Abstract
Environmental agents and genetic variants can induce heritable epigenetic changes that affect phenotypic variation and disease risk in many species. These transgenerational effects challenge conventional understanding about the modes and mechanisms of inheritance, but their molecular basis is poorly understood. The Deadend1 (Dnd1) gene enhances susceptibility to testicular germ cell tumors (TGCTs) in mice, in part by interacting epigenetically with other TGCT modifier genes in previous generations. Sequence homology to A1cf, the RNA-binding subunit of the ApoB editing complex, raises the possibility that the function of Dnd1 is related to Apobec1 activity as a cytidine deaminase. We conducted a series of experiments with a genetically engineered deficiency of Apobec1 on the TGCT-susceptible 129/Sv inbred background to determine whether dosage of Apobec1 modifies susceptibility, either alone or in combination with Dnd1, and either in a conventional or a transgenerational manner. In the paternal germ-lineage, Apobec1 deficiency significantly increased susceptibility among heterozygous but not wild-type male offspring, without subsequent transgenerational effects, showing that increased TGCT risk resulting from partial loss of Apobec1 function is inherited in a conventional manner. By contrast, partial deficiency in the maternal germ-lineage led to suppression of TGCTs in both partially and fully deficient males and significantly reduced TGCT risk in a transgenerational manner among wild-type offspring. These heritable epigenetic changes persisted for multiple generations and were fully reversed after consecutive crosses through the alternative germ-lineage. These results suggest that Apobec1 plays a central role in controlling TGCT susceptibility in both a conventional and a transgenerational manner.
Keywords: epigenetics, testicular cancer, gametogenesis, epistasis
Establishing the genetic basis and mode of inheritance for most traits and diseases has proven to be remarkably elusive (1). Susceptibility to testicular germ cell tumors (TGCTs) is illustrative (2). Testicular cancer is the most common malignancy affecting young men (3), more than 90% of testicular cancers result from TGCTs (4), TGCTs rank third in heritability among all cancers (5), and family history is the strongest known risk factor with a two- to sixfold increase among sons and a 5- to 19-fold increase among brothers of affected individuals (6–9). Despite the strong evidence for heritability, the only TGCT susceptibility factors identified in genome-wide association studies are the gr/gr deletion on the Y chromosome and autosomal variants in KITLG, SPRY4, BAK1, and DMRT1, which together account for less than 10% of risk (10–14). The substantial difference in prevalence between sons and brothers of cases and the epidemiological evidence for maternal estrogens, birth order, birth weight, and other factors (15–17) together raise the possibility that maternal conditions, epigenetic effects, and perhaps unconventional modes of inheritance also contribute to TGCT risk (18).
Only strains in the 129 inbred family of mice develop spontaneous TGCTs at an appreciable rate, with ∼7% of males presenting at least one TGCT by 4 wk of age (19, 20). These TGCTs are teratomas and teratocarcinomas that most closely resemble pediatric TGCTs in humans (21). In mice and humans, TGCTs arise from primordial germ cells (PGCs), which are the embryonic precursors of gametes in adults (21, 22). PGC transformation occurs during embryonic days 13.5–16.5 (E13.5–E16.5) when several critical events occur, including gonad determination, epigenetic reprogramming to replace most parent-of-origin molecular marks with those reflecting fetal sex, and meiotic arrest of PGCs in females and mitotic arrest in males (23–26).
Genetic variants that modify TGCT susceptibility provide insights into both the development of the PGC lineage and the regulation of TGCT pathogenesis (27, 28). Studies of interactions between these modifier genes in mice reveal remarkable evidence for transgenerational genetic effects on TGCT risk (29). In these cases, genetic variants in ancestral generations induce heritable epigenetic changes that lead to phenotypic variation in subsequent generations (18). These epigenetic changes can affect a remarkable variety of traits including behaviors, blood chemistry, pigmentation, heart development, embryonic viability, feeding habits, and susceptibility to diet-induced obesity (30–35). These epigenetic effects can be as strong and as common as those that are inherited in a conventional manner (30–35) and can persist for generations after exposure to the original genetic variant (30, 35). Evidence for transgenerational genetic effects on TGCTs involves wild-type sons of Kitl-deletion heterozygotes that are tumor-free instead of showing the 7% risk expected based on their genotype (31). Other evidence includes interactions across generations between TGCT modifiers such as Trp53, Kitlg, and the Eif2s2 translation initiation factor in the parental generation with the Ter mutation in the Deadend1 (Dnd1) gene in the offspring generation to increase the number of TGCT-affected males and the number of bilateral cases (29).
A major unresolved question is the nature of the molecular mechanism that mediates these transgenerational effects. Dnd1 shares sequence similarity with Apobec1 complementation factor (A1cf) (36), the RNA-binding component of the ApoB RNA-editing complex (37, 38). This similarity raises the possibility that DND1, like A1CF, functions in RNA editing and in turn that RNA-editing mutants affect TGCT susceptibility. Apobec1 encodes a cytidine deaminase, which is the catalytic component of the ApoB RNA-editing complex (37, 38). Recent evidence suggests that several deaminases in the Apobec1 family also have DNA demethylase activity (39–41). Targeted deletion of Apobec1 (abbreviated ko hereafter) results both in homozygous mice that are healthy and fertile and in knockout heterozygotes that show tissue-specific abnormalities in ApoB RNA editing (42). The present study focused on tests to determine whether Apobec1 deficiency affects TGCT susceptibility in a conventional or transgenerational manner, either alone or in combination with Dnd1Ter.
Results
Effects of Complete and Partial Apobec1 Deficiency on TGCTs.
The first task was to test the impact of complete and partial APOBEC1 deficiency on TGCT susceptibility with respect to both conventional inheritance and transgenerational effects. We began with a test for effects of ko homozygosity on ko/ko male offspring in a pure-breeding strain (test 1). Then we tested the effects of parental ko/+ heterozygosity on ko/+ and +/+ male offspring (test 2), parental homozygosity versus heterozygosity on genetically identical ko/+ heterozygous sons (test 3), ancestral genotype and switching the parent of origin (test 4), and finally long-term epigenetic effects of parental APOBEC1 deficiency on wild-type progeny (test 5).
Test 1: Parental and offspring ko/ko homozygosity.
