Abstract
Cells respond to changes in environment by shifting their gene expression profile to deal with the new conditions. The cellular response to changes in metal homeostasis is an important example of this. Transition metals such as iron, zinc, and copper are essential micronutrients but other metals such as cadmium are simply toxic. The cell must maintain metal concentrations in a window that supports efficient metabolic function but must also protect against the damaging effects of high concentrations of these metals. One way a cell regulates metal homeostasis is to control genes involved in metal mobilization and storage. Much of this regulation occurs at the level of transcription and the protein most responsible for this is the conserved metal responsive transcription factor 1 (MTF-1). Interestingly, the nature of the changes in the gene expression profile depends on the type of exposure. The cell somehow senses the kind of the metal challenge and responds appropriately. We have been using the Drosophila system to try to understand the mechanism of this metal discrimination. Using genome-wide mapping of MTF-1 binding under different metal stresses we find that, surprisingly, MTF-1 chooses different DNA binding sites depending on the specific nature of the metal insult. We also find that the type of binding site chosen is an important component of the capability to induce the metal-specific transcription activation.
Keywords: heavy metal, MED26
DNA recognition by sequence-specific DNA binding transcription factors is the basic mechanism that links the cis-regulatory information encoded in the genome with transcriptional control of gene expression. Although it is the DNA binding domain of these factors that dictates the sequence element bound by the protein, it is now clear that, at least for some factors, the DNA sequence of the binding site itself can influence the activation potential of the transcription factor (1, 2). Clearly the process of reading the genome and activating transcription is more intricate than a simple DNA binding event.
Some transcription factors respond to signaling events and activate the transcription of different sets of genes in a signal-specific manner. The cellular response to changes in metal homeostasis is one example of this observation. The major transcription factor involved in metal homeostasis is the metal responsive element (MRE) binding transcription factor-1 (MTF-1) (for a recent review, see ref. 3). MTF-1 is required for both constitutive and metal inducible transcription of some genes; it is also required for activated transcription of at least one gene in low metal conditions (4, 5). Thus, it responds to both excess and limiting metal conditions by activating different sets of genes. MTF-1 contains a single DNA binding domain consisting of six zinc fingers and it is structurally and functionally conserved from Drosophila to humans (6, 7). Cross-species comparison between human and Drosophila MTF-1 reveals considerable identity in the DNA binding domain (66% amino acid identity) indicating that the proteins likely interact with DNA in a conserved mode (8, 9).
One class of conserved MTF-1 targets is the metallothionein proteins (MT), a family of small, cysteine-rich proteins that are thought to sequester excess metals. They function as metal homeostasis regulators for both essential and toxic metals (10–12). MT knockout mice are viable but sensitive to cadmium (Cd) and, to a lesser degree, copper (Cu), zinc (Zn), and mercury (13). By contrast, the MTF-1 knockout is embryonic lethal, indicating that MTF-1 is important for the transcription of targets other than the MTs.
An interesting aspect of MT regulation is that the level of expression varies depending on the specific metal insult (14, 15). As an example, in Drosophila cells, exposure to heavy metals results in increased expression of the MT genes MtnA and MtnB. However, MtnA expression is stimulated more effectively by Cu than by Cd. By contrast, MtnB expression is stimulated more by Cd than by Cu (14, 16). The same type of metal-specific activation occurs in mammalian cells and likely reflects a fundamental aspect of the metal discrimination system (17–19). This selective induction mirrors the protective effects of the genes. MtnA is more protective against Cu exposure, and MtnB is more protective against Cd (14). Similar selective protective effects are evident from human toxicity data where mutations in the MTIIA gene increase susceptibility to Cd toxicity, but not Zn or Cu (20, 21).
An important DNA sequence element that stimulates transcription of nearby genes during metal shock is the MRE. The MRE was initially identified in the mouse MT gene promoters (22). A consensus sequence for the central core of this element (TGCRCNC) has been identified by examination of a number of known metal-responsive genes (3). A search for the factor that directly binds the MRE in a sequence-specific manner identified MTF-1.
The current model of metal-induced transcriptional regulation by MTF-1 suggests that excess metals displace Zn from MT (23). This free Zn fills zinc finger(s) within the DNA binding domain of cytosolic MTF-1 (24, 25). MTF-1 then moves to the nucleus where it binds to MRE sequences and activates transcription.
This model can explain some aspects of MTF-1 regulation. However, in addition to the metal-induced activation, MTF-1 also activates the transcription of genes required under low Cu concentrations and is required for basal transcription of MTF-1 regulated genes in vivo (26). Furthermore, a significant amount of MTF-1 can be found in the nucleus at target promoters in the absence of metal (16, 27). In addition, the cell, using MTF-1, is capable of tuning the transcriptional activation of metal responsive genes depending on the specific metal encountered. These findings indicate that there must be additional levels of regulation, specifically those allowing for variation in gene activation based on the stimulating metal.
Here we report the use of chromatin immunoprecipitation coupled with Drosophila genomic tiling arrays (ChIP-chip) to identify genomic regions directly bound by MTF-1 in untreated and metal (Cu and Cd) challenged cells. Surprisingly, we find MTF-1 at a large set of nonoverlapping binding sites under each of these conditions. We find that MTF-1 has a unique binding site preference that depends on the specific type of metal treatment. We identified MRE consensus sequences for both Cu and Cd preferred sites. Our results show that both sequences are similar to published core MRE sequences; however, there is a single metal-specific variable nucleotide. Reporter assays using the identified sequences confirmed metal-specific activation by these sites. Furthermore, changing the variable nucleotide in the Cd derived sequence negates the activation preference. This work reveals MTF-1 binding site preference is influenced by the specific metal insult and can result in a unique response based on slight changes in MTF-1 binding sequence preference.
Results
Initial Assay Conditions.
