Abstract
Gene regulatory networks (GRNs) control the dynamic spatial patterns of regulatory gene expression in development. Thus, in principle, GRN models may provide system-level, causal explanations of developmental process. To test this assertion, we have transformed a relatively well-established GRN model into a predictive, dynamic Boolean computational model. This Boolean model computes spatial and temporal gene expression according to the regulatory logic and gene interactions specified in a GRN model for embryonic development in the sea urchin. Additional information input into the model included the progressive embryonic geometry and gene expression kinetics. The resulting model predicted gene expression patterns for a large number of individual regulatory genes each hour up to gastrulation (30 h) in four different spatial domains of the embryo. Direct comparison with experimental observations showed that the model predictively computed these patterns with remarkable spatial and temporal accuracy. In addition, we used this model to carry out in silico perturbations of regulatory functions and of embryonic spatial organization. The model computationally reproduced the altered developmental functions observed experimentally. Two major conclusions are that the starting GRN model contains sufficiently complete regulatory information to permit explanation of a complex developmental process of gene expression solely in terms of genomic regulatory code, and that the Boolean model provides a tool with which to test in silico regulatory circuitry and developmental perturbations.
Keywords: gene regulatory logic, transcriptional control system, sea urchin embryogenesis, Boolean gene expression
Gene regulatory network (GRN) models formalize the manner in which specification of cellular domains during development is controlled by spatial and temporal gene expression (1). Each cell fate depends on expression of a specific set of regulatory genes, that is, genes encoding transcription factors and signaling molecules. In each domain of the developing organism and at each point in time, the genetic activities and therefore the fates of the cells are determined directly by the regulatory gene products present in the nuclei. The regulatory states constituted by these regulatory gene products are themselves the output of transcriptional control systems encoded in the genome. GRNs thus capture the transcriptional control functions that specify the spatial regulatory states of the embryo. GRNs consist of regulatory genes and the transcriptional interactions that determine their specific patterns of expression. Models derived from experimental studies of developmental GRNs conceptually relate genomic regulatory sequence information to developmental process. Every node in such network models represents a regulatory gene, which is controlled by interactions encoded in genomic cis-regulatory binding sites.
GRN models represent intellectual syntheses of experimental gene expression and cis- and trans-perturbation data, as well as information regarding the developmental process. It is important to note that they cannot be deduced from any single source of data. For an accurate prediction of every individual causal interaction included in a network model, diverse experimental evidence is required. Ultimately, GRN models consist of a heterogeneous conglomeration of predicted interactions. The implication is that, if all regulatory genes and their interactions are known for a given process, a complete network model can be constructed that will causally explain each gene expression event as the outcome of the preceding regulatory states. To test the consistency and conceptual sufficiency of these models is a nontrivial problem in view of their complexity, i.e., the number of players and interactions. The objective of generating GRN models is to achieve a complete explanation of how the developmental process is genomically controlled and this objective can only be attained if the model is accurate and reasonably complete. Here we develop a means of evaluating the sufficiency of a GRN model to explain the progression of regulatory states in time and space observed during a large-scale developmental process.
In all forms of embryonic development, specific regulatory gene readout appears in sharply defined spatial patterns. Thousands of in situ hybridization observations show that each domain of the embryo and each future fate is associated with a specific regulatory state detectably expressed there and not elsewhere. Irrespective of sometimes graded initial inputs, the regulatory state output in space is discrete and discontinuous, that is, Boolean in nature. Fundamentally, this underlies the basic phenomenon that given domains of the embryo generate different defined parts of the organism. Therefore, a primary requirement of a successful GRN model of developmental control must succeed in explaining the Boolean spatial expression of regulatory genes.
Here we construct a Boolean computational model based on one of the most complete GRN models presently available, that encompassing endomesoderm specification in the sea urchin embryo (2–4). The current version of the endomesoderm GRN model contains ∼50 regulatory genes plus the regulatory interactions controlling the specification of endoderm and mesoderm from early cleavage stages (6 h postfertilization) up to the onset of gastrulation (30 h). The Boolean model formalizes all aspects of biological information included in the endomesoderm GRN model, and its design recapitulates the organization and operation of the genomic control system. In the model, as in the genome, each gene has its individual control system; and as is the regulatory information encoded in the genome, the regulatory system of each gene in our computational model is exposed to the active regulatory factors at all times and places. In the model, the gene is turned “on” only if the correct combination of regulatory inputs is present and not otherwise. The nonzygotic inputs are provided as initial inputs in the model just as they are provided maternally in life; they must be sufficient to initiate a regulatory program that afterward runs autonomously. The spatial coordinates in which this model runs are provided by a geometrical definition drawn from the sea urchin embryo. To model the temporal progression of developmental events, our model incorporates the kinetics of gene expression, which were established earlier for this embryo (5).
The Boolean computational model we present here provides a direct test of whether the observed dynamic sequence of spatial and temporal gene expression can be computed by using the information included in the GRN model. Indeed we find that, with a few exceptions, the Boolean computation suffices to reconstruct the observed spatial and temporal gene expression patterns, and this supports the idea that GRN models may contain the necessary information to operate large-scale developmental spatial specification systems. Furthermore, as we show later, the model provides a powerful tool for exploring the consequences of developmental perturbations. The general framework of this computational model should be widely applicable to other systems as well.