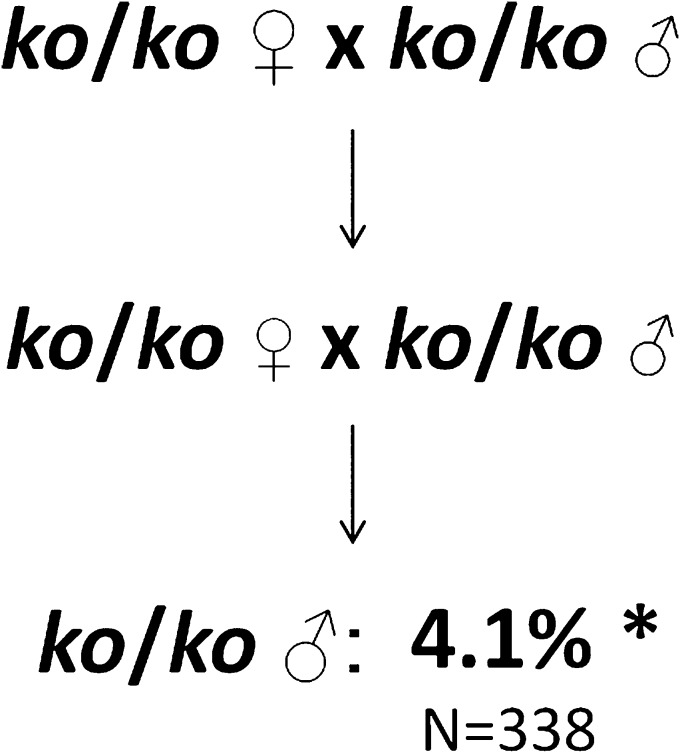
To test whether complete deficiency of Apobec1 affects TGCT susceptibility, we surveyed homozygous ko/ko males that were derived from at least two generations of ko/ko intercrosses (Fig. 1). Results were compared with both the concurrent wild-type 129 control [(+/+)c] and published reports for wild-type [(+/+)p] mice. For these two control populations, the proportion of affected (+/+)c and (+/+)p males did not differ significantly (Table 1, cross 4), suggesting that current results reflect the long-term average TGCT prevalence in 129 males (19–22, 36, 43–46). Interestingly, affected Apobec1-deficient males occurred at a frequency (4.1%) that represents an ∼43% reduction (P < 0.05) compared with that for the (+/+)c and (+/+)p control populations [Table 1, cross 1 and (+/+)p]. The finding of reduced testicular cancer risk is reminiscent of the effects of Apobec1 deficiency on small intestinal adenoma formation in ApcMin/+ mice (47). Moreover, the magnitude of the reduction is similar to that reported for males that are partially deficient for the Eif2s2 translation initiation factor, which is an established TGCT suppressor (48).
Fig. 1.
Effects of Apobec1 deficiency on TGCTs. Parental and offspring Apobec1ko/ko homozygosity (test 1). *P < 0.05.
Table 1.
Effects of complete and partial Apobec1 deficiency on occurrence of TGCT-affected males
| Cross | Parental cross | Offspring genotype | N | No. affected | Percent affected | χ2 | P value | Conclusion |
| 1 | ko/ko x ko/ko | ko/ko | 338 | 14 | 4.1 | 4.3 | <0.05 | Complete deficiency reduces susceptibility |
| 2a | ko/+ x +/+ | ko/+ | 186 | 0 | 0 | 14.0 | <0.0002 | Maternal heterozygosity provides complete protection |
| (+/+)w | 191 | 3 | 1.6 | 8.7 | <0.004 | Maternal heterozygosity reduces susceptibility | ||
| 2b | +/+ x ko/+ | ko/+ | 132 | 19 | 14.0 | 10.2 | <0.001 | Paternal heterozygosity increases susceptibility |
| (+/+)w | 125 | 9 | 7.2 | <0.1 | Ns | Paternal heterozygosity does not affect susceptibility | ||
| 3a | ko/ko x +/+ | ko/+ | 179 | 0 | 0 | 13.9 | <0.0002 | Maternal homozygosity provides complete protection |
| 3b | +/+ x ko/ko | ko/+ | 189 | 15 | 7.9 | <0.1 | Ns | Paternal homozygosity does not affect susceptibility |
| 4 | +/+ x +/+ | (+/+)w | 208 | 15 | 7.2 | 0.01 | Ns | Contemporaneous control was not different from published results for (+/+)p |
χ2 Goodness-of-fit tests were used to compare the prevalence of TGCT-affected test and concurrent wild-type (+/+)c control males. N is the number of males examined; no. affected is the number of males with at least one TGCT; significance was determined as P < 0.05; and Ns indicates results that did not pass the threshold of statistical significance. Because (+/+)w and (+/+)c were not different, and both were consistent with long-term rates in the 129 family of inbred strains, the 7% prevalence in (+/+)p was used as expectation for comparisons with +/+ and ko/+ male offspring.
Test 2: Parental ko/+ heterozygosity.
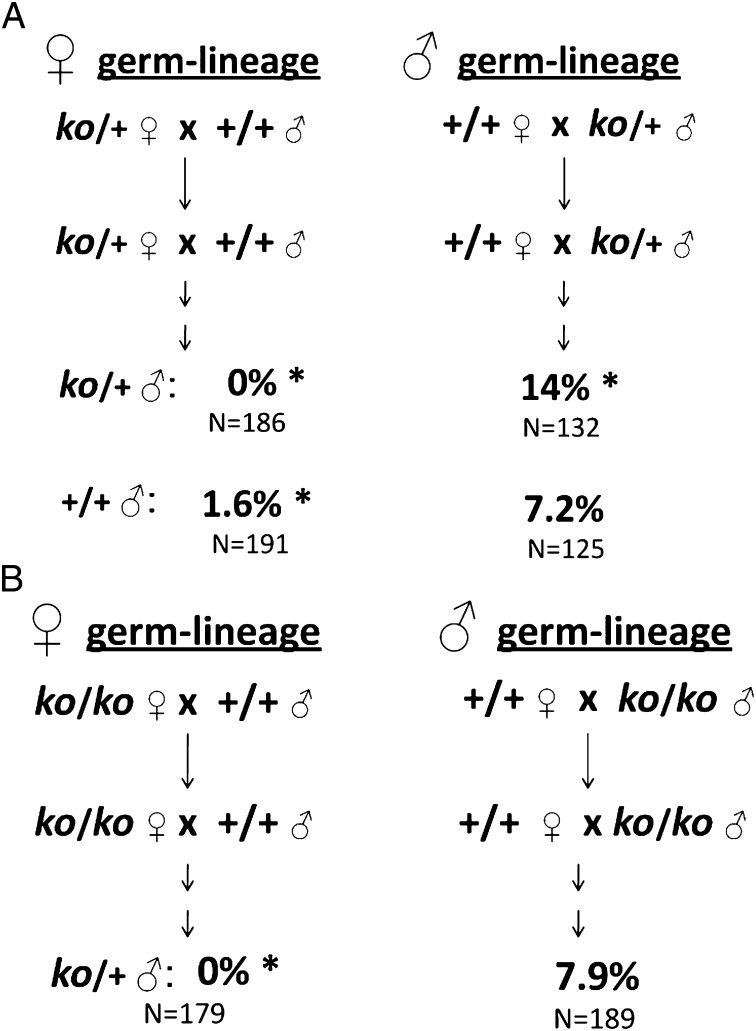
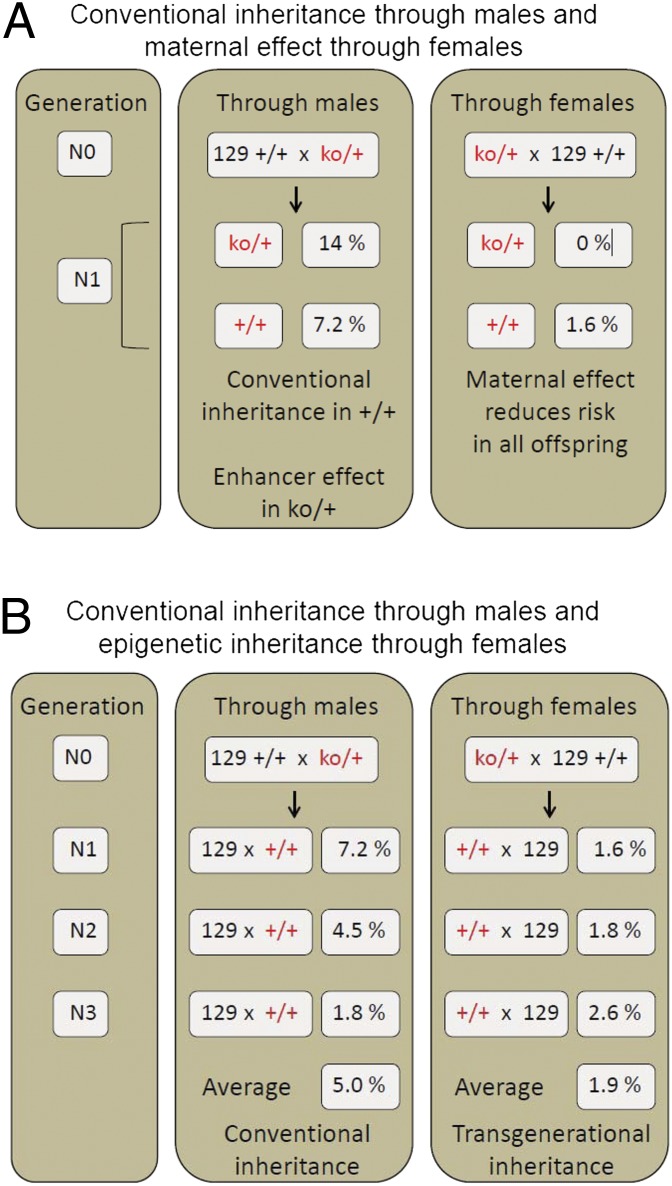
