Abstract
Precisely arranged cytoarchitectures such as layers and nuclei depend on neuronal migration, of which many in vitro studies have revealed the mode and underlying mechanisms. However, how neuronal migration is achieved in vivo remains unknown. Here we established an imaging system that allows direct visualization of cortical interneuron migration in living mouse embryos. We found that during nucleokinesis, translocation of the Golgi apparatus either precedes or occurs in parallel to that of the nucleus, suggesting the existence of both a Golgi/centrosome-dependent and -independent mechanism of nucleokinesis. Changes in migratory direction occur when the nucleus enters one of the leading process branches, which is accompanied by the retraction of other branches. The nucleus occasionally swings between two branches before translocating into one of them, the occurrence of which is most often preceded by Golgi apparatus translocation into that branch. These in vivo observations provide important insight into the mechanisms of neuronal migration and demonstrate the usefulness of our system for studying dynamic events in living animals.
Keywords: cortex, GABAergic interneuron, in utero electroporation, in vivo imaging
Neuronal migration is a critical step in the construction of the nervous system. During development, neurons migrate from the sites of their birth to their final destinations, where they form neuronal architectures, such as laminated structures and nuclei, that are necessary for information processing in the nervous system.
Because neuronal migration is a dynamic phenomenon, real-time imaging could provide important insight into its mechanisms. Studies using real-time imaging of neurons in dissociated culture have demonstrated various aspects of neuronal migration, including the fact that radial fibers (1) and neighboring cells (2) act as the substrate for migration and the molecular mechanisms driving nucleokinesis (3, 4). Real-time imaging has also helped reveal the dynamics and roles of cytoplasmic organelles and cytoskeletal components, such as the centrosome (5–9), microtubules (9), and even of some molecules like calcium (10) and actin (7, 11, 12) during migration. In addition, slice and explant preparations have demonstrated aspects of migratory behaviors, including locomotion and translocation (13), branch-induced changes in migratory direction (14), multidirectional migration (15), random-walk-like behavior (16), pausing behavior, and the transition from tangential to radial migration (17).
However, as informative as these neural migration studies are, they are limited because they are all in vitro and do not necessarily show neuronal behaviors in vivo. Here, we observed migrating neurons in the brain of living mouse embryos and analyzed their behavior, including the dynamics of the nucleus and the Golgi apparatus. We took advantage of the fact that cortical interneurons tangentially migrate near the surface of the cortex. These neurons are generated in the ganglionic eminences (GEs) in the basal forebrain and migrate along specific tangential pathways to the cortex (for review, see refs. 18–20). After arrival in the cortex, interneurons in the intermediate and subventricular zones translocate to the marginal zone (MZ), where they execute multidirectional tangential migration (15, 16, 21). We labeled cortical interneurons by in utero electroporation and imaged them through the skull of living mouse embryos at the stage when they migrate within the cortical MZ.
We found two distinct modes of nucleokinesis: Although nucleokinesis was often preceded by translocation of the Golgi apparatus, it took place in parallel with the nucleus, implying the occurrence of Golgi apparatus/centrosome-dependent and -independent nucleokinesis. We also obtained evidence suggesting that prior translocation of Golgi apparatus/centrosome into a leading process (LP) branch may be a critical determinant for the nucleus to move into that branch during change of migratory directions. These in vivo observations provide important insight into the role of the Golgi apparatus/centrosome complex in forward movement of neurons and regulation of the direction of migration.
Results
Multidirectional Migration of Cortical Interneurons in Living Embryos.
The dam was anesthetized with urethane and placed onto a metal plate. An incision was made in the abdomen of the pregnant mouse to partially expose one uterine horn, and an embryo was removed from the uterus while keeping its umbilical cord attached to the dam (Fig. S1). The embryo was then immobilized in an agarose-filled plastic container. After gelling of the agarose, the scalp of the embryo was removed. The stability of the recordings was ensured by monitoring the body temperature (33.6–37.9 °C), heartbeat and blood flow in the cortex, and the average rate of neural migration.