Logical Relations Among Computational Model, Data, and Solved GRNs
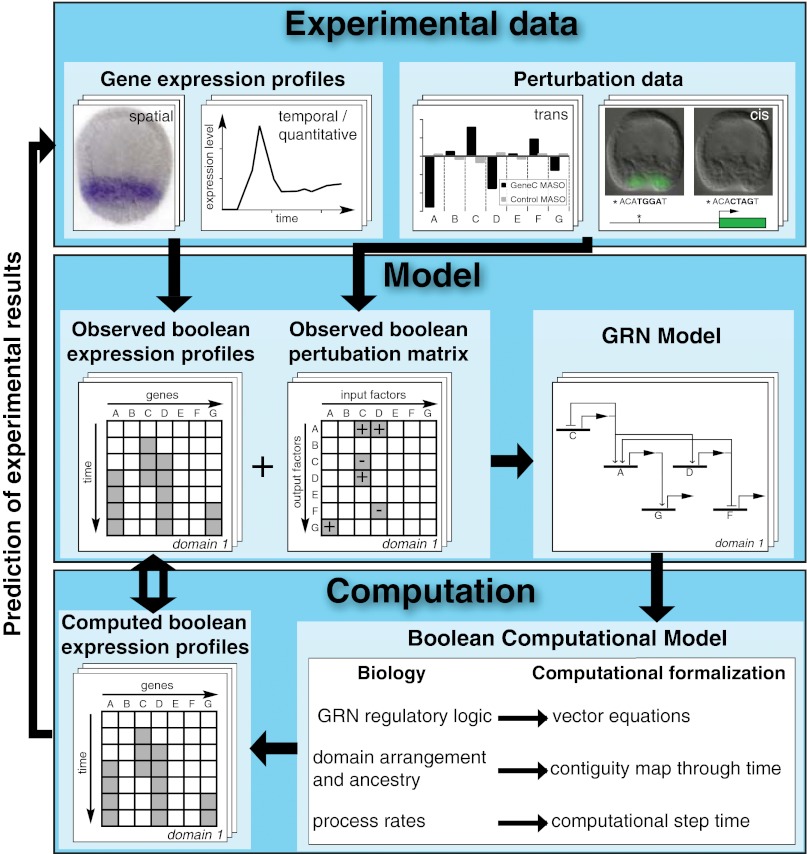
The transitions leading from experimental data to the Boolean computational model are summarized in Fig. 1. Shown in Fig. 1 (Top) are four types of experimental data: spatial and temporal data on regulatory gene expression; results of trans perturbation experiments in which the activity of each regulatory gene is interrupted and the effects on other regulatory genes measured; and results of functional cis-regulatory analyses of specific interactions. In previous studies, the expression of every regulatory gene in the endomesoderm GRN model has been perturbed and the effects on expression of every other gene in the system measured (2–4). The experimental data give rise to two kinds of abstraction, both Boolean in form. Fig. 1 (Middle) shows that these are a matrix of spatial and temporal Boolean gene expression and a matrix of Boolean perturbation results. These matrices provide the nodes and interactions, which, by application of appropriate logic rules, are then integrated into the GRN model. The Boolean computational model we have constructed here represents a further level of abstraction, as shown in Fig. 1 (Bottom). Here we see that the predicted interactions at each gene in the GRN model were used to generate the logic driving the Boolean computational model. The principle used was as follows: the input regulatory information given in the GRN model at each node was used to formulate a logic equation that would express the activity of the gene as a function of its regulatory inputs. In the Boolean model, the regulatory state is thus the output of all logic equations at each point in time and in each spatial domain considered. Thus, were the GRN model essentially sufficient, the logic equations for all network nodes would ideally contain the necessary information to reproduce all observed regulatory states. As we discuss in the following sections, formalisms were developed for the computation of regulatory interactions between network nodes, for the relative positions of different embryonic domains, for the functions of transcription factors responding to signaling cascades, and for kinetic aspects of gene expression. The Boolean model computes expression of genes at every network node through time and space. The result permits a direct comparison between the observed and the computed Boolean gene expression profiles.
Fig. 1.
Diagram summarizing the flow of information and logic relations between experimental data, the GRN model deduced from it, and the Boolean computational model. Gene expression data, systemwide perturbation experiments, and cis-regulatory analyses (Top), combined with the basic facts of the embryonic process, are used to generate abstract time-space expression pattern and the interaction matrices (Middle). These results in turn underlie the design of the deduced genetic circuitry captured in the GRN model. Inputs to the Boolean computational model (Bottom) are the vector equations derived directly from each node of the GRN model; the relative geometry of the interacting embryonic spatial domains considered in the model; and the kinetics with which gene cascades operate in this embryo. The model incorporates essential aspects of the temporal and spatial biology of the embryo, and gives rise to a matrix of specific predictions of where, when, and for how long every gene is individually expressed over a 24-h period. This computed matrix of expression results can now be compared directly with the observed matrix of gene expressions, in normal or perturbed conditions.