Because partial Apobec1 deficiency can lead to tissue-specific dysfunction (42, 47), we asked whether parental ko/+ heterozygosity affected TGCT susceptibility (cross 2). For this test, we measured the frequency of affected ko/+ (test) wild type and (+/+)w (control) male offspring from reciprocal crosses between ko/+ heterozygotes and strain 129 wild-type (+/+)c homozygotes. The direction of ko inheritance was maintained independently through the female and male lineages for at least three generations before the test crosses, and results were analyzed separately (Fig. 2A). Surprisingly, with maternal heterozygosity, TGCTs were absent in ko/+ male offspring, a highly significant reduction relative to (+/+)p (Table 1, cross 2a). Interestingly, the 1.6% prevalence in (+/+)w male offspring also was significantly reduced relative to wild-type controls (Table 1, cross 2a), even though these wild-type (+/+)w males were genetically identical to strain 129 (+/+)c and (+/+)p control males. By contrast, with paternal heterozygosity, 14% of heterozygous ko/+ male offspring were affected, which represents a twofold increase relative to (+/+)p controls (Table 1, cross 2b), an increase that was similar to published values for established heterozygous TGCT enhancers including Dnd1Ter (17%), KitlgSlJ (14%), and Trp53tm1Tyj (15%) (27, 31, 36, 43–46, 49, 50). In contrast, (+/+)w male offspring resulting from paternal heterozygosity showed the baseline prevalence typical of strain 129 males (Table 1, cross 2b). Thus, maternal partial deficiency reduced risk in a parent-of-origin manner among both heterozygous and wild-type sons, whereas paternal partial deficiency enhanced tumorigenesis only in ko/+ sons in a manner that showed conventional inheritance.
Fig. 2.
Effects of partial deficiency of Apobec1 on TGCTs. (A) Parental Apobec1ko/+ heterozygosity (test 2). (B) Parental Apobec1ko/ko homozygosity and offspring Apobec1ko/+ heterozygosity (test 3). *P < 0.05.
Test 3: Parental homozygosity versus heterozygosity effects on ko/+ offspring.
Next we tested whether TGCT prevalence among ko/+ male offspring depended on parental homozygosity (ko/ko) versus heterozygosity (ko/+) (Fig. 2B). Occurrence of TGCT-affected males among genetically identical heterozygous (ko/+) progeny derived from reciprocal ko/ko × (+/+)c intercrosses (cross 3) was compared with those described above for ko/+ × (+/+)c backcrosses (Table 1, crosses 2 and 3, respectively). To control for lineage-specific effects, parents were obtained from lineages in which the parental germ-lineage had been maintained consistently for at least three prior generations.
When the ko mutation was inherited through the female germ-lineage, heterozygous ko/+ sons of ko/ko females were fully protected (0% affected) from TGCTs (Table 1, crosses 2a and 3a). By contrast, when the ko mutation was inherited through the male germ-lineage, the prevalence of affected males (7.9%) did not differ significantly from the (+/+)p baseline (Table 1, cross 3b) but was significantly less than the 14% prevalence found in genetically identical ko/+ offspring of ko/+ heterozygous males (χ2 = 5.8, P < 0.02; Table 1, cross 2b). Thus, TGCT prevalence differed dramatically among genetically ko/+ identical male offspring depending on parental sex and genotype, with complete protection among sons of ko/+ heterozygous and ko/ko homozygous females and with increased risk in ko/+ heterozygous sons of ko/+ heterozygous but not ko/ko homozygous males.
Test 4: Ancestral genotype and switching parent of origin.