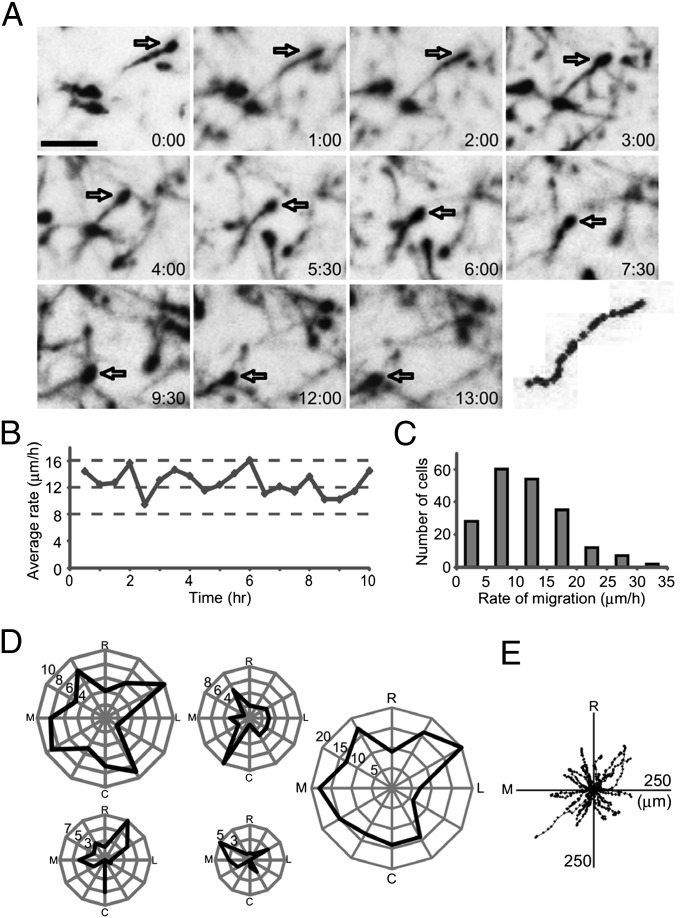
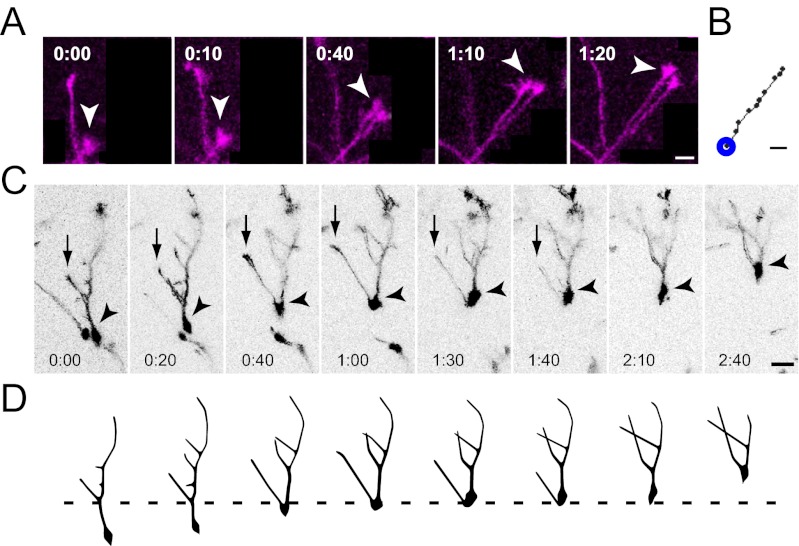
First, we visualized migrating interneurons in the cortical MZ of living embryos. We found that tdTomato-labeled interneurons extended a LP while migrating actively (Fig. 1A and Movie S1). Interneuron migration was recorded for a long period (up to 13 h) in stable conditions (Fig. 1B). The migration rate varied from one cell to another, with an average of 12 ± 6 μm/h (mean ± SD, n = 198; Fig. 1C), although some cells were stationary during the recording period (Movie S1, blue arrow). These results demonstrate that interneuron migration can be stably visualized through the skulls of living embryos by using our system.
Fig. 1.
Migration of interneurons in living embryos. (A) Time-lapse sequence of migrating neurons recorded from an embryonic day (E) 16.5 mouse embryo. Neurons were labeled by electroporation of tdTomato at E12.5. Open arrows point to a neuron tracked up to 13 h in the MZ. Elapsed time is indicated in the lower right of each image (h:min). The track of this interneuron is shown in the bottom-right panel in which cell position is plotted at 30-min intervals. (B) Average rate of migration during recording showing stability. Each dot indicates the average migration rate of the neuronal population (n = 31–38 cells). (C) Mean rate of migration for individual neurons estimated from the initial 3 h [n = 4 embryos, 198 cells, 12 ± 6 μm/h (mean ± SD)]. (D) Polar plot of interneuron migration. The tangential plane was divided into 12 sectors, and the number of neurons migrating in each sector was plotted. Data from four embryos are shown independently. Right shows pooled data (n = 4 embryos, 162 cells). Interneurons migrated in all directions in the MZ. R, rostral; L, lateral; C, caudal; M, medial. (E) Direction of migration for neurons shown in D Upper Middle. Black dots show cell body positions at each time point of recording, with the initial position of migration at the center (n = 36 cells). (Scale bar, 50 μm.)
Analyses of the direction of migration showed that interneurons migrated in all directions in the MZ (Fig. 1 D and E), and the migratory trajectories varied from one neuron to another (Fig. 1E). These findings support previous observations of interneurons in the MZ of cortical explants (15, 16, 21).
Dynamics of Cellular Organelles and LPs.
Two aspects of neuronal migration have been highlighted by in vitro studies: its association with changes in the cytoplasmic localization of cellular organelles and formation of LP branches (22). To analyze these aspects in vivo, we used a two-photon microscope equipped with a water-immersion lens (N.A. = 1.0). In addition, we used membrane-targeted EGFP (GAP–EGFP) or tdTomato (GAP–tdTomato) to better visualize neuronal processes.
Nucleokinesis.