Conceptual Architecture of the Model
Our object was to generate a stepwise computation in which, at every hour in real time, the transcriptional states—on or off—of all regulatory genes in the system are assessed, based on the computed outputs at the previous step. To this end, a computational application (GeNeTool) was developed. A flowchart describing the use of the logic equations in GeNeTool is shown in Fig. S1. Boolean computational models have been built to analyze other specific developmental processes, including expression patterns of segment polarity genes in the late cleavage Drosophila embryo (6, 7), and in mouse midhindbrain boundary formation (8). The model we describe here has many features that allow us to capture the causal impact of cis-regulatory function, signaling interactions, and cell lineage geometry, all of which contribute to the regulatory developmental biology of the sea urchin embryo. In brief, the model operates as follows: each node of the GRN model is computationally represented in the computational model by explicit statements, vector equations, capturing its cis-regulatory interactions. At each node, these statements are used to compute the nodal output at each step in time and in each spatial domain. These outputs are then fed to all the nodes in the system at the next step, and the computation is repeated. It is important to realize that this is a synthetic de novo computation and not a simulation in the sense that it simply restates in different language what is already included in the previous model. Thus, the starting GRN has no dynamics, nor does it encompass the geometrical features required for integration of signaling with gene expression as we describe later; nor is the output iteratively fit to the observed expression patterns as in a typical simulation. The goal here is not to simply reproduce observed expression patterns with a specific set of network interactions. The goal is instead to test the experimentally derived GRN topology and identify lacunae in its informational content. The outcome is comparable to an automaton that, when it has been fed the initial conditions, runs by itself until the end of the process considered.
The features of the developmental regulatory system served as direct guiding principles for the organization of our model, as follows.
GRNs.
The sources were for the skeletogenic GRN (2); for the mesodermal GRN (3, 9), and additional data published online (http://sugp.caltech.edu/endomes/#BioTapestryViewer); and for the anterior and posterior endoderm GRNs (3, 4). A current version of the GRN models for the domains considered here shortly before gastrulation is reproduced in Fig. S2. The essential point is that we used the informational content of the GRNs as the source of the specific relations stated in each vector equation in the Boolean model. All nodes in the GRN models are represented by individual vector equations. GRN models differ from the Boolean model in that they explicitly display subcircuit topology with its discrete functionality (10, 11), whereas these features are implicit in the Boolean model.
cis-Regulatory Function.
In the genome, the sequences that determine the expression or silence of regulatory genes are located in their cis-regulatory modules. Inputs may function together on a given cis-regulatory module according to “and,” “or,” or “not” logic, or combinations of these (12). Experimentally, “and” logic is detected by perturbation or cis-regulatory results, which show that interference with either single input of two severely depresses output relative to the level obtained when both are present. “Not” (i.e., repression) functions are typically revealed by spatially ectopic expression when a spatial repressor is made to be absent. To represent cis-regulatory function, we constructed a Boolean logic statement for every regulatory gene encompassed in the respective GRN models. All inputs that regulate a respective gene, and their logical relation to one another, are captured in vector equations (the term “vector equation” reflects the matrices of gene expression in space and time that these equations generate). Where alternatively acting cis-regulatory modules must be taken into account, each is represented by a separate vector equation. For genes with multiple modules/vector equations, the intermodular logic is explicitly stated. Thus, each logic statement computes the output of a network node given the availability of the specific required inputs and the resulting logic functions. For example (in words), a vector equation for gene Y might state that if an input A is 1 and another input B is 1 and if the input (from a repressor) C is 0, then the output of Y is 1, else 0. The inputs are, of course, the outputs of other genes in the system. The vector equations directly represent, in logic processing terms, the hardwired identity, the input repertoire, and the genomic logic processing functions of the cis-regulatory systems controlling these genes in life. The complete set of 75 vector equations can be seen in Fig. S3.
Computation of Regulatory States.
Relevant maternal inputs provide the initial conditions and are used as the starting regulatory state. Beginning at 5 h, when the only inputs are the initial conditions, every vector equation is operated and the outputs (0 or 1) are computed. These outputs provide the inputs for the next step in the computation. The computation was carried out at 1-h intervals in each of the four domains. At each point in time, the regulatory state in each spatial domain is the sum of the computed outputs of all vector equations. In sum, the inputs at each step in the computation are the computed outputs of the previous steps.
Signaling.
In development, after very early stages, intercellular signaling is required for the specification of regulatory state domains. Spatial expression of ligands by given cells is a result of genomically encoded GRN linkages that result in the transcription of a gene encoding a signaling ligand. Recipient cells activate novel regulatory genes, thus altering the local regulatory states. A mechanistic feature of many—perhaps all—signaling systems used in embryonic development for this inductive purpose is that there is always a responsive transcription factor present in the cells anyway, the state of which is changed by the signal transduction biochemistry. Despite the variety of these biochemistries, the response factor can generally be considered to exist in one of two states at all times: either it becomes activated in cells receiving the signal, whereupon it interacts with its target sites in the cis-regulatory apparatus of the target regulatory genes; or, in the absence of the signal, the same factor acts as an obligatory repressor of those same target genes (11, 13). Thus, in our model, for every inductive signaling interaction, we defined a “J” (Janus) factor, the state of which is 1 in cells receiving the ligand that triggers the signal transduction system activating that factor, but 0 in other cells. For example, Suppressor of Hairless [Su(H)], the Notch signaling responsive transcription factor, is represented as J[Su(H)], the state of which can be 1 or 0, depending on whether a given cell is receiving a Delta/Notch signal. The vector equations for signal responsive genes thus incorporate the value of the J-factors in given times and places. This computational apparatus can accommodate signals at any range, but, in the sea urchin embryo before gastrulation, the relevant signals operate only at short range (14, 15).
Spatial Domains.