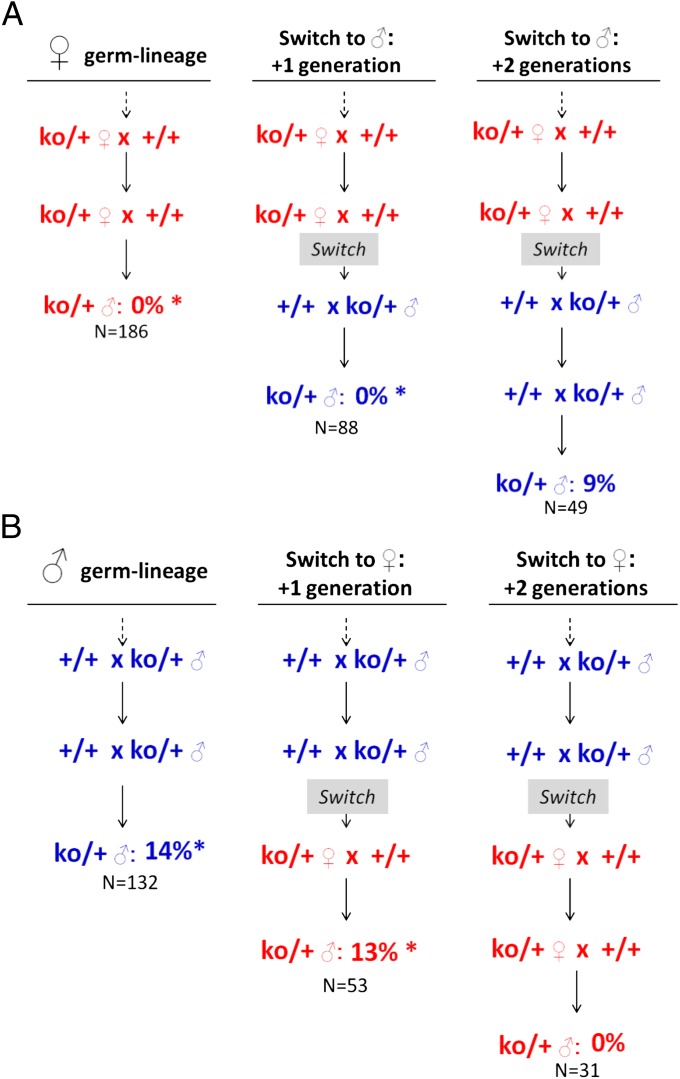
The next test involved determining whether the special physiological relationship between mother and fetus during the critical developmental period for PGC transformation to TGCTs contributed to the transgenerational genetic effects observed in cross 2a (Table 1). In the first cross, ko/+ male offspring that inherited the ko deletion through the female germ-lineage for at least two previous generations were mated to wild-type females, thereby switching the ancestral-female mode of inheritance to transmission through males (Fig. 3A). Heterozygous ko/+ sons that inherited this ancestral-female allele from their father instead of their mother were screened for TGCTs, and the prevalence was compared with that observed for ko/+ males that inherited the deficiency through the female germ-lineage. Surprisingly, none of the ko/+ males from this switched mating were affected with TGCTs, and this prevalence was significantly reduced relative to both the increased TGCT prevalence (14%) observed in ko/+ males inheriting the ancestral-male ko allele through the male germ-lineage and to the baseline rates in (+/+)c and (+/+)p controls (Fig. 3A). In the reciprocal cross, we tested the effect of transmitting the ancestral-male ko allele through females (Fig. 3B). In this case, the 13% TGCT prevalence was not significantly different from direct inheritance through the male lineage but was significantly increased over the expected rate based on direct maternal effects. Together these reciprocal crosses showed that ancestral rather than parental genotype underlies the lineage-dependent effects of partial Apobec1 deficiency on TGCT prevalence.
Fig. 3.
Effects of switching parent of origin on TGCTs. χ2 contingency tests were used to determine whether the two groups were derived from populations with similar properties, whereas χ2 goodness-of-fit tests were used to determine whether a given result was consistent with the baseline prevalence of 7%. (A) Ancestral genotype and switched transmission from ancestral females to males (test 4). Details for comparisons: 0% before switch versus 0% one generation after switch: not significant; 0% one generation after switch versus 9% two generations after switch: χ2 = 6.6, P < 0.02; 0% one generation after switch versus 7% baseline expectation: χ2 = 6.4, P < 0.02; 9% two generations after switch versus 7% baseline expectation: χ2 = 0.8, not significant. (B) Ancestral genotype and switched transmission from ancestral males to females (test 4). Details for comparisons: 14% before switch versus 13% one generation after switch: not significant; 13% one generation after switch versus 0% two generations after switch: χ2 = 3.3, P < 0.07; 13% one generation after switch versus 7% baseline expectation: χ2 = 5.7, P < 0.02; 0% two generations after switch versus 7% baseline expectation: χ2 = 0.1, not significant. * indicates a significant difference relative to the baseline TGCT prevalence in 129 (+/+)p control males.
We then asked whether maintaining the new direction of inheritance of the ko allele for another generation was sufficient to reverse lineage-specific effects to reflect the grandparental direction of inheritance. In the first cross, the ancestral-female allele was inherited for a second generation through the male germ-lineage (Fig. 3A), and in the reciprocal cross, the ancestral-male allele was inherited for a second generation through the female germ-lineage (Fig. 3B). In contrast to results for the ancestral-female allele in the previous generation, we found that the prevalence of affected ko/+ males now was switched in the second generation, showing a significant increase over the absence of affected males in the previous generation and a nonsignificant difference from the expected prevalence of 7% based on their genotype and the direction of the cross (Fig. 3A). Similarly, the occurrence of affected ko/+ males with the ancestral-male allele also was switched in the second generation, with a significant decrease from the previous generation and a nonsignificant difference from the rate expected with maternal inheritance (Fig. 3B). Thus, lineage-specific factors that control the TGCT phenotype were reset after transmission for two consecutive generations through the alternative germ-lineage and corresponded with the grandparental rather than the parental direction of inheritance.
Test 5: Persistence of transgenerational effects in wild-type (+/+)w males.
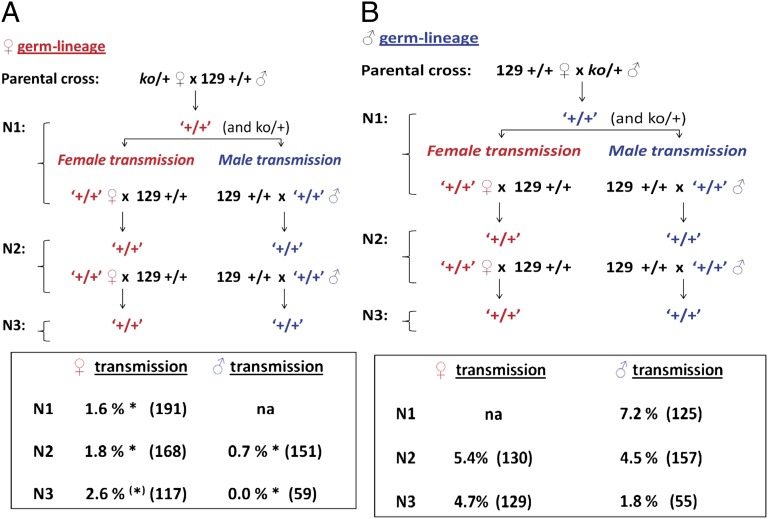
Finally, we asked whether the transgenerational effects of Apobec1 on wild-type progeny persisted to subsequent generations, independently of maternal ko/+ heterozygosity. If the observed effect among wild-type (N1) sons resulted from in utero exposure to the consequences of partial deficiency of maternal APOBEC1, reduced prevalence would be found among sons (N1) and perhaps grandsons (N2), but not among great-grandsons (N3), where prevalence should return to baseline values. Alternatively, if TGCT prevalence found among N1 males resulted from heritable epigenetic changes in N0 females, a similarly reduced prevalence might be expected among N1, N2, and N3 male offspring.
For this test, we backcrossed wild-type (+/+)w N1 offspring from reciprocal ko/+ × (+/+)c N0 crosses (Table 1, with cross 2a as the test and cross 2b as the control) to 129 (+/+)c mice (Fig. 4). We then surveyed (+/+)w N2 males and, after an additional backcross, (+/+)w N3 males for TGCTs. In these tests, the ko allele was present only in the N0 parents and N1 littermates. The N2 and N3 males that were surveyed for TGCTs did not inherit the ko/+ allele and were genetically, but perhaps not epigenetically, identical to the 129 (+/+)c and (+/+)p controls.