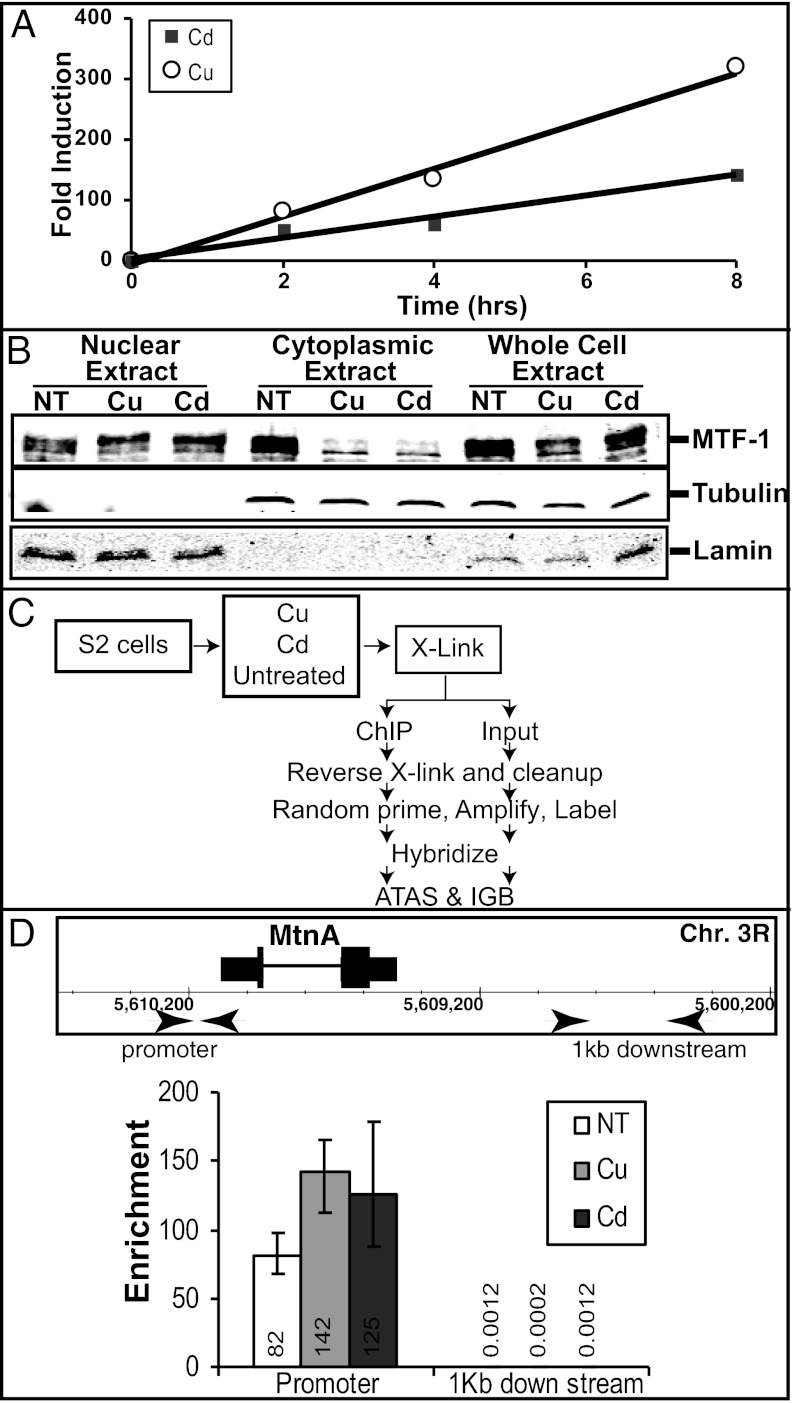
To determine the linear range of metal-dependant transcription activation in the Drosophila S2 cells used in our experiments, we treated the cells with Cu or Cd for increasing amounts of time. The amount of mature MtnA mRNA was measured by quantitative RT-PCR (RT-qPCR), and the fold activation was determined by comparison with a housekeeping gene (Rp49). As previously observed, both Cu and Cd induce MtnA expression several hundred-fold, although Cu is the more potent activator (Fig. 1A) (7). MtnA has a linear range of activation up to at least 8 h after treatment with either Cu or Cd. In addition, the majority of MTF-1 has moved to the nucleus in 4 h under either Cu or Cd induction (Fig. 1B). Due to the maximum translocation of MTF-1 and dynamic range of expression, the 4-h time point was chosen for subsequent assays designed to identify MTF-1 targets.
Fig. 1.
Initial assay conditions. (A) MtnA expression in Drosophila S2 cells following treatment by Cu or Cd measured by RT-qPCR. Error bars are too small to visualize on the graph, largest error is ± 8. (B) Western blot of extracts prepared from Drosophila S2 cells; cells were nontreated (NT), Cu treated (Cu), or Cd treated (Cd) for 4 h. Tubulin and lamin are loading and separation controls. Multiple bands of MTF-1 are detected because of phosphorylation (33). (C) Diagram of processing for analysis. (ATAS, Affymetrix Tiling Analysis software; IGB, Integrated Genome Browser). (D) Enrichment, relative to rRNA gene, of MTF-1 at the MtnA promoter and a region 1 kb downstream. A map of the region is shown at top with primers indicated by arrows.
MTF-1 binding sites were identified by immunoprecipitation of crosslinked genomic DNA fragments from S2 cells following Cu or Cd treatment as well as untreated cells (Fig. 1C). We analyzed the binding of MTF-1 to the MtnA promoter region to evaluate the assay. Using quantitative PCR (qPCR), we find >100-fold enrichment for the precipitated samples under metal conditions at the MtnA promoter (Fig. 1D). A control region 1 kb downstream exhibited no enrichment after IP (Fig. 1D). In agreement with previous work, there is only a twofold increase in signal from the MtnA promoter region after metal treatment (16). This finding is consistent with the notion that MTF-1 is bound to this region in untreated cells. These results indicate that our MTF-1 antibodies are capable of precipitating MTF-1 when it is crosslinked to DNA providing high levels of enrichment with low background from adjacent DNA.