The model encompasses four spatial domains as they can be perceived based on differential gene expression in the sea urchin (Strongylocentrotus purpuratus) embryo. These are the skeletogenic domain, the nonskeletogenic mesoderm domain, the future anterior endoderm domain, and the future posterior endoderm domain (a summary of the developmental fates and spatial locations of these domains is shown in Fig. S4). Needless to say, each domain arises from precursor cells that traverse several stages of specification. In the model, each domain encompasses the terminal cell fate domain and all its precursors at earlier times in development.
Embryonic Geometry.
The sea urchin embryo generates a canonical cell lineage and cleavage pattern, which essentially means that cells of given lineage and regulatory state are located in canonical positions with respect to one another at each stage. The spatial disposition of cell lineages in the developing sea urchin embryo, and their developmental fates, are summarized digitally in Fig. S5A. The relative positions of the four domains changes in the course of development, determining the potential target of interdomain signals. The Boolean model incorporates the changing embryonic geometry by use of the relationships shown in Fig. S5B. Briefly, for every domain in the model, the relative position to all other domains is defined at every point in time by using two positional terms: contiguous (CC) for adjacent domains and noncontiguous (NCC) otherwise. From the beginning to the end of the period considered, information is available to the model on which cells are immediately adjacent to which other cells (for very short range signals such Delta), or which cells are close to given other cells, for signals that (in sea urchin embryo) diffuse only a few cell diameters from the cells where the ligand is transcribed. The range of target domains for given signaling interactions is incorporated in the vector equations for every J factor. For example, the activity of J(SuH) depends on the expression of the delta ligand gene in a CC domain (Fig. S3).
Real-Time Kinetics of Sea Urchin Embryo Development.
The basic metric of regulatory progress in development is the time interval between activation of a given regulatory gene and the activation of an immediate downstream target regulatory gene. This interval, which we shall term the step time, is a function of the basic kinetics of the molecular processes of transcription, RNA turnover, protein synthesis and turnover, transcription factor–DNA interaction, and cis-regulatory activity. In the sea urchin embryo, which develops at 15 °C, biosynthetic processes are much slower than, for example, in Drosophila, and it requires several hours for successive changes of regulatory states to occur. In 2003, Bolouri and Davidson (5) modeled these kinetics for sea urchin embryos living at 15 °C, using a large set of kinetic parameters previously measured for this system. This model was based on a first-principles treatment of cis-regulatory occupancy, a probabilistic mathematical argument that the rate of transcription relative to the measured maximum possible rate depends on the cis-regulatory occupancy, and standard synthesis/turnover kinetics (Fig. S6). The step time that emerged was approximately 3 h. Many subsequent direct observations on S. purpuratus embryos made in the course of GRN analysis confirmed that this canonical computation approximates reality for specific cases (e.g., refs. 2, 4). Recently, we also noticed that rates of regulatory gene transcription are remarkably similar to one another in the sea urchin embryo, varying by only a factor of approximately two from ∼100 molecules per embryo-hour (16).
In the present automaton model, we imposed a priori the canonical step time obtained for the general case in the earlier calculation (Fig. S6). As the results discussed later show, this assumption works remarkably well. In the model, dynamic animation of the computed expression patterns was achieved by assuming the canonical 15° sea urchin embryo step times (5). Throughout most of the pregastrular period, the step time was set at 3 h, in accord with the results of this calculation, although, as empirical evidence suggested, the step time is a little faster very early in development, when in the model was set at 2 h. However, because signal transduction biochemistry is relatively rapid, in signaling interactions, the downstream transcriptional effects were assumed to begin without delay when the signal has become available. The step times were included in the vector equations for each gene. As, in the model, we are concerned only with transcriptional output of each gene in consequence of the transcription of its regulatory inputs, a time lag is required to account for the real-time pace of transcription, translation, accumulation of biosynthetic products, and occupancy at the cis-regulatory system, which, as described earlier, constitutes the step time. For every input variable stated in the vector equations, the time required for the output gene to be affected after the input gene is turned on is stated in hours, i.e., the step time (e.g., a 3-h step time for an input from gene A would be stated as “if AT-3 gene A = 1…”; Fig. S3). Although the step time is approximately 3 h, of course, the regulatory events occur asynchronously, and so it was necessary in the model to compute the regulatory states at intervals that are only a fraction of the step time, and, as noted earlier, the computation was performed at 1-h intervals. No finer assessment interval is warranted by the pace of developmental events in this embryo.