Fig. 4.
Ancestral Apobec1 genotype controls TGCT susceptibility across multiple generations. (A) Female germ-lineage transmission of Apobec1 deficiency before switching the direction of the cross. (B) Male germ-lineage transmission of Apobec1 deficiency before switching the direction of the cross. Red text indicates crosses where the ko allele was maternally transmitted, and blue text indicates crosses where the ko allele was paternally transmitted. *P < 0.05. ‘+/+’ indicates a genotypically wild-type mouse that had at least one ancestor that was ko; this designation is used to distinguish these mice from genotypically similar mice from a conventional wild-type (+/+) strain.
Remarkably, maternal Apobec1 deficiency significantly reduced TGCT prevalence, not only among N1 males but also among N2 and N3 (+/+)w males relative to (+/+)p controls (Fig. 4A). Inheritance of this suppressive effect from the ancestral female lineage was independent of parental direction in subsequent generations and did not require the ko allele to maintain the phenotype, suggesting that the observed TGCT suppression in sons of Apobec1-deficient females results from a stable, inherited epigenetic factor rather than direct exposure to the maternal deficiency in utero. For partial Apobec1 deficiency in males, TGCT prevalence in N1 (+/+)w sons did not differ from the expected baseline prevalence in wild-type sons (Fig. 4B). As expected, with continued backcrossing, TGCTs were observed in wild-type sons at the baseline rate regardless of whether the ancestral-male allele was transmitted through a male or female wild-type parent in subsequent generations.
Transgenerational Interactions Between Apobec1ko and Dnd1Ter
Previous studies revealed transgenerational genetic interactions that led to both an increased prevalence of affected Dnd1Ter (Ter) males and an increased proportion of bilateral cases in Ter/+ males when any of six TGCT modifier genes were present in the parental generation, compared with results for Ter/+ and these modifiers in separate single-gene control crosses (29). We therefore sought to test whether ko interacts with Ter in a similar manner. For this study, we focused on three specific ko and Ter interaction effects. We also asked whether ko and Ter have additive effects in double-heterozygous males. The study design of Lam et al. (29) was used for these tests (Fig. S1), with the expected frequency of affected males based on results from corresponding single-modifier tests (Table S1).
Test 1: Dnd1Ter sons.
To determine whether partial deficiency of Apobec1 in the parental generation affected TGCT risk in Ter/+ +/+ sons, we compared the TGCT prevalence in Ter/+ +/+ males obtained from the interaction test cross with the prevalence in Ter/+ control males (31.5%) from a separate Ter-only colony. Male offspring were affected with TGCTs at a rate of 21.0%, which represents a significant 40% reduction relative to the prevalence observed in Ter/+ controls (Table 2). These results with ko differed from those with all six previously tested modifiers in that the parental modifier, in this case Apobec1ko, reduced rather than increased prevalence in Ter/+ +/+ sons (cf ref. 29).
Table 2.
Occurrence of TGCT-affected males in transgenerational interaction tests between Apobec1ko and Dnd1Ter
| Purpose | Test | Offspring genotype | N | No. affected | No. expected | Percent affected | Percent expected | χ2 | P |
| Test for transgenerational effects | 1 | Ter/+ +/+ | 124 | 26 | 39.0 | 21.0 | 31.5 | 6.3 | <0.02 |
| 2 | +/+ ko/+ (m) | 163 | 14 | 0 | 8.6 | 0 | 12.6 | <0.0004 | |
| +/+ ko/+ (f) | 159 | 17 | 15.1 | 10.7 | 9.5 | 0.3 | Ns | ||
| Internal wild-type (w) control | 3 | +/+ +/+ | 213 | 14 | 15.4 | 6.6 | 7.2 | 0.1 | Ns |
| Test for epistasis | 4 | Ter/+ +/+ | 189 | 52 | 56.6 | 27.5 | 29.0 | 0.5 | Ns |
Mice in the contrast group were obtained from contemporaneous crosses in which only the designated genetic variant was segregating. The expected effects for the additive model were obtained by summing the independent effects for ko/+ and Ter/+. (m) and (p) refer to maternal and paternal ko/+ heterozygosity, respectively. Several considerations are relevant: We found a TGCT prevalence of 31.5% in Ter/+ control males on the 129S1/SvImJ background, which represents a significant increase (P < 10−5) compared with the published values (30, 38, 39). However, the previous studies were performed with a different 129 substrain (129T1/Sv). Therefore, throughout the present study, we used the control values observed in the concurrent surveys because these mice were bred on the same 129 background, screening was performed concurrently with test males, and mice used in the interaction test and in control crosses were on the same inbred genetic background.
Because partial Apobec1 deficiency acted as a TGCT suppressor in the female germ-lineage and as an enhancer in the male germ-lineage (Table 1, crosses 2a and 2b), we tested whether the sex of the Apobec1-deficient parent in the interaction test-cross affected TGCT prevalence in Ter/+ sons by comparing the prevalence in sons of Apobec1-deficient mothers versus sons of Apobec1-deficient fathers. However, the prevalence in Ter/+ +/+ sons of partially deficient mothers (22.0%) and partially deficient fathers (19.1%) was not significantly different (not shown), indicating that, although parental Apobec1 deficiency acted in a sex-specific manner in independent crosses, deficiency in either parent interacted with Ter in sons to reduce TGCT risk.
Test 2: Apobec1ko sons.
We compared the TGCT prevalence in ko/+ +/+ sons from the interaction test cross with the expected prevalence based on a separate ko-only colony. Because Apobec1 deficiency had lineage-specific effects on the frequency of TGCT-affected males in separate crosses (Table 1), sons inheriting the ko allele through the maternal versus paternal germ-lineage were analyzed separately. The expected values for this analysis took into account the parent of origin and the Apobec1 genotype of the parents. Although Apobec1 deficiency had divergent effects in the maternal and paternal lineage (Table 1), TGCT prevalence in ko/+ +/+ males from the interaction test cross did not vary significantly with the direction of inheritance (Table 2, test 2). Interestingly, although maternal Apobec1 deficiency was protective in ko-only crosses (Table 1, crosses 2a and 3a), an appreciable number of affected males was observed among those that inherited the ko allele maternally when Ter also was present in the parental generation [Table 2, test 2, +/+ ko/+ (m)]. In the reciprocal cross, in contrast, the prevalence of affected ko/+ males did not differ significantly from wild-type strain 129 controls [Table 2, test 2, +/+ ko/+ (p)], suggesting that presence of Ter in the parental generation negated the protective parent-of-origin effect of Apobec1 deficiency in the female lineage.
Test 3: Wild-type sons.
Although wild-type offspring from Apobec1-deficient females showed a significant reduction in TGCT prevalence in separate crosses (Table 1, cross 2a), wild-type males in the interaction test crosses were affected at the expected baseline rate (6.6%) (Table 2), suggesting that the increased prevalence in Ter/+ sons and the reversal of the protective effect of maternal Apobec1 deficiency in ko/+ sons result from specific interactions between the parental and inherited mutations rather than being attributable solely to premeiotic effects of the parental mutations that would manifest in all offspring regardless of their genotype.