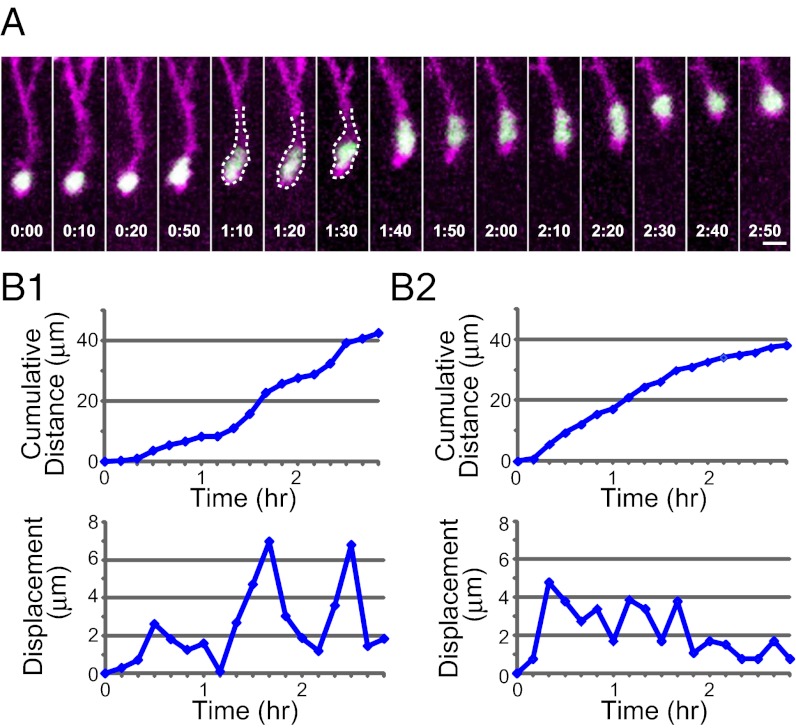
One notable feature of neuron migration in vitro is saltatory movement in which migrating neurons show alternating periods of cell soma/nucleus advancing and resting (2–4, 23, 24). To examine whether saltatory movement also occurs in vivo, we visualized the nucleus and cell outline of migrating interneurons by nuclear localization sequence (nls)–EGFP and GAP–tdTomato, respectively, and found that some neurons displayed cyclic advancing/resting phases of cell soma/nucleus (Fig. 2 A and B1 and Movie S2), consistent with a saltatory mode of migration. However, this mode was not always seen, but, rather, quite often we found interneurons migrating at a relatively constant rate (Fig. 2B2). Thus, it appears that some interneurons migrate in a saltatory manner, although saltatoriness is not obvious in others in vivo.
Fig. 2.
Nucleokinesis of interneurons. (A) Time-lapse sequence of nucleokinesis. Interneurons were labeled by coelectroporation of nls–EGFP (green) and GAP-tdTomato (magenta). The dashed lines partially outline the cell body. Elapsed time is indicated at the bottom of each image (h:min). (B) The cumulative distance (Upper) and displacement at each step (Lower) of the two migrating neurons are plotted against time. B1 corresponds to the neuron shown in A. Although displacement in each step in B2 was not constant, saltatoriness was not obvious. (Scale bar, 10 μm.)
Deformation of the nucleus during nucleokinesis.
Previous studies have indicated that cytoplasmic swelling or dilation moves into the LP before nucleokinesis occurs (3, 4, 6). However, we rarely observed occurrence of clear cytoplasmic dilation, even in interneurons migrating in a saltatory fashion. The cyclic change in cell morphology that we did observe was the changing shape of cell nucleus. Before nucleokinesis, the shape of the nucleus changed from round (Fig. 2A; t = 0:00–0:20) to elongated (Fig. 2A; t = 1:10–1:20, 1:40–2:20; see also Fig. 5 A and B). This elongated shape was maintained during nucleokinesis but became round again when the nucleus was stationary (Fig. 2A; t = 2:30–2:50; see also Fig. 5B; t = 1:40).
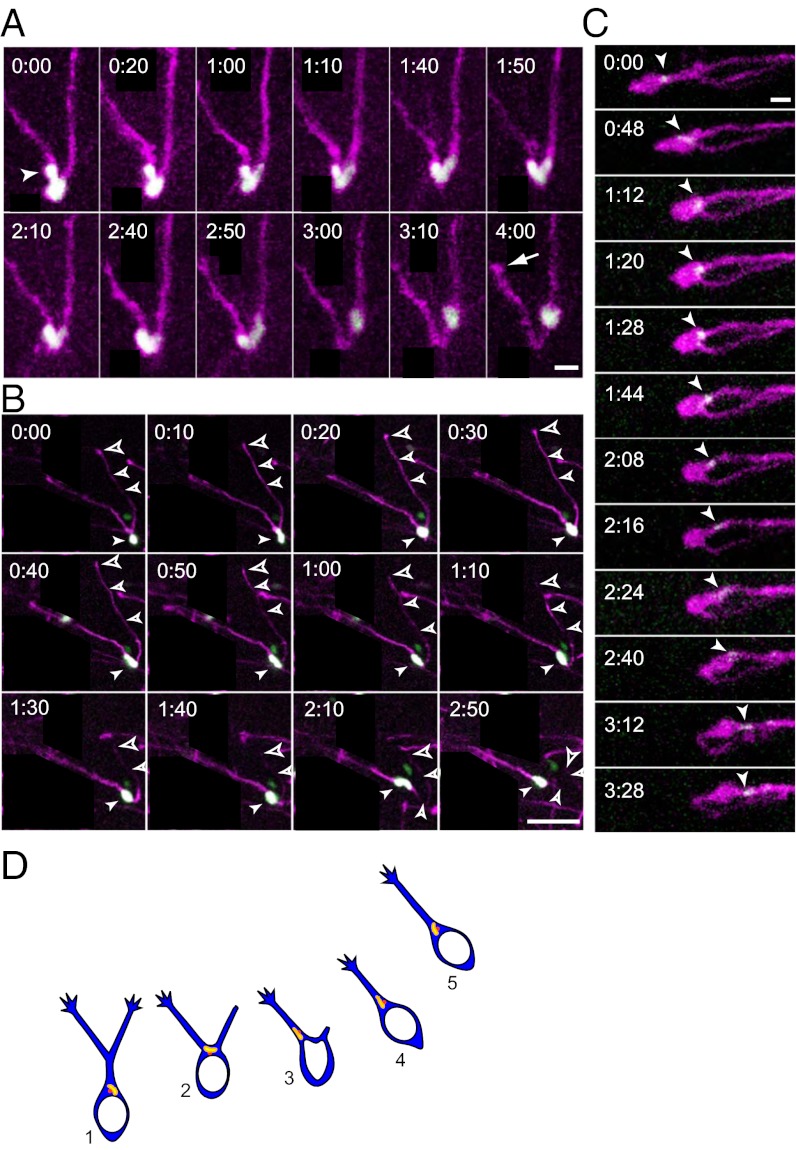
Fig. 5.