MTF-1 Binds Distinct Regions of Known Targets Depending on Metal Treatment.
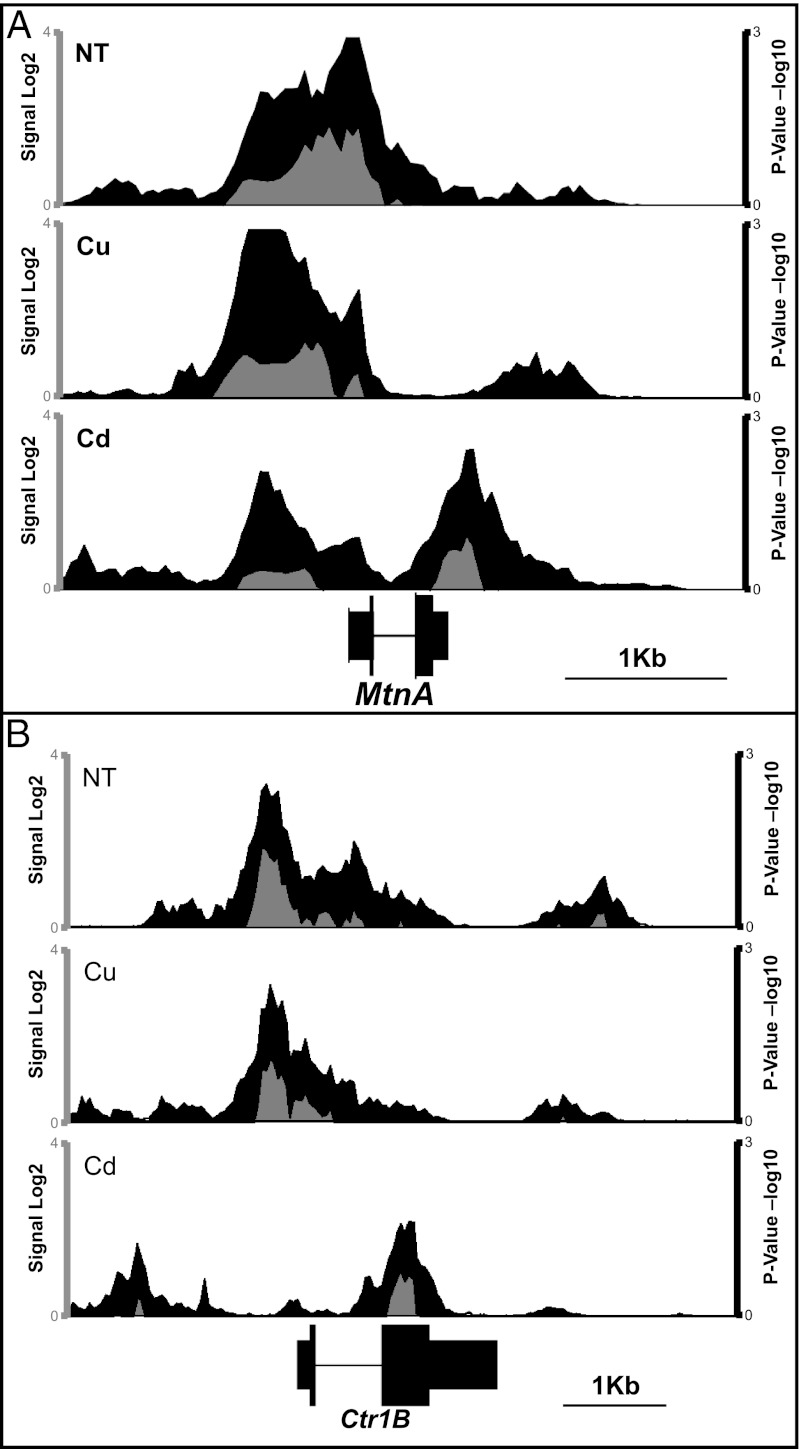
Having established the assay conditions, Affymetrix Drosophila tiling arrays were used to identify regions bound by MTF-1 genome-wide. Triplicate data sets for each condition were combined and analyzed using the Affymetrix Tiling Analysis software. Labeled input DNA was used as a control. MTF-1 binding sites were defined as regions enriched over the input samples. To access the validity of the approach, the MTF-1 binding profiles for known targets were visually evaluated using the Integrated Genome Browser (IGB) (28). MTF-1 binding is detected at the promoter regions of MtnA and Ctr1B substantiating the tiling array results (Fig. 2 A and B). As observed by our laboratory and others, MTF-1 is present in both the untreated and metal treated conditions (16) (Fig. 1D).
Fig. 2.
MTF-1 binding at MtnA and Ctr1b. (A) IGB view of the region surrounding the MtnA gene with signal and P value plotted. Signal is shown in gray, and P value is shown in black. x-axis is local genomic coordinates in bases. (Scale bar: 1 kb.) The y axis is log2 for signal (Left) and −log10 for P value (Right). (B) Region surrounding the Ctr1b gene with signal and P value analysis plotted as in A.
Surprisingly, in addition to the binding in the MtnA promoter region, MTF-1 is bound further downstream specifically in the Cd treated samples. A similar pattern is observed at Ctr1B (4). MTF-1 is present in the Ctr1B promoter in untreated and Cu-treated samples, but the pattern of binding is changed with binding observed further downstream under Cd treatment (Fig. 2B). These observations indicate that MTF-1 is capable of binding to different regions of the same gene depending on which metal stimulates MTF-1.
Identification of MTF-1 Binding Sites and Associated Genes Genome-Wide.
The binding observed at MtnA and Ctr1B represent the top 1% of signal detected on the microarrays. Using this benchmark, we used the top 1% signal from each set of arrays to identify MTF-1 binding sites under these conditions. The analysis identified 2,026 sites bound by MTF-1 under Cu treatment, 1,629 under Cd treatment, and 1,404 in untreated cells. The overlap between these three regions is small. There are 143 sites bound in both Cu and Cd treatment, 115 sites bound in both Cd-treated and untreated cells, and 260 sites bound in both Cu-treated and untreated cells. There are only 34 sites that have overlap under all three conditions (Dataset S1).