Test 4: Apobec1ko and Dnd1Ter sons.
To test for traditional genetic interactions between Apobec1 and Dnd1, double-mutant male offspring (ko/+, Ter/+) were surveyed for TGCTs with the expected prevalence (29.9%) based on an additive model of gene interaction and calculated using the observed frequencies for single-mutant and wild-type littermate controls (29). The observed TGCT prevalence in double-mutants (27.5%) did not differ significantly from the predicted prevalence and therefore fits an additive model of gene interaction. Apobec1ko therefore is the only genetic variant that does not interact with Ter to modulate TGCT susceptibility.
Lineage-specific ko–Ter interactions and embryonic viability.
In the ko–Ter interaction test, we noted that segregation ratios for single- and double-mutant males did not appear to be normal (Table 2). We therefore examined this question directly (Table 3). In fact, these ratios deviated dramatically from expectations, depending on parental and progeny genotypes. When ko was inherited through the paternal germ-lineage, segregation ratios at 3–4 wk of age did not differ significantly from Mendelian expectations (χ2 = 1.0, not significant). However, when ko was inherited through the maternal germ-lineage, these ratios showed a highly significant deviation (χ2 = 58.5, P < 0.0001). If we accept that the observed number of wild-type males (n = 64) was the number expected in each genotypic class, then a total of 192 (3 × 64) males would be expected across the three other genotypes. However, with a total of only 51 observed, ∼75% (n = 141) of the single- and double-heterozygotes were missing. We next asked when these deviations arose, focusing first at ∼E12.5 and then at E3.5 (Table S2). Interestingly, segregation ratios were already biased at ∼E3.5 in maternal ko–paternal Ter crosses but not in the reciprocal cross. Litter sizes did not differ significantly across the entire study (Table S3).
Table 3.
Non-Mendelian segregation in ko–Ter interaction tests
| Ter/+ ♀ x ko/+ ♂ | Offspring genotype (♀,♂) | ko/+ ♀x Ter/+ ♂ | ||||
| Nobs | Nexp | %obs | Nobs | Nexp | %obs | |
| 27 | 32 | 24 | ko/+, Ter/+ | 13 | 4 | 11 |
| 30 | 32 | 26 | +/+, Ter/+ | 19 | 64 | 17 |
| 25 | 32 | 22 | ko/+, +/+ | 19 | 64 | 17 |
| 32 | 32 | 28 | +/+, +/+ | 64 | 64 | 56 |
| 114 | 128 | Total males | 115 | 256 | ||
Nobs is the number of males of each genotype observed, and Nexp is the number of males of each genotype expected based on Mendelian expectations, i.e., 25% of the total number of males in each genotypic class n the reciprocal crosses; %obs is the percent observed; and %exp is the percent expected based on Mendelian expectations, i.e., 25% of the total number of males in each genotypic class.
Discussion
Many environmental factors and genetic variants are known to induce heritable epigenetic changes that can persist for multiple generations, affecting a broad range of traits, and that often are as frequent and strong as direct environmental exposures and conventional genetic inheritance (18, 51–57). These transgenerational effects challenge our understanding of the modes and mechanisms for inherited phenotypic variation and disease risk, as well as the premise of most genetic studies in which causal DNA sequence variants are sought within the genome of affected individuals. Several molecular mechanisms have been implicated, ranging from inherited RNAs to chemically modified DNA and proteins (51–61). These transgenerational effects have important implications for our understanding of adaptation and evolution, the origins of phenotypic variation and disease risk, and the molecules in addition to DNA that can be the basis for inheritance (18, 62, 63).
Studies of transgenerational genetic effects on testicular cancer in mice show that Dnd1 is an essential component of a cross-generation system, with genes such as Trp53, Kitl, and Eif2s2 in the parental generation acting with Dnd1 in male offspring to increase significantly the number of TGCT-affected males and the proportion of bilateral cases (29). Dnd1 encodes an RNA-binding protein that controls access of specific microRNAs to their mRNA targets in testicular cancer cell lines (64) and also transports mRNAs from the nucleus to perinuclear RNA granules under stress conditions (65). We reasoned that sequence similarity between Dnd1 and A1cf, which is the RNA-binding subunit of the APOBEC1 RNA-editing complex (37, 38), raised the possibility that Dnd1 acts in a similar manner with Apobec1 and therefore that Apobec1 also might control TGCT risk and transgenerational effects, either alone or in combination with Dnd1. We tested this hypothesis by studying effects of Apobec1 deficiency and its interactions with the Dnd1Ter mutation on TGCT susceptibility in 129/SvImJ mice.
Several findings in this study document the conventional and transgenerational effects of Apobec1 deficiency on TGCT susceptibility, both alone and in combination with Dnd1. In the paternal germ-lineage, the increased number of affected ko/+ but not +/+ sons (Table 1, crosses 2b and 3b) shows that Apobec1 acts as TGCT enhancer and that this effect is inherited in a conventional manner. By contrast, ko/+ sons of ko/+ and ko/ko females were not affected with a TGCT (Table 1, crosses 2a and 3a; cf. refs. 28, 66). In addition, maternal ko/+ heterozygosity significantly reduced the prevalence of affected +/+ wild-type sons, not only in the first generation (Table 1, cross 2a) but also for at least two subsequent generations (Fig. 4), suggesting both that reduced dosage of maternal Apobec1 alone was sufficient to induce a heritable epigenetic change that persisted for multiple generations and that direct maternal–fetal interactions were not essential. Interestingly, parental Dnd1Ter/+ heterozygosity does not induce transgenerational effects (29). Finally, these lineage-specific effects were fully reversed after transmission for two consecutive generations through the alternative parental germ-lineage (Fig. 3), suggesting that the heritable epigenetic change was not permanent and could be reversed under specific conditions. The ability of Apobec1 to modulate TGCT risk in both a conventional and a transgenerational manner, depending on parental germ-lineage, raises important questions concerning the mechanisms by which Apobec1 may function in PGC biology, testicular cancer etiology, and, most importantly, in heritable epigenetic changes.
Tests for interactions between Apobec1ko and Dnd1Ter provided contrasting results, depending on parental genotypes. The combination of maternal ko/+ and paternal Ter/+ heterozygosity neutralized each other’s effects on TGCTs among male offspring, significantly increasing or reducing risk in the direction of the baseline rate that is characteristic of the 129 strain (Table 2). In contrast, in the reciprocal cross with maternal Ter/+ heterozygosity and paternal ko/+ heterozygosity, a striking deviation from Mendelian expectations was found at embryonic day E3.5 (Table S2), with substantial loss of both double-heterozygous and single-heterozygous mutant mice (Table 3). Thus, Apobec1 and Dnd1 interact, albeit in unconventional and complex manners, to modulate TGCT risk and also, unexpectedly, to control the occurrence of genotypic classes among early embryos.