Dynamics of the nucleus and Golgi apparatus at the bifurcating point of the LP. (A) Time-lapse sequence showing swinging behavior of a nucleus with V-shaped morphology at a LP bifurcating point. Neurons were labeled by nls–EGFP (green) and GAP–tdTomato (magenta). The arrowhead points to a budding form of the nucleus, and the arrow points to the tip of the retracting process. (B) Time-lapse sequence showing nucleokinesis near a branching point. Filled arrowheads indicate the nucleus, and open arrowheads point to the branch that will eventually retract, with the larger arrowheads pointing to its tip. Both process branches were extending when the nucleus reached the branching point (t = 0:20). Subsequently, the nucleus directly entered one (t = 0:30), followed by the retraction of the other (t = 0:50). (C) Time-lapse sequence showing the dynamics of the Golgi apparatus at the bifurcating point of the LP. The Golgi apparatus is shown as a white arrowhead. In most cases, the Golgi apparatus moved into a branch (t = 1:44) before the nucleus did (n = 14/16). (D) Model of a migratory behavior at the branching point. At the LP branching point (2), the Golgi/centrosome complex moved into a LP branch (3) before the nucleus did (4). Translocation of the Golgi/centrosome complex into a LP appears to be a key step for selecting the new direction of migration. Elapsed time is indicated at the upper left corners (h:min) in A–C. Neurons or their processes irrelevant to our analysis are covered by black rectangles. [Scale bars, 10 μm (A and C); 50 μm (B).]
Because the nucleus occupies most of the somatic region, the terms “nucleokinesis” and “somal translocation” will be used interchangeably hereafter.
Dynamics of the Golgi apparatus and nucleokinesis.
Cell migration requires polarization of the cell, resulting in a leading edge and a trailing edge. This polarized cell shape is underlined by a reorientation of the centrosome, which is the microtubule organizing center, and by the Golgi apparatus moving toward the direction of migration (reviewed in ref. 25), both of which are frequently found within cytoplasmic swellings in the LP of interneurons in vitro (3). The observation that the forward movement of the cytoplasmic swelling precedes that of the cell soma suggests that the Golgi/centrosome complex and cytoplasmic swelling exert a pulling force mediated by microtubules to drive nucleokinesis (3, 22).
To study the role of the Golgi/centrosome complex in interneuron nucleokinesis in vivo, we analyzed the localization and dynamics of the Golgi apparatus in migrating interneurons. We chose the Golgi apparatus as being representative of the Golgi/centrosome complex because of its relatively larger size and, therefore, brighter fluorescence. The close association of Golgi apparatus and centrosome was validated in fixed preparations (Fig. S2).
We first analyzed the localization of the Golgi apparatus in fixed preparations where it was found most often within the front of the cell soma (Fig. S3, front) and in the LP (Fig. S3, LP), consistent with the proposed role of the Golgi/centrosome complex. However, in a minor population, localization in the middle and even in the rear one-third of the soma was also observed (Fig. S3, rear).
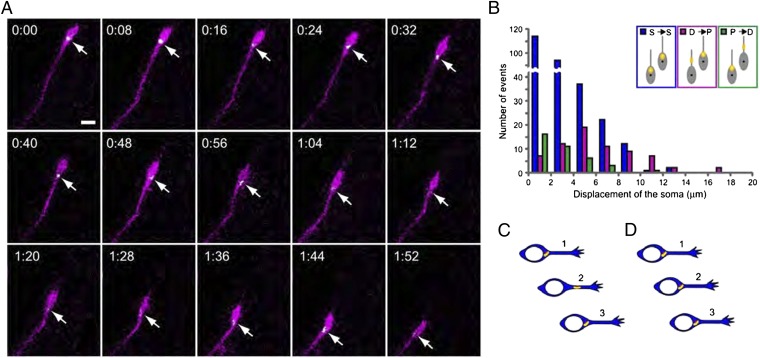
Next, we examined the localization of the Golgi apparatus and its dynamics in migrating interneurons in vivo. The established model of nucleokinesis predicts that, in cells at the resting phase, one would see the Golgi/centrosome complex and nucleus moving apart, whereas in cells during active nucleokinesis, one would see the two cellular compartments moving closer (26). Therefore, we analyzed the distance between the Golgi and nucleus within two successive movie frames 8 min apart and plotted these data in relation to the rate of soma translocation (Fig. 3B Inset). Fig. 3A and Movie S3 show a representative example in which the Golgi apparatus remains at a stable distance from the nucleus during nucleokinesis. This type was more frequently associated with small somal translocation steps (3 ± 2 μm, mean ± SD; n = 281 pairs; Fig. 3B, blue columns). Less commonly, however, we also observed instances with an increase (Fig. 3B, magenta columns) or a decrease (Fig. 3B, green columns) in the distance between the Golgi and the nucleus. The former tended to occur during larger steps of somal translocation (6 ± 4 μm, mean ± SD; n = 68 pairs), whereas the latter occurred during small steps of somal translocation (3 ± 2 μm, mean ± SD; n = 37 pairs). Such nucleokinesis associated with changes in soma–Golgi distance is reminiscent of the classical mode of nucleokinesis during saltatory migration. These findings suggest that interneurons display at least two types of nucleokinesis in vivo, a conventional type associated with changes in the distance between the soma to the Golgi apparatus and an unconventional type in which the distance between them is stable.