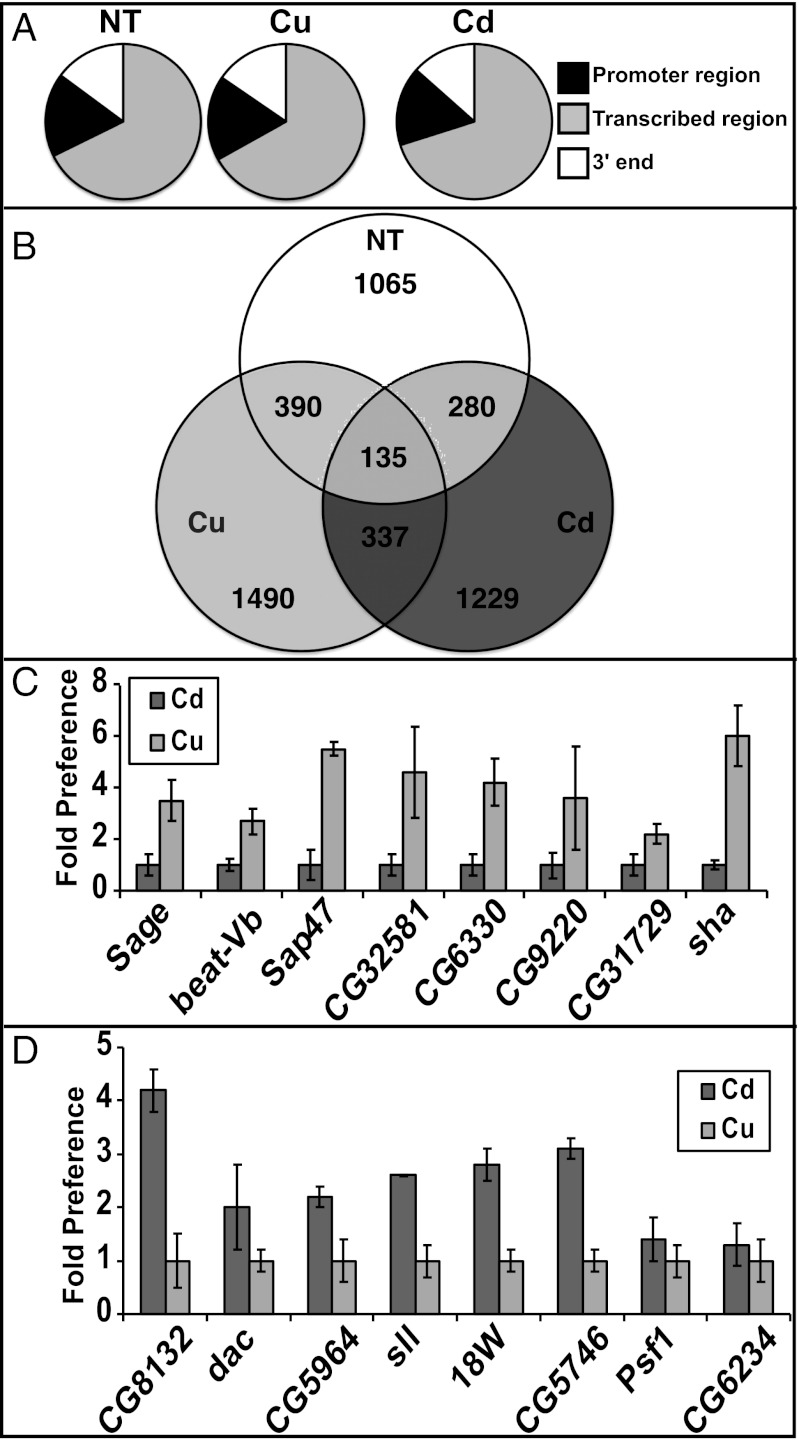
Despite the difference in genomic position of binding sites, the distribution relative to the transcribed unit remained similar in all three conditions (Fig. 3A). The majority of the sites (70%) were in regions annotated as transcribed units. A fraction of the sites (17%) were found upstream of transcribed units, presumably in the proximal promoter. The remaining sites were found in the region downstream of the transcribed unit.
Fig. 3.
Genome-wide characterization of MTF-1 binding sites. (A) Charts showing distribution of binding sites relative to local genes. Black indicates promoter bound MTF-1, and gray indicates transcribed region, and white indicates 3′ end of the gene. (B) Venn diagram of the number of genes within 250 bases of MTF-1 in no treatment, Cu treatment, or Cd treatment. (C) Enrichment levels of sites predicted to be preferentially bound under Cu. In each case, the data are plotted relative to the nonpreferred condition. (D) As in C, but plotted for Cd-preferred sites.
To identify genes associated with these sites, we extended the regions defined above by 250 bp on each side and screened for overlap with known genes (Dataset S1). Using these parameters, there are 1,490 genes bound under Cu treatment, 1,229 under Cd treatment, and 1,065 in untreated cells. Interestingly, only 135 genes are present in all three data sets (Fig. 3B). This finding indicates that, despite the fact MTF-1 has a single DNA binding domain, MTF-1 has metal-specific preferences for binding.
Validation of MTF-1 Preferential Binding by Quantitative-PCR.
Eight sites bound by MTF-1 in a metal-specific manner were selected from each condition for direct qPCR assay to validate the differential binding seen in the array data. Sites preferentially bound by MTF-1 in the Cu data are between two- and sixfold more enriched for MTF-1 binding under Cu treatment (Fig. 3C). Six sites preferentially bound by MTF-1 in Cd conditions are between two- and fivefold enriched for MTF-1 binding under Cd treatment. The other two genes from the Cd set were also enriched in MTF-1 binding when treated with Cd, but by only 30–50% (Fig. 3D). The results of the direct qPCR analysis of the Cd- and Cu-specific genes agree with the genome-wide profiles, substantiating the existence of metal-induced preferential MTF-1 binding sites.