The strong but unexpected bias in segregation ratios in maternal ko–paternal Ter intercrosses has several unusual features. The effects depended completely on the direction of the cross (Table 3), with Apobec1 acting in the female germ-lineage, and Dnd1 acting in the male germ-lineages. Loss of substantial numbers of single and double heterozygotes in the interaction test is remarkable, given that both ko/ko and Ter/Ter homozygotes are fully viable in separate crosses (36, 42). Normal segregation also is found in mutant heterozygotes in separate crosses to wild-type mice (29; also see Table S3), showing that gametes are produced in normal ratios and functionality. Moreover, modest differences in litter size do not account for the substantial genotypic bias (Table S3). Together these observations suggest that strong segregation bias results from a preference for specific combinations of gametes at fertilization rather from than selective embryo loss after conception. Interestingly, segregation ratios for heterozygous A1cf knockout mice also are strongly skewed (10:1) toward mutant heterozygotes versus wild type (67). These results are reminiscent of non-Mendelian segregation in males that are heterozygous for t-haplotypes in mice (68, 69) but differ in that both maternal and paternal factors are required. Together these observations suggest that Apobec1, Dnd1, and perhaps A1cf have important but as yet unknown functions during gametogenesis and fertilization.
Questions that emerge from this work include whether these genetic effects act directly on the germ-line or indirectly through the soma to the germ-line, perhaps through signaling pathways such as Kit ligand from somatic cells and Kit receptor on germ cells (18, 31); whether heritable epigenetic changes in the germ-line involve methylation, histone modifications, RNAs, or perhaps other molecules (18, 51–61); whether the relevant Apobec1 functions involve RNA editing or DNA demethylation (37–41, 60, 61); and whether the same epigenetic changes are transmitted to subsequent generations or are responsible for reversing heritable epigenetic changes.
Materials and Methods
Mice.
Mice from the 129S1/SvImJ inbred strain (“129”, previously known as 129/SvJ and 129S3/SvImJ; stock number JR002448) were obtained from the Jackson Laboratory. A targeted deletion of Apobec1 (herein abbreviated ko) was made with the RF8 ES cells (cell line from 129/SvImJ mice) and then maintained on the 129/SvImJ inbred genetic background (42 and this study). Mice were genotyped using a PCR-based assay as described previously (42), with wild-type homozygotes, heterozygotes, and mutant homozygotes designated as (+/+)w, ko/+, and ko/ko, respectively. Two control groups were included, (+/+)c and (+/+)p, which correspond respectively to a contemporaneous (c) survey of wild-type 129/Sv males from an independent colony and to a review of TGCT incidence in wild-type 129/Sv reported in the literature (p) (19–22, 36, 43–46). All mice in this study were maintained on a standard 5010 rodent chow diet (PMI Nutrition International), housed in the Case Western Reserve University Animal Resource Center, and maintained on a 12/12-h light/dark cycle. The Case Western Reserve Institutional Animal Care and Use Committee approved all mouse work described in this report.
TGCT Survey.
Male mice (3- to 5-wk-old) were killed, and incisions were made in the abdomen to expose the testes. TGCTs were identified through macroscopic inspection of the testis for abnormalities in size, color, shape, or texture (19–22, 36, 43–46). Mice in the various test and control groups were scored for the presence of unilateral or bilateral TGCTs. The frequency of affected males with either unilateral or bilateral TGCT was used as a measure of prevalence. χ2 tests were used to test for significant differences in the frequency of affected males.
Supplementary Material
Acknowledgments
We thank members of the J.H.N. group for ideas, comments, and discussions. This work was supported by National Institutes of Health (NIH)/National Cancer Institute Grant CA75056 and NIH Pioneer Award DP1OD006911 (to J.H.N.) and by NIH/National Heart, Lung and Blood Institute Grant HL-38180, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant DK-56260, and NIDDK Grant DK-52574 (to N.O.D.). P.J.T. is funded as a Robertson Investigator of the New York Stem Cell Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 16400.
See Author Summary on page 16414 (volume 109, number 41).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207169109/-/DCSupplemental.
References
- 1.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert D, Rapley E, Shipley J. Testicular germ cell tumours: Predisposition genes and the male germ cell niche. Nat Rev Cancer. 2011;11:278–288. doi: 10.1038/nrc3021. [DOI] [PubMed] [Google Scholar]
- 3.Buetow SA. Epidemiology of testicular cancer. Epidemiol Rev. 1995;17:433–449. doi: 10.1093/oxfordjournals.epirev.a036202. [DOI] [PubMed] [Google Scholar]
- 4.Bahrami A, Ro JY, Ayala AG. An overview of testicular germ cell tumors. Arch Pathol Lab Med. 2007;131:1267–1280. doi: 10.5858/2007-131-1267-AOOTGC. [DOI] [PubMed] [Google Scholar]
- 5.Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer. 2002;99:260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- 6.Heimdal K, et al. Familial testicular cancer in Norway and southern Sweden. Br J Cancer. 1996;73:964–969. doi: 10.1038/bjc.1996.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindelöf B, Eklund G. Analysis of hereditary component of cancer by use of a familial index by site. Lancet. 2001;358:1696–1698. doi: 10.1016/S0140-6736(01)06721-6. [DOI] [PubMed] [Google Scholar]
- 8.Bromen K, et al. Testicular, other genital, and breast cancers in first-degree relatives of testicular cancer patients and controls. Cancer Epidemiol Biomarkers Prev. 2004;13:1316–1324. [PubMed] [Google Scholar]
- 9.Chia VM, et al. Risk of cancer in first- and second-degree relatives of testicular germ cell tumor cases and controls. Int J Cancer. 2009;124:952–957. doi: 10.1002/ijc.23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nathanson KL, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am J Hum Genet. 2005;77:1034–1043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanetsky PA, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rapley EA, et al. UK Testicular Cancer Collaboration A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbull C, et al. UK Testicular Cancer Collaboration Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanetsky PA, et al. A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. Hum Mol Genet. 2011;20:3109–3117. doi: 10.1093/hmg/ddr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michos A, Xue F, Michels KB. Birth weight and the risk of testicular cancer: A meta-analysis. Int J Cancer. 2007;121:1123–1131. doi: 10.1002/ijc.22771. [DOI] [PubMed] [Google Scholar]
- 16.McGlynn KA, Cook MB. Etiologic factors in testicular germ-cell tumors. Future Oncol. 2009;5:1389–1402. doi: 10.2217/fon.09.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook MB, et al. A systematic review and meta-analysis of perinatal variables in relation to the risk of testicular cancer—experiences of the son. Int J Epidemiol. 2010;39:1605–1618. doi: 10.1093/ije/dyq120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson VR, Nadeau JH. Transgenerational genetic effects. Epigenomics. 2010;2:797–806. doi: 10.2217/epi.10.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens LC, Little CC. Spontaneous testicular teratomas in an inbred strain of mice. Proc Natl Acad Sci USA. 1954;40:1080–1087. doi: 10.1073/pnas.40.11.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens LC, Hummel KP. A description of spontaneous congenital testicular teratomas in strain 129 mice. J Natl Cancer Inst. 1957;18:719–747. [PubMed] [Google Scholar]