Computed Spatial Gene Expression
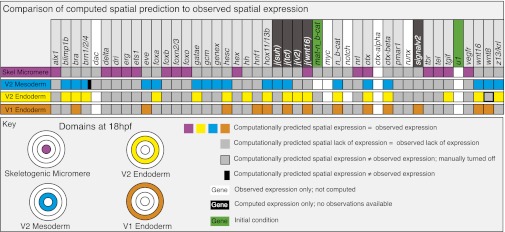
The main function of the genomic regulatory system for development is specification of spatial patterns of gene expression. The first test for the usefulness of the Boolean computational model is therefore its ability to reproduce the correct regulatory state for each regulatory domain. The regulatory state was defined for each of the four endomesodermal regulatory domains as the sum of all regulatory genes expressed in that domain for at least one time interval between 18 and 30 h, except for the early mesoderm GRN, for which a 12- to 16-h time window was used because the later GRN model is not complete. The comparison between computed and observed spatial regulatory states is shown in Fig. 2. Transcriptional control is not yet understood for a few regulatory genes in the GRN model, and no vector equations could be formulated for these genes (Fig. 2, genes on white background). They were nonetheless incorporated into the Boolean model as they are known to provide inputs for other genes. For all J factors functioning downstream of signaling interactions, plus for genes predicted to be included in the GRN circuitry but not yet identified, there are no observed gene expression data available (Fig. 2, genes on black background). However, we have generated observed and computed gene expression data for 33 regulatory genes in the four domains, allowing 132 direct comparisons (Fig. 2, genes on gray background). Of these comparisons, all but two showed a perfect match between observed and computed spatial expression (Fig. 2). One exception was brn1/2/4, for which the computational model predicted expression in the early mesoderm, where expression of this gene has not been observed, in addition to the correctly computed expression in the veg2 endoderm. The reason is that all currently known regulatory inputs into brn1/2/4 are present in the early mesoderm, predicting that additional factor(s) must regulate brn1/2/4. The second deviation between computation and observation is caused by wnt8, which fails to clear from the veg2 endoderm domain in the computational model, indicating a missing repressor in the GRN model. These two exceptions aside, Fig. 2 proves that the regulatory information included in the model suffices to explain almost every known spatial regulatory gene expression pattern in the endomesoderm up to 30 h of development.
Fig. 2.
Comparison of computed and observed spatial expression patterns. Upper: All regulatory genes included in the model. Experimental expression data are summarized digitally online (http://sugp.caltech.edu/endomes/#BioTapestryViewer). Expression in this chart indicates presence of the computed or observed transcript at any time between 18 and 30 h, except for the V2 mesoderm domain, where the relevant interval is 12 to 16 h (see Computed Spatial Gene Expression above). As indicated in the key, the actual comparison between observed and computed results, i.e., for genes in which both types of data exist, pertains to the colored and gray squares. A discrepancy between computed and observed expression is indicated by a black bar. For genes or regulatory operators shown on black backgrounds, no observational data are available; for genes on white backgrounds, no upstream regulatory information is available, and the activation of these genes was not computed. Where such genes are expressed, or for maternal nuclear β-catenin, in which it is present, the time/space domain is shown in white; where they are observed not to be expressed is shown in gray. The disposition of the four spatial domains in the chart, in embryos viewed from the vegetal pole, is color-coded (Fig. S4 shows a general diagram of embryogenesis).
Clocklike Dynamics of Spatial Gene Expression
The comparison of computed and observed spatial expression of the many regulatory genes in Fig. 2 extends over a broad temporal window (18–30 h). However, when examined in detail, each gene displays its own individual temporal pattern of activation and sometimes repression. The temporal resolution of the observed expression patterns is based on whole mount in situ hybridization data, which exist at a time resolution of approximately 3 h for most genes included up to the 30 h developmental stage (data provided at http://sugp.caltech.edu/endomes/#BioTapestryViewer). In addition, a NanoString data set provides transcript accumulation measurements for all these genes at 1-h resolution (16).
The computed results are shown with respect to the observed expression patterns in Fig. 3, for every gene at every hour for every domain. Discrepancies between computed and observed expression in real time are considered significant if greater than the step time of 3 h (Fig. 3, solid black bars), whereas deviations less than 3 h (Fig. 3, open black bars) may not be real, as this is the limit of resolution of observed spatial data. We see, perhaps surprisingly, that the assumption of a uniform, canonical step time reproduces the observed dynamics within the experimental resolution for the vast majority of genes with no significant discrepancies from data (Fig. 3, colored and gray rectangles). In the skeletogenic domain, aside from a few 1-h discrepancies (beyond the 3-h observational variance), the model turned on foxb 3 h too early and failed to turn off foxn2/3 for 4 h when it should have; in the mesoderm domain, over the 6- to 18-h period, there was only one 1-h discrepancy; in the veg2 endoderm, brn1/2/4 was briefly turned on when it should not have been, and was later turned on a couple of hours too early, and tgif was turned on 3 h early; in veg1 endoderm, the only serious discrepancies were that otxβ was turned off 6 h too early and z13 was turned on for 4 h when it should not have been. For three genes, manual corrections were imposed to avoid promulgating downstream effects (Fig. 3, boxes with heavy lined black borders): delta and wnt8 cease to be expressed in veg2 endoderm after certain times for reasons not yet known, and these genes were turned off manually at the observed times; similarly, the known inputs into foxa (17) do not indicate why this gene continues to be transcribed after 24 h, and this activity was set manually for the 24- to 30-h interval. However, considering that there are 2,772 time/space gene expression domains computed in the model and shown in Fig. 3, this is a remarkably small frequency of significant temporal discrepancies, all of relatively brief duration, and there was only one spatial error.
Fig. 3.
Computed vs. observed spatial expression in real time. The genes are indicated at the top and the comparison symbolism is as in Fig. 2; i.e., plain filled colored or gray squares indicate exact congruence between computed and predicted expression. The four embryonic spatial domains (A–D) are separately portrayed, each against 1- to 30-h time scales. Open rectangles indicate discrepancies <3 h; as described in the text, as the observations have only 3-h resolution, these discrepancies may not be real. Discrepancies >3 h are indicated by black solid rectangles. The green striped bars indicate maternal mRNAs that are not part of the zygotic GRN model and therefore not computationally predicted. Some of these maternally supplied factors provide initial inputs to the computation. The three places where the whole column is striped (delta, foxa, and wnt8) indicate phases of expression or nonexpression known to occur, but not explained, and set manually in the computation.