Analysis of Cu- and Cd-Induced MTF-1 Binding Sequence Preference.
Our microarrays indicate that there are specific sets of sites bound by MTF-1 under different metal insults. To determine whether the Cu or Cd treatment leads to an alteration in the sequence-specific binding preference of MTF-1, the regions bound under Cu or under Cd treatment by MTF-1 were pooled into separate data sets for analysis. To identify any sequence motifs that are overrepresented in each dataset, we calculated a ratio of significance. This ratio is the frequency of a specific sequence within the datasets divided by the frequency of the same sequence throughout the entire genome. A high ratio of significance means that a sequence of DNA appears preferentially within our MTF-1 data sets compared with the entire genome.
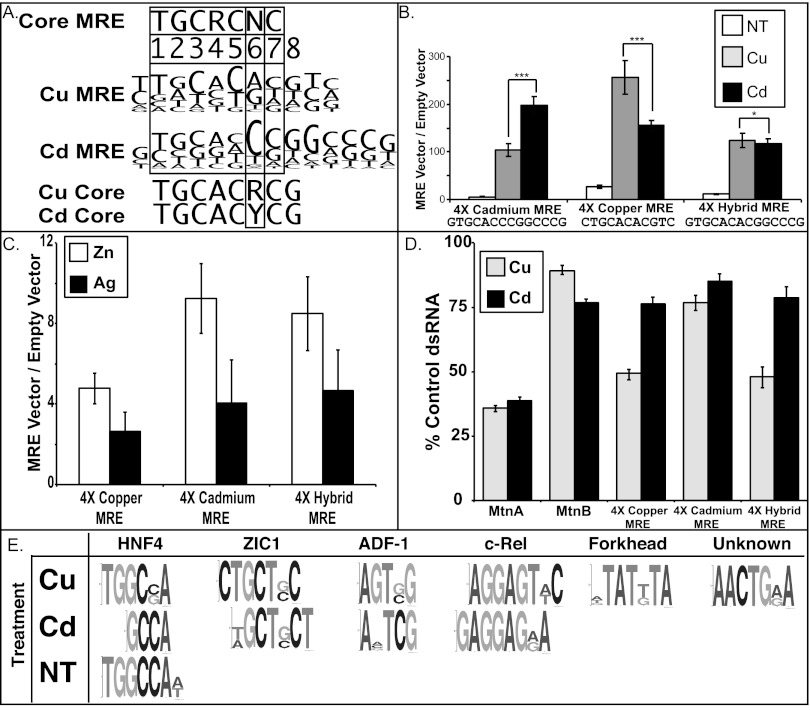
We calculated the ratio of significance for all 7-, 8-, 9-, and 10-bp-length sequences. Alignments of the top 100 sequences in the 262,144 total sequences for the 9-bp set and 1,048,576 sequences for the 10-bp set produced a consensus sequence similar to the established MRE core sequence (TGCRCNC) (Fig. 4A). In addition, the new alignments have a conserved G in position 8. Once separated into Cu- and Cd-preferred binding sites, the nonspecific nucleotide in the previously defined MRE consensus core exhibits specificity. It appears that a pyrimidine is preferred for Cd, and a purine is preferred for Cu (Fig. 4A). This finding indicates a bias to MTF-1 binding under specific metal inductions.
Fig. 4.
Sequence preference based on ratio of significance alignment. (A) Cu MRE and Cd MRE from ratio of significance. Previously defined MRE consensus is shown at the top for comparison. (B) Metal-specific MREs derived above tested using a firefly luciferase reporter assay. Luciferase levels are normalized to an empty vector containing only the minimal promoter. ***, statistically different (P < 1 × 10−7); *, not statistically different P > 0.1. (C) The MRE-containing reporters treated with Zn or Ag and assayed for differential response to these metals. (D) The MRE-containing reporters were induced by Cu or Cd in cells that were treated with dsRNA against lacI or MED26. The data are plotted as response in the MED26 cells divided by the response in control dsRNA (lacI). (E) DREME identified motifs for Cu-treated, Cd-treated, and nontreated MTF-1–bound regions. Association across datasets is based on a similarity greater than 60% using the Motif Alignment and Search Tool (MAST). Possible DNA binding factor based on a TOMTOM sequence alignment (Upper).
In addition to the core binding sequence for Cu, the alignment of top significant sequences identified three favored flanking nucleotides: one upstream and two downstream of the core. The Cd sequence alignment also generated additional flanking nucleotides downstream of the binding site, consisting of a cluster of Gs and Cs. Combined with the previously derived MRE core, these results indicate that there may be key differences in the MRE flanking sequences as well as the sixth nucleotide position that was previously thought to contain little or no sequence preference.
Newly Identified Motifs Confer Metal-Specific Transcription Activation.
To confirm the metal-specific nature of our defined Cu and Cd sequences, we created transcriptional reporters containing the Cu or Cd defined sequences. Four direct repeats of the Cu- and Cd-specific sequences were inserted upstream of a minimal Adh promoter driving firefly luciferase. The reporter constructs were transfected into S2 cells and challenged with Cu or Cd. The response of the reporter to metal was defined as a ratio of the Cu or Cd reporter construct to a construct containing only the Adh core promoter (Fig. 4B). Comparison between Cd and Cu induction levels shows the metal-induced luciferase levels for the Cd and Cu sequence reporters correspond to the metal-induced sequence preference defined from the MTF-1 ChIP.
To determine the importance of the variable nucleotide identified in the MRE core defined in this work, we created an additional reporter construct containing a hybrid Cd-specific sequence with a purine instead of a pyrimidine at the critical nucleotide. Comparisons between the original Cd sequence and the hybrid sequence indicate a loss of metal discrimination (Fig. 4B). The change does not generate the strong Cu preference seen with the sequence identified in the Cu dataset, but it eliminates the metal induction preference, substantiating the idea that the variable nucleotide in the MRE core plays a key role in the metal-specificity of the genes regulated by MTF-1.