- 21.Stevens LC. The biology of teratomas. Adv Morphog. 1967;6:1–31. doi: 10.1016/b978-1-4831-9953-5.50005-6. [DOI] [PubMed] [Google Scholar]
- 22.Stevens LC. Origin of testicular teratomas from primordial germ cells in mice. J Natl Cancer Inst. 1967;38:549–552. [PubMed] [Google Scholar]
- 23.Munger SC, Capel B. Sex and the circuitry: Progress toward a systems-level understanding of vertebrate sex determination. Wiley Interdiscip Rev Syst Biol Med. 2012;4:401–412. doi: 10.1002/wsbm.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weaver JR, Susiarjo M, Bartolomei MS. Imprinting and epigenetic changes in the early embryo. Mamm Genome. 2009;20:532–543. doi: 10.1007/s00335-009-9225-2. [DOI] [PubMed] [Google Scholar]
- 25.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seydoux G, Braun RE. Pathway to totipotency: Lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 27.Stevens LC. Genetic influences on teratocarcinogenesis and parthenogenesis. Prog Clin Biol Res. 1981;45:93–104. [PubMed] [Google Scholar]
- 28.Carouge D, Nadeau JH. 2012. Mouse models of testicular germ cell tumors, Germ Cell Tumors, ed. Matin A (In Tech Publishing, Rijeka, Croatia)
- 29.Lam MY, Heaney JD, Youngren KK, Kawasoe JH, Nadeau JH. Trans-generational epistasis between Dnd1Ter and other modifier genes controls susceptibility to testicular germ cell tumors. Hum Mol Genet. 2007;16:2233–2240. doi: 10.1093/hmg/ddm175. [DOI] [PubMed] [Google Scholar]
- 30.Rassoulzadegan M, et al. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 31.Heaney JD, Lam MY, Michelson MV, Nadeau JH. Loss of the transmembrane but not the soluble kit ligand isoform increases testicular germ cell tumor susceptibility in mice. Cancer Res. 2008;68:5193–5197. doi: 10.1158/0008-5472.CAN-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner KD, et al. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Grandjean V, et al. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 34.Nelson VR, Spiezio SH, Nadeau JH. Transgenerational genetic effects of the paternal Y chromosome on daughters’ phenotypes. Epigenomics. 2010;2:513–521. doi: 10.2217/epi.10.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yazbek SN, Spiezio SH, Nadeau JH, Buchner DA. Ancestral paternal genotype controls body weight and food intake for multiple generations. Hum Mol Genet. 2010;19:4134–4144. doi: 10.1093/hmg/ddq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youngren KK, et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2010;2:594–602. doi: 10.1002/wsbm.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol. 2012;23:258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan HD, et al. Activation-induced cytidine deaminases deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues. J Biol Chem. 2004;279:52352–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 40.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano K, et al. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J Biol Chem. 1996;271:9887–9890. doi: 10.1074/jbc.271.17.9887. [DOI] [PubMed] [Google Scholar]
- 43.Stevens LC. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J Natl Cancer Inst. 1973;50:235–242. doi: 10.1093/jnci/50.1.235. [DOI] [PubMed] [Google Scholar]
- 44.Noguchi T, Noguchi M. A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J Natl Cancer Inst. 1985;75:385–392. [PubMed] [Google Scholar]
- 45.Asada Y, Varnum DS, Frankel WN, Nadeau JH. A mutation in the Ter gene causing increased susceptibility to testicular teratomas maps to mouse chromosome 18. Nat Genet. 1994;6:363–368. doi: 10.1038/ng0494-363. [DOI] [PubMed] [Google Scholar]
- 46.Matin A, Collin GB, Asada Y, Varnum D, Nadeau JH. Susceptibility to testicular germ-cell tumours in a 129.MOLF-Chr 19 chromosome substitution strain. Nat Genet. 1999;23:237–240. doi: 10.1038/13874. [DOI] [PubMed] [Google Scholar]
- 47.Blanc V, et al. Deletion of the AU-rich RNA binding protein Apobec-1 reduces intestinal tumor burden in Apc(min) mice. Cancer Res. 2007;67:8565–8573. doi: 10.1158/0008-5472.CAN-07-1593. [DOI] [PubMed] [Google Scholar]
- 48.Heaney JD, Michelson MV, Youngren KK, Lam MY, Nadeau JH. Deletion of eIF2beta suppresses testicular cancer incidence and causes recessive lethality in agouti-yellow mice. Hum Mol Genet. 2009;18:1395–1404. doi: 10.1093/hmg/ddp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 50.Harvey M, McArthur MJ, Montgomery CA, Jr, Bradley A, Donehower LA. Genetic background alters the spectrum of tumors that develop in p53-deficient mice. FASEB J. 1993;7:938–943. doi: 10.1096/fasebj.7.10.8344491. [DOI] [PubMed] [Google Scholar]
- 51.Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 52.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 54.Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: More questions than answers. Genome Res. 2010;20:1623–1628. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuzin F, Rassoulzadegan M. Non-Mendelian epigenetic heredity: Gametic RNAs as epigenetic regulators and transgenerational signals. Essays Biochem. 2010;48:101–106. doi: 10.1042/bse0480101. [DOI] [PubMed] [Google Scholar]
- 56.de Boer P, Ramos L, de Vries M, Gochhait S. Memoirs of an insult: Sperm as a possible source of transgenerational epimutations and genetic instability. Mol Hum Reprod. 2010;16:48–56. doi: 10.1093/molehr/gap098. [DOI] [PubMed] [Google Scholar]
- 57.Guerrero-Bosagna C, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of phenotype and disease. Mol Cell Endocrinol. 2012;354:3–8. doi: 10.1016/j.mce.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 59.Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 62.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Day T, Bonduriansky R. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am Nat. 2011;178:E18–E36. doi: 10.1086/660911. [DOI] [PubMed] [Google Scholar]
- 64.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 65.Slanchev K, et al. Control of Dead end localization and activity—implications for the function of the protein in antagonizing miRNA function. Mech Dev. 2009;126:270–277. doi: 10.1016/j.mod.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 66.Anderson PD, Nelson VR, Tesar PJ, Nadeau JH. Genetic factors on mouse chromosome 18 affecting susceptibility to testicular germ cell tumors and permissiveness to embryonic stem cell derivation. Cancer Res. 2009;69:9112–9117. doi: 10.1158/0008-5472.CAN-09-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blanc V, et al. Targeted deletion of the murine apobec1 complementation factor (acf) gene results in embryonic lethality. Mol Cell Biol. 2005;B25:7260–7269. doi: 10.1128/MCB.25.16.7260-7269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrmann BG, Koschorz B, Wertz K, McLaughlin KJ, Kispert A. A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature. 1999;402:141–146. doi: 10.1038/45970. [DOI] [PubMed] [Google Scholar]
- 69.Véron N, et al. Retention of gene products in syncytial spermatids promotes non-Mendelian inheritance as revealed by the t complex responder. Genes Dev. 2009;23:2705–2710. doi: 10.1101/gad.553009. [DOI] [PMC free article] [PubMed] [Google Scholar]