Fig. 3.
Changes in Golgi apparatus localization before and after nucleokinesis. (A) Representative time-lapse sequence showing the localization of the Golgi apparatus in migrating neurons, labeled by GalT–EGFP (green) and GAP–tdTomato (magenta). Arrows point to the Golgi apparatus (white). Elapsed time is indicated at the upper left corners of each image (h:min). (Scale bar, 10 μm.) Neurons irrelevant to our analysis are covered by black rectangles. (B) Relationship between localization of the Golgi apparatus and soma displacement of migrating interneurons between two successive movie frames. In the case shown by the blue column, the Golgi apparatus does not change its position relative to the soma of migrating neurons (S→S); in the magenta column case, the Golgi apparatus changes its relative position from the distal to the proximal portion of migrating neurons (D→P), and in the green column case, translocation from the proximal to the distal portion of migrating neurons occurs (P→D). These are schematically shown in Inset. The Golgi apparatus is shown in yellow. Data were analyzed from 386 pairs of movie frames in 39 cells from three embryos. The interval of movie frames was 8 min. Average soma displacements between two successive movie frames in each case were 3 ± 2 μm, 6 ± 4 μm, and 3 ± 2 μm (mean ± SD) for S→S, D→P, and P→D, respectively. The ratio of these events was 73% for S→S, 18% for D→P, and 9% for P→D. (C and D) Schematic representations of the relationship between the nucleus and the Golgi/centrosome complex during nucleokinesis. Translocation of the Golgi/centrosome complex occurred mostly in synchrony with that of the nucleus (D1→D2→D3), although the former preceded the nucleus in some instances (C1→C2→C3).
Dynamics of LP extension and branching.
We observed growth cone-like swellings at the edges of LPs (Fig. 4A, arrowhead) and bifurcations of LPs (Fig. 4 A and C). LP growth cones most often advanced in a straight line without any notable turning (Fig. 4 A and B). This result was further confirmed by the value of growth path linearity being close to 1 (0.95 ± 0.01, mean ± SD; n = 25 processes, 25 cells, and 3 embryos), which was calculated by the sum of each step divided by the distance from the initial (indicated by a blue circle in Fig. 4B) and final points. These findings suggest that turning of LP growth cones may not be important for the navigation of migrating interneurons in vivo, unlike axonal growth cones (reviewed in ref. 27).
Fig. 4.
Dynamics of the LP and its branches. (A) Time-lapse sequence showing extension of growth-cone-like swellings of LP tips. Arrowheads indicate process tips. Neuronal processes irrelevant to our analysis are covered by black rectangles. (B) Track of the process tip indicated by the arrowheads in A. The blue circle illustrates the initial position of tracking. (C) Time-lapse sequence showing the dynamics of branch formation during migration. Arrowheads indicate the soma. Arrows indicate a retracting process. (D) Drawings depicting the branching pattern of the neuron shown in C. The soma translocated to the branching point and then moved into one branch, followed by retraction of the other. [Scale bars, 10 μm (A and B); 20 μm (C).]
Previous in vitro studies indicated that selection of a LP branch leads to changes in direction of neuronal migration (14, 16, 28). We found that the LP was usually branched either in the vicinity of or away from the soma (Fig. 4 C and D). In some cases, a branch of the LP formed secondary branches (Fig. 4 C and D). The nucleus usually moved to the branching point (Fig. 4C; t = 0:00–0:40), where it paused (t = 0:40–1:00; see below). Sometimes a complete retraction of a LP branch took place, a phenomenon associated with an extension of the other branch, eventually causing the nucleus to migrate to the direction of a remaining branch (Fig. 4C; t = 1:40–2:10; see also Fig. 5B). Collectively, these findings indicate that the selection of a LP branch, rather than the turning of a LP tip, is critical for the navigation of migrating interneurons in vivo.
Behavior of the nucleus and Golgi apparatus at the branching point of the LP.