The discrepancies are individually valuable items of information, for they pinpoint exactly what remains to be solved in terms of transcriptional causality. However, the main conclusion is that imposition of a uniform step time in the computation results in a generally accurate representation of the dynamics of the spatial expression patterns. The regulatory genes of the sea urchin embryo operate more or less similarly to one another, and, like a clock, the pace of which is set by the kinetics of the basic processes of transcription molecular biology. The application of a correct step time is important for the overall performance of the Boolean model. To test the implications of variations in the step time, we ran the model by using different step times. The catastrophic performance of the model when the step time was changed from 3 h to 4 h is shown in Fig. S7. Thus, we discovered that the embryo regulatory system operates largely like a clock by using a 3 h step time. This fact obviates detailed, gene-by-gene synthesis/turnover kinetic analysis. The few discrepancies most likely indicate missing regulatory inputs, rather than specific kinetic deviations from this general conclusion.
In Silico Perturbations
Sea urchin embryos have been experimentally exposed to various perturbations. These types of experiment offer an opportunity to challenge the completeness of the explanatory power of the Boolean model, again by comparing computational prediction to observed gene expression or phenotypic results, but now under specifically altered conditions that mimic in silico well studied experimental perturbations. Here we consider four such projects.
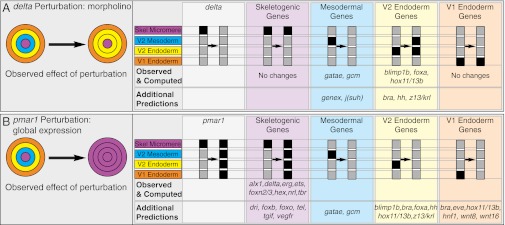
In the first of these, we mimicked extinction of delta expression by injection into the egg of delta morpholinos (4, 18, 19). The experimental result is loss of almost all mesodermal gene expression from the veg2 cells, which normally give rise to all nonskeletogenic mesoderm (i.e., the ring of cells shown in blue in Figs. 2 and 4). Instead, as symbolized in Fig. 4A (which represents the embryo at approximately 18 h), these cells express veg2 endodermal genes, which are now transcribed ectopically in a double rather than single ring of cells (4). When delta gene expression is manually turned off in the model, the computational result is precisely consistent with the experimental result (Fig. 4A; Fig. S8A shows complete detailed results). Only the two early mesoderm genes gcm and gatae have been examined in delta morpholino embryos, affording a direct comparison with data, and the expected loss of expression of these genes was obtained in silico. Similarly, although only hox11/13b, foxa, and blimp1 among veg2 endodermal genes have been studied experimentally in this regimen, the Boolean model again produced the expected result of predicted ectopic expression in the presumptive mesodermal ring of cells. The model also predicted that the same happens for the other veg2 endoderm genes in the model that have not yet been examined experimentally.
Fig. 4.
In silico perturbation experiments. Left: Experimental perturbation mimicked in the computation and diagram of its general result. The five panels containing gray and black columns (Right) indicate respectively the in silico perturbation and its computational effects in the four embryonic domains. The specific genes affected in the computation are shown below: “Observed & Computed” indicates genes with a comparable experimentally observed change in expression; “Additional Predictions” indicate genes that have not been experimentally studied in these perturbations. (A) Interference with skeletogenic micromere expression of delta and (B) global overexpression of Pmar1. The detailed results as computed for all genes in the model are shown in Fig. S8 A and B.
In a second perturbation, we mimicked experimentally induced global expression of the Pmar1 repressor by injection of pmar1 mRNA into the egg (2, 20, 21). The observed experimental result is dramatic: the whole embryo is converted into a ball of mesenchymal cells expressing skeletogenic genes, as indicated diagrammatically (Fig. 4B). This is also just what occurred in silico when pmar1 is manually activated in all domains from 5 h on: eight skeletogenic genes have been shown experimentally to be globally expressed in pmar1 embryos, and, in the computation, all eight were expressed in all domains (Fig. 4B). An additional prediction is that the same should be true of the remaining skeletogenic genes as well (Fig. S8B shows detailed results). Furthermore, the Boolean model predicted that expression of mesodermal genes, veg2 endoderm genes, and veg1 endoderm genes would be turned off. This result is certainly consistent with the powerful global mesenchymal phenotype (20).
In a third perturbation, we directly mimicked a recent molecular biology experiment (4) in which the quantitative and spatial effects on the whole GRN of blocking hox11/13b expression with hox11/13b morpholino was determined throughout the pregastrular period. This was done by manually extinguishing hox11/13b expression in veg2 and veg1 endoderm (Fig. S8C). With the exception of two discrepancies that were merely 1-h temporal deviations, every observed change in gene expression was generated computationally.