To determine whether the motifs confer specificity for other metal ions, we tested Zn and Ag in our transient transfection assay. Although the metals weakly induce the reporters, there is no difference in metal specificity with either of the motifs (Fig. 4C). This result indicates that the motifs act as generic MREs but provide specificity for Cu or Cd.
Single Nucleotide Change Dictates Coactivator Requirements.
Previously, we showed the requirement for individual subunits of the mediator coactivator complex in response to Cu stimulation varied between MtnA and MtnB (16). One of the subunits, MED26, was required for full activation of the endogenous MtnA promoter but not for MtnB. To investigate the role of the MRE sequence in coactivator requirements, we depleted MED26 from cells with double-stranded RNA (dsRNA) directed against MED26. We then compared the response of our reporters in cells treated with a control dsRNA to the response in the MED26-depleted cells. The data are plotted as percent response relative to control treatment in Fig. 4D. We also tested the full length MtnA and MtnB reporters. In agreement with our previous data, MtnA had a strong requirement for MED26 for Cu activation, whereas MtnB was resistant to the depletion of MED26 (16). The same MED26 requirements were observed for Cd activation. This result indicates that the system is responding appropriately. All three synthetic MREs were equally resistant to MED26 depletion in Cd treatment. Differences were apparent when we treated with Cu. The Cu-derived MRE required MED26 for full response, whereas the Cd-derived MRE had a lower requirement for this subunit. Interestingly, the mutated Cd MRE, containing a single nucleotide change, now had a requirement for MED26 that paralleled the Cu-derived MRE. This finding indicates that small changes in MRE sequence can change coactivator requirements for activation.
Discovering Motifs Associated with the ChIP-Chip Datasets Using DREME.
In addition to binding site preference, sequence-specific DNA binding proteins often partner with DNA binding proteins to change gene expression profiles. In an attempt to identify possible MTF-1 partners we analyzed the data sets of the Cu, Cd, and nontreated sites by DREME, the Discriminative DNA Motif Discovery tool. To identify only significant motifs that appear within our datasets, the analysis limits were set to an E-value ≤ 0.01. This analysis resulted in the identification of a single enriched motif common to all three datasets. Using a motif comparison tool, the motif was similar to an Hnf4 binding site. Additional motifs were identified in the Cu- and Cd-bound datasets (Fig. 4E). All identified motifs had P values ≤ 10−6. Motif comparison of sites identified in treated cells suggest the sites resemble Zic1-, Adf1-, and c-Rel-binding sites. Of the two Cu-specific sites, one shows no similarity to known factor binding sites and one aligns with a forkhead binding motif.
Discussion
In order for a cell to mount an appropriate response to environmental insults, there must be a mechanism for discrimination of the nature of the insult. Cells that are exposed to transition metals mount a suitable metal-specific response. Our analysis shows that the genes bound by MTF-1 are largely metal specific (Fig. 3B). The selectivity of binding appears to include a single nucleotide difference within the core MRE. What was previously thought to be a variable nucleotide shows a preference for a C or T under Cd treatment and a G or A under Cu exposure. Our reporter assays show that this sequence variation alters the transcription activation potential of MTF-1 in a metal dependent manner. Changing this nucleotide was enough to abolish metal-specific transcription activation without activation per se. This finding suggests that the way genes respond to the presence of different metals can vary significantly with MRE sequence.
The ability of a sequence-specific DNA binding protein to have variant sequence affinity in response to alternative ligands is not unprecedented. Other transcription factors, such as nuclear hormone receptors, have been shown to exhibit binding site diversity that dictates the fine-tuning of transcriptional activation and interactions with ligands and cofactors (2, 29). The basis for this selective interaction appears to result from conformational changes in the protein. Estrogen receptor binds a diverse set of sites and, based on the sequence of that site, interacts selectively with cofactors (29). Likewise, glucocorticoid receptor transcriptional output and structure are influenced by the exact sequence of the bound glucocorticoid response element (2, 30). Previous work on nuclear receptors used a combination of natural and synthetic ligands. The work presented here is unique in that it shows natural ligands (i.e., Cu and Cd) changing the activation potential and binding site preference of a sequence-specific DNA binding protein.
The activation domain of MTF-1 also has metal-specific conformations (27). There are two distinct regions within the activation domain of the protein that are differentially required for Cu and Cd transcription activation (27, 31–33). Protease accessibility of the MTF-1 activation domain in vitro show different patterns in the presence of Cu and Cd, diagnostic of a structural difference under each metal insult (27). Taking into account the alteration in MTF-1 structure under metal treatment, the metal-specific binding site preferences and the differential coactivator requirement shown here, MTF-1 may behave much like the nuclear receptor family in its mechanisms of transcription activation.
An allosteric change in MTF-1 modifies its activation potential dependent on the nature of the metal treatment. In previous work designed to look at the cofactor requirements of MTF-1 under Cu treatment it was found that MTF-1 has a promoter-specific requirement for the repertoire of coactivators needed for efficient transcription activation (16). This could reflect differences in the MREs associated with each of the promoters tested. Perhaps each metal-specific conformation of MTF-1 requires a different set of coactivators for transcription activation.
Motif analysis of the MTF-1–bound sites reveals additional DNA motifs associated with all three conditions. If subsets of genes have MRE sequences preferred by MTF-1 in Cu or Cd treatment, or if there are alternative MREs within the same gene, as appears to be the case for MtnA and Ctr1b, then there may be specific DNA binding factors associated with each of the different MRE variants. These factors could influence the selective response under different metal conditions, providing an additional level of metal-specific control. We are currently investigating this possibility.
Materials and Methods
Time Course of MtnA Expression Following Cu and Cd Induction.