Because the choice of the LP branch appears to be important for whether interneurons take a new migration direction, we observed the nucleus and Golgi apparatus in more detail at the branching point. Although on some occasions the nucleus entered one of the branches directly (Fig. 4C; t = 1:30), this entrance was sometimes delayed due to wavering behaviors of the nucleus (Fig. 5 A and B). In these instances, the nucleus paused at the branching point, exhibited a deformed shape, and then moved into one of the branches. In the example shown in Fig. 5A, the nucleus formed a budding into left branch of the LP (t = 0:00, arrowhead) and then took a swinging behavior. Thereafter, the major part of the nucleus stayed in the left branch (t = 2:10–2:40) until it suddenly moved into the right branch (t = 2:50–3:10), an action that was associated with the retraction of the LP branch initially chosen (Movie S4, arrow). One possibility is that the selection of a new LP branch might be instigated by the retraction of the other LP. However, this possibility, too, does not appear to be the case (Fig. 5B and Movie S5). In this example, the nucleus initially stayed at the LP bifurcating point, alternating between an elongated and spherical shape (t = 0:00–0:20). Then, the nucleus (arrowhead) entered one of the processes (t = 0:30) to resume nucleokinesis and display an elongated shape (t = 0:30–1:00). However, the LP that was not chosen (open arrowheads) retained its length (t = 0:30–040). Only ∼30 min later did it begin to clearly retract (t = 1:10). Once the nucleus entered into a LP completely, the neuron usually initiated migration in the direction of that LP (t = 2:10; n = 8). Thus, retraction of a LP branch might be a consequence of the selection of the other branch.
Furthermore, we found that the translocation of the Golgi apparatus into the LP branch preceded that of the nucleus in most cases (n = 14/16) (Fig. 5C and Movie S6). At time 0, the Golgi apparatus was localized at the leading head of the soma. When the soma translocated and reached the branching point (t = 1:20), the Golgi apparatus (arrowhead) moved into one of the branches ahead of the soma (t = 1:44). As the Golgi apparatus moved further into that branch (t = 2:16), the nucleus remained stationary at the branch point until the nucleus also began to move into that branch (t = 2:24). Retraction of the unchosen branch was observed even later (t = 3:12). Thus, the Golgi apparatus appears to pilot the nucleus to the LP branch.
Discussion
A number of studies have investigated the dynamics of neural migration, including those that have observed dissociated neurons on 2D substrates (1), neurons emigrating from an explant (2), and those migrating within a slice culture (14, 17, 29) or explant (15, 16, 21, 30). However, as informative as those studies are, their conclusions would not be supported if they were found to disagree with in vivo results. Although in vivo observations have been made in zebrafish (31–34), similar studies have proven far more difficult in mammals. One report showed in vivo analysis of interneuron migration in mouse embryos (35), but analyses in this report were of low resolution, unable to provide detailed description of the dynamics of individual neurons and their organelles. Therefore, the present study reveals detailed migratory behavior of mammalian neurons in vivo. In particular, we illuminate the dynamics of the LPs, nucleus and Golgi apparatus.
Multidirectional Migration.
Previously, we demonstrated that interneurons located in the MZ exhibit a peculiar type of migration, migration in all directions (15, 16, 21). The present study confirms that this type of neuronal migration occurs in vivo, too. We also found that several labeled interneurons occasionally changed their direction of migration. These findings are consistent with our previous study that demonstrated random-walk-like behavior of cortical interneurons in the MZ (16).
Nucleokinesis of Interneurons.
Currently, the following sequence of events is thought to be necessary for neural migration: (i) extension of the LP in the direction of migration, (ii) repositioning of the centrosome in the LP, and (iii) nucleokinesis through the LP (26). In this model, nucleokinesis is thought to depend on the centrosome, which links the microtubule-based pulling forces generated by the LP and the microtubule network surrounding the nucleus (8, 26, 36). Additionally, the centrosome, along with the Golgi, is frequently found within swellings of fixed neurons (3, 6, 37). These swellings have been seen to move forward in migrating neurons before nucleokinesis (3, 4, 38, 39), suggesting that cytoplasmic swellings associated with the Golgi/centrosome complex are involved in pulling the nucleus (3, 22).
Conversely, evidence suggesting other force-generating mechanism(s) for nucleokinesis is also accumulating. Studies using cerebellar granule cells have indicated that the LP myosin II near the nucleus may function to pull the centrosome and the soma (7), whereas another using dissociated granule cells showed evidence that the tip of the LP pulls the soma forward during neuronal migration via a myosin II-dependent forward F-actin flow (11). Other reports have suggested that the activity of the actomyosin system at the rear of the soma generates the forward force (3, 4, 9, 12).
We have found two types of dynamics for migrating interneurons in living embryos: (i) those that exhibit conventional saltatory nucleokinesis (Figs. 3C and 2B1) and (ii) those that exhibit a nonsaltatory continuous movement at a moderate rate (Figs. 3D and 2B2). In the first type, the distance between the Golgi and nucleus during active somal translocation decreases, whereas in the second, it remains relatively constant. Occurrence of two types of migration mode can be explained by assuming that the molecular mechanisms underlying these two might be different. The first type can be explained by assuming operation of pulling force by microtubules emanating from the centrosome. In contrast, given that the Golgi apparatus is associated with the centrosome (refs. 3, 37, 40, and 41; Fig. S2), the second type challenges any theory that postulates pulling forces generated by centrosome-nucleated microtubules within the LP predominantly drives the nucleus (26) and requires other mechanisms. Our results regarding centrosome localization are further supported by two in vitro studies that have shown that the centrosome is located near the nucleus in migrating cerebellar granule neurons (36, 42). Involvement of actomyosin system might be a plausible explanation.
The question of which of these alternative mechanisms might be predominantly used and thus observed may be highly dependent on context, such as the types of neurons and the physical environment through which these neurons are migrating. Our in vivo study raises the possibility that multiple mechanisms may coordinate in vivo, resulting in different modes of nucleokinesis observed in the population of cortical interneurons.