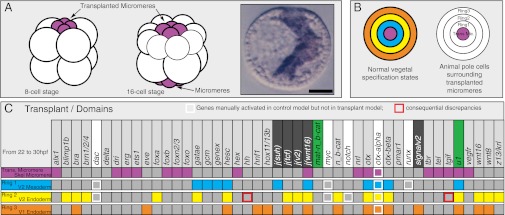
A fourth perturbation was more challenging. Here we attempted to use the Boolean model to determine whether the computed regulatory relationships suffice to predict the results of a famous blastomere transplantation experiment reported by Hoerstadius in 1935 (22) and, 58 y later, further explored by us by using a molecular marker (23). In this experiment, the four fourth cleavage skeletogenic micromeres are removed from a donor embryo and transplanted to the animal pole of an otherwise normal early cleavage embryo possessing its own set of vegetal micromeres. The result is that a complete second gut, plus a second set of nonskeletogenic mesoderm lineages (18), is induced to form from the cells at the animal pole of the embryo (Fig. 5A, diagram). Essentially, this means that the signals emitted from the transplanted skeletogenic micromeres elicit the encoded developmental GRNs, and this suffices to recreate all the regulatory and spatial relationships that normally generate the concentric regulatory states of the vegetal pole of the embryo, as shown diagrammatically in Fig. 2. Were we to mimic the micromere transplantation experiment in our model, would it regenerate in a computationally naive patch of cells the same set of concentric mesodermal and endodermal regulatory states as it generates (Figs. 2 and 3) at the normal vegetal end? We modeled these naive cells by endowing them with the global maternal factors, but without the localized nuclear β-catenin that normally occurs in veg2 and veg1 lineages (24, 25). The “transplanted” micromeres were allowed all their own normal specification functions, as these are autonomous with respect to the rest of the embryo. The surrounding naïve cells were considered as forming three domains (Fig. 5B), the immediately adjacent cells (ring 1), the cells surrounding these (ring 2), and the cells surrounding ring 2 (ring 3). We now asked whether the in silico regulatory states of ring 1 would equal those of the mesoderm, and those of rings 2 and 3 the regulatory states of veg2 and veg1 endoderm, respectively. Fig. 5C shows that this is exactly what occurred; the only discrepancies are two genes that are dependent on inputs from two genes for which vector equations are lacking (dac, myc). If we manually activate dac and myc expression, there are no discrepancies.
Fig. 5.
Computational recreation of the micromere transplantation experiment. (A) Diagrammatic summary of experiment in which a set of micromeres is transplanted to the animal pole of an eight-cell embryo; by the next cleavage, the endogenous set of micromeres has also appeared; and, ultimately, a complete second gut and mesoderm are induced to form. (B) Conceptual organization of animal pole cells surrounding the descendants of the transplanted micromeres, at ∼200- to 400-cell stage, in similar geometric relation as at the vegetal pole. (C) Chart comparing computed regulatory states of ring 1 vs. V2 mesoderm, ring 2 vs. V2 endoderm, and ring 3 vs. V1 endoderm; symbols are as in Figs. 2 and 4 and analysis was carried out for the 22- to 30-h interval. The comparison between computed expression patterns in the normal vegetal region of the embryo and computed expression patterns surrounding the in silico transplanted micromeres is almost perfect.
In considering the import of these in silico perturbations, we note that, except for the hox11/13b case, none of the experimental perturbation results we sought to reproduce were used in any specific way to construct the linkages in the GRN models on which the vector equations of Fig. S3 were based. Thus, a model based on functional analysis in normal development correctly predicts independent experimental results in unnatural contexts: this provides a proof of principle that the genetic interaction system in the model operates essentially as in the embryo. Furthermore, in each case, the model extends our spotty understanding of the consequences of the perturbations, which is limited by the genes the experimentalists happen to have investigated, to all of the genes in the whole system, producing, for example, the additional predicted consequences indicated in Fig. 4.
Discussion
Spatial gene expression is both the driver and product of developmental mechanisms. Here we show that observed embryonic regulatory gene expression in space and time can be recreated, systemwide, by reducing these mechanisms to the underlying gene regulatory interactions. We apply a Boolean interpretation to regulatory gene expression and regulatory gene interaction. This approach is consistent with the visibly apparent Boolean patterns of spatial gene expression, a fundamental property of embryonic development. Given cells at given times express given regulatory genes at detectable levels or do not significantly express them. Diverse mechanisms conspire to produce this basic property.
First, in sea urchin embryos, the transcriptional cascade kinetics themselves contribute in a crucial way. Our earlier analyses showed that the point at which a downstream gene is activated with respect to the upstream drivers long precedes the attainment of driver steady state (5), and this means that the kinetics of gene cascades in the embryo are relatively level-insensitive and insensitive to synthesis and decay rates. The next gene in a sequence goes on as soon as there is sufficient driver transcription factor in the system. In accord with this, measurements show that levels of regulatory gene transcripts in different individual batches of eggs in these polymorphic animals frequently vary by twofold or more (16): the developmental gene regulatory system cares only about on and off, and level control is sloppy. Of course, as our high-resolution time courses for all genes included in this model show (16), the expression levels change through time. However, our model is not aimed at explication of expression kinetics, but rather at explication of spatial domain of expression, and, for this, what is causal is the availability of activators and repressors. As we have seen, in this system, it takes approximately 3 h from the time a gene is detectably activated to the time its product can affect an immediate target gene. Thereafter, the levels of the gene product usually continue to increase within the same spatial domain without change in the set of affected target genes.
Second, the boundaries of Boolean gene expression territories are in embryonic development usually set by GRN subcircuits that use repression. A variety of subcircuits of different design are known that accomplish this function (10, 11). Although repression itself is usually modeled as a continuous equilibrium function, this is a severe oversimplification for animal cells because, following binding of the repressor, secondary changes in chromatin structure and composition follow that may produce irreversible repression states that can last for the life of the cell. Thus, per se, repression behaves in a Boolean manner in space and in time.