Drosophila S2 cells were 80% confluent before inducing with Cu and Cd separately. At indicated time points, cells were harvested, and RNA isolated by trizol. cDNA was prepared using the SuperScript III First-Strand Synthesis protocol and random hexamers. RNA levels were measured by qPCR with the Promega GoTaq system. Changes in MtnA were determined relative to the housekeeping gene RP49.
Chromatin Immunoprecipitation Conditions.
One six-well plate with 2 × 106 S2 cells per well were used for each condition (nontreated, Cu treated, or Cd treated). After cells adhered, media was aspirated and 3 mL of fresh media with 500 µM Cu, 50 µM Cd, or water (NT) was added. After 4 h, ChIP was carried out essentially as described (16). RNA samples were taken in parallel and processed as described above. MtnA induction was confirmed by qPCR in both the Cu- and Cd-induced cells. Fold enrichment in the pilot experiments was determined relative to the 28s rRNA gene.
Array Hybridization and Analysis.
Input and precipitated DNA were amplified as described (34) and then biotin labeled (35). Labeled samples were hybridized to the Affymetrix GeneChip Drosophila Tiling 2.0R Array and processed using the Affymetrix Hybridization Protocol. Three replicates were completed for each condition. Profiles for each replicate were combined using the Affymetrix Tiling analysis software. Combined input arrays were used to set the background signal intensities for the ChIP arrays. The Integrated Genome Browser (28) was used to establish a binding threshold and peak distribution for MTF-1 binding sites based on the binding profile at the well characterized MtnA gene locus. The parameters used to define the top 1% of the signal for untreated and Cu-treated were a run length of 150 and a gap of 70. For Cd-treated samples, the parameters were run length of 100 and gap length of 180. All data sets are available in the Gene Expression Omnibus (GEO) database.
Sites bound by dMTF-1 were categorized as either promoter region, transcribed region or 3′ end region. Transcribed regions were defined by the D. melanogaster NCBI RNA reference sequence collection. Promoter region was defined as 1 kb upstream of the transcriptional start site, and 3′ end was defined as 1 kb downstream of the polyadenylation site. Motif analysis was performed using the MEME-ChIP suite (http://meme.nbcr.net/meme/intro.html) (36).
Ratio of Significance and Sequence Alignment.
Ratio of Significance = (Frequency of n-mer sequences within the regions of interest/Length of region)/(Frequency of n-mer sequences within the entire genome/Length of genome). All possible 7-, 8-, 9-, and 10-bp sequences were tested. The top 100 sequences, based on their ratio of significance, were aligned using ClustalW version 2 (37) and visualized using WebLogo (38) to establish the Cu and Cd sequences.
Reporter Assays.
Four direct repeats of the metal-specific MRE sequences were inserted upstream of an alcohol dehydrogenase core promoter driving firefly luciferase (39). MRE reporter constructs and an Actin 5C renilla luciferase control vector were transfected into Drosophila S2 cells (5 × 105 cells per well, 24-well plate) using effectene (Qiagen). Twenty-four hours later, the cells were treated with 500 µM CuSO4, 50 µM CdCl2, 2 mM ZnCl2, 50 µM AgNO3, or water (NT) and incubated for 4 h. Expression levels were determined as a ratio of firefly to renilla activity measured using the dual luciferase assay (Promega). The expression level for MRE-containing vectors were divided by the expression level for of vector without MREs. Results presented are the average of three replicate experiments. The MtnA reporter has been described (16). The MtnB reporter contains a 1351-bp fragment spanning nucleotides −1264 to +87 relative to the transcription start site. For RNAi assays, S2 cells (200 µL per well, 24-well plate, at 2.5 × 106 cells per mL) were combined with dsRNA for MED26 or Escherichia coli lacI at 40 µg/mL in serum free Schneider’s media for 1 h at room temperature. Transfections were carried out as above. Seventy-two hours later, the cells were treated as above.
MTF-1 Antibodies.
Rabbit antisera were raised against amino acids 1–113 and amino acid 406–540 of MTF-1 fused to GST.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health Grant R01GM085250 (to M.T.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE40535).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207737109/-/DCSupplemental.
References
- 1.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunther V, Lindert U, Schaffner W. The taste of heavy metals: Gene regulation by MTF-1. Biochim Biophys Acta. 2012;1823:1416–1425. doi: 10.1016/j.bbamcr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Selvaraj A, et al. Metal-responsive transcription factor (MTF-1) handles both extremes, copper load and copper starvation, by activating different genes. Genes Dev. 2005;19:891–896. doi: 10.1101/gad.1301805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heuchel R, et al. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 1994;13:2870–2875. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balamurugan K, et al. Metal-responsive transcription factor (MTF-1) and heavy metal stress response in Drosophila and mammalian cells: A functional comparison. Biol Chem. 2004;385:597–603. doi: 10.1515/BC.2004.074. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Egli D, Georgiev O, Schaffner W. The Drosophila homolog of mammalian zinc finger factor MTF-1 activates transcription in response to heavy metals. Mol Cell Biol. 2001;21:4505–4514. doi: 10.1128/MCB.21.14.4505-4514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giedroc DP, Chen X, Apuy JL. Metal response element (MRE)-binding transcription factor-1 (MTF-1): Structure, function, and regulation. Antioxid Redox Signal. 2001;3:577–596. doi: 10.1089/15230860152542943. [DOI] [PubMed] [Google Scholar]
- 9.Potter BM, et al. The six zinc fingers of metal-responsive element binding transcription factor-1 form stable and quasi-ordered structures with relatively small differences in zinc affinities. J Biol Chem. 2005;280:28529–28540. doi: 10.1074/jbc.M505217200. [DOI] [PubMed] [Google Scholar]
