equipped with a Turbo V ion source (Applied Biosystems Inc, Foster City, CA). Detailed protocols and reagents used and growth conditions are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the C.R.H.R. laboratory for helpful suggestions and experimental assistance. This research was partly supported by Lipid Maps Large Scale Collaborative Grant GM-069338 from the National Institutes of Health (NIH), which supports B.K.T. and Z.G. and the mass spectrometry facility at Duke University. In addition, partial support for these studies came from NIH Grant R37 GM 20478 and the John Dunn Research Foundation (W.D.).
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212797109/-/DCSupplemental.
See Commentary on page 16402.
References
- 1.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: Implications for lipid-linked disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haines TH, Dencher NA. Cardiolipin: A proton trap for oxidative phosphorylation. FEBS Lett. 2002;528:35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- 3.Schlame M, et al. Phospholipid abnormalities in children with Barth syndrome. J Am Coll Cardiol. 2003;42:1994–1999. doi: 10.1016/j.jacc.2003.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Vreken P, et al. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279:378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 5.Bione S, et al. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12:385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 6.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together: cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acehan D, et al. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys J. 2011;100:2184–2192. doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pluschke G, Hirota Y, Overath P. Function of phospholipids in Escherichia coli: characterization of a mutant deficient in cardiolipin synthesis. J Biol Chem. 1978;253:5048–5055. [PubMed] [Google Scholar]
- 11.Shibuya I, Miyazaki C, Ohta A. Alteration of phospholipid composition by combined defects in phosphatidylserine and cardiolipin synthases and physiological consequences in Escherichia coli. J Bacteriol. 1985;161:1086–1092. doi: 10.1128/jb.161.3.1086-1092.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiraoka S, Matsuzaki H, Shibuya I. Active increase in cardiolipin synthesis in the stationary growth phase and its physiological significance in Escherichia coli. FEBS Lett. 1993;336:221–224. doi: 10.1016/0014-5793(93)80807-7. [DOI] [PubMed] [Google Scholar]
- 13.Gold VAM, et al. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci USA. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romantsov T, Guan Z, Wood JM. Cardiolipin and the osmotic stress responses of bacteria. Biochim Biophys Acta. 2009;1788:2092–2100. doi: 10.1016/j.bbamem.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arias-Cartin R, et al. Cardiolipin-based respiratory complex activation in bacteria. Proc Natl Acad Sci USA. 2011;108:7781–7786. doi: 10.1073/pnas.1010427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mileykovskaya E, Dowhan W. Role of membrane lipids in bacterial division-site selection. Curr Opin Microbiol. 2005;8:135–142. doi: 10.1016/j.mib.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y-H, Guan Z, Zhao J, Raetz CRH. Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J Biol Chem. 2011;286:5506–5518. doi: 10.1074/jbc.M110.199265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirschberg CB, Kennedy EP. Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc Natl Acad Sci USA. 1972;69:648–651. doi: 10.1073/pnas.69.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandoval-Calderón M, Geiger O, Guan Z, Barona-Gómez F, Sohlenkamp C. A eukaryote-like cardiolipin synthase is present in Streptomyces coelicolor and in most actinobacteria. J Biol Chem. 2009;284:17383–17390. doi: 10.1074/jbc.M109.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo D, Tropp BE. A second Escherichia coli protein with CL synthase activity. Biochim Biophys Acta. 2000;1483:263–274. doi: 10.1016/s1388-1981(99)00193-6. [DOI] [PubMed] [Google Scholar]
- 22.Hiraoka S, Nukui K, Uetake N, Ohta A, Shibuya I. Amplification and substantial purification of cardiolipin synthase of Escherichia coli. J Biochem. 1991;110:443–449. doi: 10.1093/oxfordjournals.jbchem.a123600. [DOI] [PubMed] [Google Scholar]
- 23.Michaelis AM, Gitai Z. Dynamic polar sequestration of excess MurG may regulate enzymatic function. J Bacteriol. 2010;192:4597–4605. doi: 10.1128/JB.00676-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction: The Phobius web server. Nucleic Acids Res. 2007;35:429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karras GI, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsatskis Y, et al. The osmotic activation of transporter ProP is tuned by both its C-terminal coiled-coil and osmotically induced changes in phospholipid composition. J Biol Chem. 2005;280:41387–41394. doi: 10.1074/jbc.M508362200. [DOI] [PubMed] [Google Scholar]
- 28.Ito T, et al. The implication of YggT of Escherichia coli in osmotic regulation. Biosci Biotechnol Biochem. 2009;73:2698–2704. doi: 10.1271/bbb.90558. [DOI] [PubMed] [Google Scholar]
- 29.Nishijima S, et al. Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J Bacteriol. 1988;170:775–780. doi: 10.1128/jb.170.2.775-780.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serricchio M, Bütikofer P. An essential bacterial-type cardiolipin synthase mediates cardiolipin formation in a eukaryote. Proc Natl Acad Sci USA. 2012;109:E954–E961. doi: 10.1073/pnas.1121528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang S-C, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem. 1998;273:14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- 32.Chen D, Zhang X-Y, Shi Y. Identification and functional characterization of hCLS1, a human cardiolipin synthase localized in mitochondria. Biochem J. 2006;398:169–176. doi: 10.1042/BJ20060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlame M, Zhao M, Rua D, Haldar D, Greenberg ML. Kinetic analysis of cardiolipin synthase: a membrane enzyme with two glycerophospholipid substrates. Lipids. 1995;30:633–640. doi: 10.1007/BF02537000. [DOI] [PubMed] [Google Scholar]
- 34.Heacock AM, Agranoff BW. CDP-diacylglycerol synthase from mammalian tissues. Biochim Biophys Acta. 1997;1348:166–172. doi: 10.1016/s0005-2760(97)00096-9. [DOI] [PubMed] [Google Scholar]
- 35.Kawai F, et al. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol. 2004;186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koprivnjak T, et al. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J Bacteriol. 2011;193:4134–4142. doi: 10.1128/JB.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen D, et al. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J Biol Chem. 2011;286:13261–13271. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim KS, Manasherob R, Cohen SN. YmdB: A stress-responsive ribonuclease-binding regulator of E. coli RNase III activity. Genes Dev. 2008;22:3497–3508. doi: 10.1101/gad.1729508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.