Third, even when there are graded upstream driver inputs, Boolean gene expression outputs occur downstream as a result of particular GRN subcircuit designs that function hysteretically (10). To take one example, in the vertebrate neural tube, a dynamically changing Sonic Hedgehog gradient input is interpreted by the encoded GRN to produce sharp stripes of regulatory gene expression (26). Thus, whereas the input is graded, the spatial regulatory output is Boolean. The majority of such mechanisms basically devolve from the input/output functions generated by the designs of a variety of particular GRN subcircuits, which can be said to perform “Booleanization” functions. In general, signaling interactions in development produce essentially Boolean regulatory outputs. Most known developmental signaling pathways function by use of a response system that, in cells receiving the signal, promote target gene expression, and, in cells not receiving it, prevent it, by repressing the same target genes. This is a feature captured directly in the structure of our model, as described earlier.
The aim of this computational exercise was to assess the sufficiency of a system-level causal explanation for development. This explanation, couched in the format of the GRN model, provided the crucial logic functions of the computational model. The success of the computational model in reproducing developmental gene expression, shows that our knowledge of the underlying GRNs is relatively complete. What we do not know is specifically circumscribed. Lacunae in the model fall into two classes. For the several “white genes” of Fig. 2, we lack sufficient experimental cis-regulatory and perturbation data on which to base vector equations that could explain their activation, but because their downstream effects are known, these genes are included subsequently in the model. Second, the model executes a few miscomputations, in which the control operations built in fail to explain why a given gene turns off at a certain time, or why it continues to be transcribed (Fig. 3). Identification of these gaps in the GRN model is one of the useful outcomes of the computation. In addition, as we have seen, the GeNeTool Boolean model lends itself readily to predictive exploration of cis- and trans-perturbations of the GRN.
The most important general outcome of this work is that an assumed mechanism, based exclusively on experimentally established control functions, suffices to explain specifically almost all observed regulatory gene expression in space and time. This model contributes a step-by-step mechanistic understanding of how the complicated GRN shown in Fig. S2 actually works. There remains little room for causal explanation of spatial regulatory gene expression by any other mechanism. The formalism presented here provides a general means of representing in silico the genomic regulatory system underlying animal body plan development and further applications may be anticipated, ranging from evolution to synthetic developmental biology.
Materials and Methods
A computational platform, named GeNeTool, was constructed to enable the model computation. The graphics and design functionalities of GeNeTool are to be described in detail elsewhere. However, Fig. S1 shows a flowchart that describes the overall process of the computation, and the program can be obtained from the authors on request. A brief summary of the rules of logic operation, which accommodate temporal and relative spatial relationships in the model, is presented in Fig. S9.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants HD 037105 (to E.H.D.), GM 061005 (to E.H.D.), and HD 0656016A (to R. A. Cameron).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207852109/-/DCSupplemental.
References
- 1.Davidson EH. The Regulatory Genome. Gene Regulatory Networks in Development and Evolution. San Diego: Academic/Elsevier; 2006. [Google Scholar]
- 2.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature. 2011;474:635–639. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: Initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sánchez L, Chaouiya C, Thieffry D. Segmenting the fly embryo: Logical analysis of the role of the segment polarity cross-regulatory module. Int J Dev Biol. 2008;52:1059–1075. doi: 10.1387/ijdb.072439ls. [DOI] [PubMed] [Google Scholar]
- 7.Albert R, Othmer HG. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J Theor Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittmann DM, et al. Spatial analysis of expression patterns predicts genetic interactions at the mid-hindbrain boundary. PLOS Comput Biol. 2009;5:e1000569. doi: 10.1371/journal.pcbi.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 10.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peter IS, Davidson EH. Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett. 2009;583:3948–3958. doi: 10.1016/j.febslet.2009.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci USA. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 14.Davidson EH. How embryos work: A comparative view of diverse modes of cell fate specification. Development. 1990;108:365–389. doi: 10.1242/dev.108.3.365. [DOI] [PubMed] [Google Scholar]
- 15.Davidson EH, Cameron RA, Ransick A. Specification of cell fate in the sea urchin embryo: Summary and some proposed mechanisms. Development. 1998;125:3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- 16.Materna SC, Nam J, Davidson EH. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr Patterns. 2010;10:177–184. doi: 10.1016/j.gep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de-Leon SB, Davidson EH. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc Natl Acad Sci USA. 2010;107:10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 19.Materna SC, Davidson EH. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev Biol. 2012;364:77–87. doi: 10.1016/j.ydbio.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 21.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoerstadius S. Uber die determination im verlaufe der eiachse bei seeigeln. Pubb Staz Zool Napoli. 1935;14:251–479. doi: 10.1007/BF02569028. [DOI] [PubMed] [Google Scholar]
- 23.Ransick A, Davidson EH. A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science. 1993;259:1134–1138. doi: 10.1126/science.8438164. [DOI] [PubMed] [Google Scholar]
- 24.Logan CY, McClay DR. The allocation of early blastomeres to the ectoderm and endoderm is variable in the sea urchin embryo. Development. 1997;124:2213–2223. doi: 10.1242/dev.124.11.2213. [DOI] [PubMed] [Google Scholar]
- 25.Croce J, et al. Wnt6 activates endoderm in the sea urchin gene regulatory network. Development. 2011;138:3297–3306. doi: 10.1242/dev.058792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balaskas N, et al. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.