Changes in the Direction of Migration and Selection of a LP Branch by the Nucleus.
Migrating neurons observed in our preparation extended branched LPs. We have also found that migrating neurons change their direction of migration by choosing one of multiple branches rather than steering a single LP, suggesting that this selection is a critical step for determining the migratory direction (22). Similar behaviors of migrating neurons were observed in previous in vitro studies (14, 16, 28, 43). Our results indicate that these in vitro studies recapitulated genuine behavior of migrating neurons.
Dynamics of the Golgi Apparatus and Nucleus During Branch Selection.
The selection of a branch by the nucleus was associated with nuclear translocation into it and subsequent retraction of the unselected branch (Fig. 5 A, B, and D). The mechanism by which this choice occurs is poorly understood. One clue may come from nucleus swinging behavior at the branch point, which raises the possibility that the direction of migration is not predetermined at the time the nucleus arrives at the branching point. However, this idea is challenged by the behavior of the Golgi apparatus at the branching point. This behavior is because nucleokinesis at the selected branch was preceded by translocation of the Golgi apparatus into the branch (Fig. 5C). Furthermore, the Golgi apparatus did not show any swinging behavior, unlike the nucleus. These findings suggest that, although translocation of the nucleus may not always be under the control of the Golgi/centrosome as discussed in Nucleokinesis of Interneurons, but, rather, the movement of the Golgi/centrosome complex at the branch point appears to be a determinant step for branch selection.
How prior movement of Golgi/centrosome into a branch may influence the selection of that branch is still not clear. The fact that the nucleus swings between two branches at the branch point for some time suggested that initially the coupling between the nucleus and the Golgi/centrosome is not so tight and that the nucleus may be under multiple force influences. However, over a period after the Golgi/centrosome entering into a branch, the nucleus may eventually be “persuaded” to move into the selected branch. We speculate that the Golgi/centrosome complex may strengthen the pulling force emanating from the selected branch. It is also possible that the Golgi apparatus itself is directly involved in the selection of a new direction. This idea is supported by a recent study that showed inhibition of the expression of golgin 160 or GMAP210, which altered Golgi positioning without disrupting secretion or actin and microtubule networks, compromised HeLa cell polarization (44). Further studies are needed to validate this hypothesis.
In conclusion, we successfully visualized the dynamics of migrating cortical interneurons and their organelles using living mouse embryos. During migration, the Golgi apparatus tended to be localized near the front end of the nucleus, and its dynamics in relation to that of the nucleus suggested occurrence of two types of nucleokinesis: Golgi/centrosome-dependent and -independent. Interneurons changed the direction of migration by choosing one of the branched LPs. This process was led by translocation of the Golgi apparatus, which was followed by that of the nucleus. Our results provide important insights into the cellular mechanisms underlying neuronal migration.
Methods
Animals.
ICR mice (Japan SLC) were used. Noon on the day of vaginal plug detection was termed embryonic day (E) 0.5. All experiments followed the institutional guidelines.
In Vivo Time-Lapse Imaging.
In vivo time-lapse imaging was performed by using a confocal or a two-photon microscope through the skulls of embryos with the umbilical cord attached.
Further experimental details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Y. Komai, S. Nakamura, A. Sakakibara, and K. Kitamura for technical advice; J. Miyazaki, K. Nishida, R. Y. Tsien, F. Matsuzaki, Y. Tanabe, Mr. T. Kobayashi, and Mr. Y. Furukawa for constructs; and Drs. Y. Tanabe, H. Kobayashi, S. C. Fujita, and P. Karagiannis for critical reading of this manuscript. This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI, 22220004) from the Ministry of Education, Culture, Sports, Science and Technology (Japan).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209166109/-/DCSupplemental.
References
- 1.Hatten ME, Liem RK, Mason CA. Two forms of cerebellar glial cells interact differently with neurons in vitro. J Cell Biol. 1984;98:193–204. doi: 10.1083/jcb.98.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wichterle H, Garcia-Verdugo JM, Alvarez-Buylla A. Direct evidence for homotypic, glia-independent neuronal migration. Neuron. 1997;18:779–791. doi: 10.1016/s0896-6273(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 3.Bellion A, Baudoin JP, Alvarez C, Bornens M, Métin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: Forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci USA. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Anda FC, Meletis K, Ge X, Rei D, Tsai LH. Centrosome motility is essential for initial axon formation in the neocortex. J Neurosci. 2010;30:10391–10406. doi: 10.1523/JNEUROSCI.0381-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higginbotham H, Tanaka T, Brinkman BC, Gleeson JG. GSK3beta and PKCzeta function in centrosome localization and process stabilization during Slit-mediated neuronal repolarization. Mol Cell Neurosci. 2006;32:118–132. doi: 10.1016/j.mcn.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Solecki DJ, et al. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T, et al. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 10.Komuro H, Rakic P. Intracellular Ca2+ fluctuations modulate the rate of neuronal migration. Neuron. 1996;17:275–285. doi: 10.1016/s0896-6273(00)80159-2. [DOI] [PubMed] [Google Scholar]