- 10.Lynes MA, Kang YJ, Sensi SL, Perdrizet GA, Hightower LE. Heavy metal ions in normal physiology, toxic stress, and cytoprotection. Ann N Y Acad Sci. 2007;1113:159–172. doi: 10.1196/annals.1391.010. [DOI] [PubMed] [Google Scholar]
- 11.Miles AT, Hawksworth GM, Beattie JH, Rodilla V. Induction, regulation, degradation, and biological significance of mammalian metallothioneins. Crit Rev Biochem Mol Biol. 2000;35(1):35–70. doi: 10.1080/10409230091169168. [DOI] [PubMed] [Google Scholar]
- 12.Klaassen CD, Liu J, Choudhuri S. Metallothionein: An intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol. 1999;39:267–294. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- 13.Park JD, Liu Y, Klaassen CD. Protective effect of metallothionein against the toxicity of cadmium and other metals. Toxicology. 2001;163(2-3):93–100. doi: 10.1016/s0300-483x(01)00375-4. [DOI] [PubMed] [Google Scholar]
- 14.Egli D, et al. The four members of the Drosophila metallothionein family exhibit distinct yet overlapping roles in heavy metal homeostasis and detoxification. Genes Cells. 2006;11:647–658. doi: 10.1111/j.1365-2443.2006.00971.x. [DOI] [PubMed] [Google Scholar]
- 15.Miura N, Koizumi S. [Heavy metal responses of the human metallothionein isoform genes] Yakugaku Zasshi. 2007;127:665–673. doi: 10.1248/yakushi.127.665. [DOI] [PubMed] [Google Scholar]
- 16.Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadhu C, Gedamu L. Regulation of human metallothionein (MT) genes. Differential expression of MTI-F, MTI-G, and MTII-A genes in the hepatoblastoma cell line (HepG2) J Biol Chem. 1988;263:2679–2684. [PubMed] [Google Scholar]
- 18.Murata M, Gong P, Suzuki K, Koizumi S. Differential metal response and regulation of human heavy metal-inducible genes. J Cell Physiol. 1999;180:105–113. doi: 10.1002/(SICI)1097-4652(199907)180:1<105::AID-JCP12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Lorenzi I, Georgiev O, Schaffner W. Metal-responsive transcription factor-1 (MTF-1) selects different types of metal response elements at low vs. high zinc concentration. Biol Chem. 2004;385:623–632. doi: 10.1515/BC.2004.077. [DOI] [PubMed] [Google Scholar]
- 20.Kayaalti Z, Mergen G, Söylemezoğlu T. Effect of metallothionein core promoter region polymorphism on cadmium, zinc and copper levels in autopsy kidney tissues from a Turkish population. Toxicol Appl Pharmacol. 2010;245:252–255. doi: 10.1016/j.taap.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Nath R, Paliwal VK, Prasad R, Kambadur R. Role of metallothionein in metal detoxification and metal tolerance in protein calorie malnutrition and calcium deficient monkeys (Macaca mulatta) Experientia Suppl. 1987;52:631–638. doi: 10.1007/978-3-0348-6784-9_67. [DOI] [PubMed] [Google Scholar]
- 22.Stuart GW, Searle PF, Chen HY, Brinster RL, Palmiter RD. A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc Natl Acad Sci USA. 1984;81:7318–7322. doi: 10.1073/pnas.81.23.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 24.Bittel D, Dalton T, Samson SL, Gedamu L, Andrews GK. The DNA binding activity of metal response element-binding transcription factor-1 is activated in vivo and in vitro by zinc, but not by other transition metals. J Biol Chem. 1998;273:7127–7133. doi: 10.1074/jbc.273.12.7127. [DOI] [PubMed] [Google Scholar]
- 25.Koizumi S, Suzuki K, Ogra Y, Gong P, Otuska F. Roles of zinc fingers and other regions of the transcription factor human MTF-1 in zinc-regulated DNA binding. J Cell Physiol. 2000;185:464–472. doi: 10.1002/1097-4652(200012)185:3<464::AID-JCP18>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Selvaraj A, et al. Metal-responsive transcription factor (MTF-1) handles both extremes, copper load and copper starvation, by activating different genes. Genes Dev. 2005;19:891–896. doi: 10.1101/gad.1301805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marr SK, Pennington KL, Marr MT. Efficient metal-specific transcription activation by Drosophila MTF-1 requires conserved cysteine residues in the carboxy-terminal domain. Biochim Biophys Acta. 2012;1819:902–912. doi: 10.1016/j.bbagrm.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 30.Meijsing SH, Elbi C, Luecke HF, Hager GL, Yamamoto KR. The ligand binding domain controls glucocorticoid receptor dynamics independent of ligand release. Mol Cell Biol. 2007;27:2442–2451. doi: 10.1128/MCB.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, et al. Copper sensing function of Drosophila metal-responsive transcription factor-1 is mediated by a tetranuclear Cu(I) cluster. Nucleic Acids Res. 2008;36:3128–3138. doi: 10.1093/nar/gkn103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Günther V, Davis AM, Georgiev O, Schaffner W. A conserved cysteine cluster, essential for transcriptional activity, mediates homodimerization of human metal-responsive transcription factor-1 (MTF-1) Biochim Biophys Acta. 2012;1823:476–483. doi: 10.1016/j.bbamcr.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Günther V, Waldvogel D, Nosswitz M, Georgiev O, Schaffner W. Dissection of Drosophila MTF-1 reveals a domain for differential target gene activation upon copper overload vs. copper starvation. Int J Biochem Cell Biol. 2012;44:404–411. doi: 10.1016/j.biocel.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 34.Arneson N, Hughes S, Houlston R, Done S. Whole-genome amplification by degenerate oligonucleotide primed PCR (DOP-PCR) Cold Spring Harbor Protoc. 2008;2008 doi: 10.1101/pdb.prot4919. [DOI] [PubMed] [Google Scholar]
- 35.Roychoudhury R, Wu R. Terminal transferase-catalyzed addition of nucleotides to the 3′ termini of DNA. Methods Enzymol. 1980;65:43–62. doi: 10.1016/s0076-6879(80)65009-5. [DOI] [PubMed] [Google Scholar]
- 36.Machanick P, Bailey TL. MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heberlein U, England B, Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985;41:965–977. doi: 10.1016/s0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.