- 11.He M, Zhang ZH, Guan CB, Xia D, Yuan XB. Leading tip drives soma translocation via forward F-actin flow during neuronal migration. J Neurosci. 2010;30:10885–10898. doi: 10.1523/JNEUROSCI.0240-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martini FJ, Valdeolmillos M. Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J Neurosci. 2010;30:8660–8670. doi: 10.1523/JNEUROSCI.1962-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- 14.Martini FJ, et al. Biased selection of leading process branches mediates chemotaxis during tangential neuronal migration. Development. 2009;136:41–50. doi: 10.1242/dev.025502. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka DH, Maekawa K, Yanagawa Y, Obata K, Murakami F. Multidirectional and multizonal tangential migration of GABAergic interneurons in the developing cerebral cortex. Development. 2006;133:2167–2176. doi: 10.1242/dev.02382. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka DH, et al. Random walk behavior of migrating cortical interneurons in the marginal zone: Time-lapse analysis in flat-mount cortex. J Neurosci. 2009;29:1300–1311. doi: 10.1523/JNEUROSCI.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe H, Murakami F. Real time analysis of pontine neurons during initial stages of nucleogenesis. Neurosci Res. 2009;64:20–29. doi: 10.1016/j.neures.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Marín O, Rubenstein JLR. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 19.Métin C, Baudoin JP, Rakić S, Parnavelas JG. Cell and molecular mechanisms involved in the migration of cortical interneurons. Eur J Neurosci. 2006;23:894–900. doi: 10.1111/j.1460-9568.2006.04630.x. [DOI] [PubMed] [Google Scholar]
- 20.Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka D, Nakaya Y, Yanagawa Y, Obata K, Murakami F. Multimodal tangential migration of neocortical GABAergic neurons independent of GPI-anchored proteins. Development. 2003;130:5803–5813. doi: 10.1242/dev.00825. [DOI] [PubMed] [Google Scholar]
- 22.Valiente M, Marín O. Neuronal migration mechanisms in development and disease. Curr Opin Neurobiol. 2010;20:68–78. doi: 10.1016/j.conb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Edmondson JC, Hatten ME. Glial-guided granule neuron migration video microscopic study in vitro: A high-resolution time-lapse video microscopic study. J Neurosci. 1987;6:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komuro H, Rakic P. Dynamics of granule cell migration: A confocal microscopic study in acute cerebellar slice preparations. J Neurosci. 1995;15:1110–1120. doi: 10.1523/JNEUROSCI.15-02-01110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukata M, Nakagawa M, Kaibuchi K. Roles of rho-family GTPases in cell polarisation and directional migration. Curr Opin Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 26.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 28.Ward ME, Jiang H, Rao Y. Regulated formation and selection of neuronal processes underlie directional guidance of neuronal migration. Mol Cell Neurosci. 2005;30:378–387. doi: 10.1016/j.mcn.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 29.O’Rourke NA, Dailey ME, Smith SJ, McConnell SK. Diverse migratory pathways in the developing cerebral cortex. Science. 1992;258:299–302. doi: 10.1126/science.1411527. [DOI] [PubMed] [Google Scholar]
- 30.Ang ES, Jr, Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J Neurosci. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Distel M, Hocking JC, Volkmann K, Köster RW. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol. 2010;191:875–890. doi: 10.1083/jcb.201004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köster RW, Fraser SE. Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr Biol. 2001;11:1858–1863. doi: 10.1016/s0960-9822(01)00585-1. [DOI] [PubMed] [Google Scholar]
- 33.Köster RW, Fraser SE. FGF signaling mediates regeneration of the differentiating cerebellum through repatterning of the anterior hindbrain and reinitiation of neuronal migration. J Neurosci. 2006;26:7293–7304. doi: 10.1523/JNEUROSCI.0095-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohata S, et al. Neuroepithelial cells require fucosylated glycans to guide the migration of vagus motor neuron progenitors in the developing zebrafish hindbrain. Development. 2009;136:1653–1663. doi: 10.1242/dev.033290. [DOI] [PubMed] [Google Scholar]
- 35.Yokota Y, et al. Radial glial dependent and independent dynamics of interneuronal migration in the developing cerebral cortex. PLoS ONE. 2007;2:e794. doi: 10.1371/journal.pone.0000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi K, Kawai-Hirai R, Harada A, Takata K. Inhibitory neurons from fetal rat cerebral cortex exert delayed axon formation and active migration in vitro. J Cell Sci. 2003;116:4419–4428. doi: 10.1242/jcs.00762. [DOI] [PubMed] [Google Scholar]
- 38.Kappeler C, et al. Branching and nucleokinesis defects in migrating interneurons derived from doublecortin knockout mice. Hum Mol Genet. 2006;15:1387–1400. doi: 10.1093/hmg/ddl062. [DOI] [PubMed] [Google Scholar]
- 39.Shinohara R, et al. A role for mDia, a Rho-regulated actin nucleator, in tangential migration of interneuron precursors. Nat Neurosci. 2012;15:373–380, S1–S2. doi: 10.1038/nn.3020. [DOI] [PubMed] [Google Scholar]
- 40.Bisel B, et al. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rios RM, Bornens M. The Golgi apparatus at the cell centre. Curr Opin Cell Biol. 2003;15:60–66. doi: 10.1016/s0955-0674(02)00013-3. [DOI] [PubMed] [Google Scholar]
- 42.Umeshima H, Hirano T, Kengaku M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc Natl Acad Sci USA. 2007;104:16182–16187. doi: 10.1073/pnas.0708047104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lysko DE, Putt M, Golden JA. SDF1 regulates leading process branching and speed of migrating interneurons. J Neurosci. 2011;31:1739–1745. doi: 10.1523/JNEUROSCI.3118-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.