Abstract
Quorum sensing allows bacteria to sense and respond to changes in population density. Acyl-homoserine lactones serve as quorum-sensing signals for many Proteobacteria, and acyl-homoserine lactone signaling is known to control cooperative activities. Quorum-controlled activities vary from one species to another. Quorum-sensing controls a constellation of genes in the opportunistic pathogen Pseudomonas aeruginosa, which thrives in a number of habitats ranging from soil and water to animal hosts. We hypothesized that there would be significant variation in quorum-sensing regulons among strains of P. aeruginosa isolated from different habitats and that differences in the quorum-sensing regulons might reveal insights about the ecology of P. aeruginosa. As a test of our hypothesis we used RNA-seq to identify quorum-controlled genes in seven P. aeruginosa isolates of diverse origins. Although our approach certainly overlooks some quorum-sensing–regulated genes we found a shared set of genes, i.e., a core quorum-controlled gene set, and we identified distinct, strain-variable sets of quorum-controlled genes, i.e., accessory genes. Some quorum-controlled genes in some strains were not present in the genomes of other strains. We detected a correlation between traits encoded by some genes in the strain-variable subsets of the quorum regulons and the ecology of the isolates. These findings indicate a role for quorum sensing in extension of the range of habitats in which a species can thrive. This study also provides a framework for understanding the molecular mechanisms by which quorum-sensing systems operate, the evolutionary pressures by which they are maintained, and their importance in disparate ecological contexts.
Keywords: bacterial communication, systems biology, transcription control
Bacteria use quorum-sensing signals to communicate with each other and control gene expression in a cell density-dependent manner. Many species of Proteobacteria use diffusible acyl-homoserine lactones (AHLs) as quorum-sensing signals. AHLs are produced by signal synthase enzymes and are detected by signal-specific transcriptional regulators. AHL quorum-sensing circuits regulate a wide spectrum of phenotypes in a diverse array of α-, β-, and γ-Proteobacteria (1). Interspecies differences in quorum regulons often are a reflection of the diverse habitats that bacteria occupy, and quorum-controlled phenotypes often play a crucial role in niche persistence. The classic example is quorum control of luminescence in Vibrio fischeri, which allows this bacterium to discriminate between its free-living, low-population-density seawater habitat and its high-density symbiotic habitats, the light organs of certain fish and squid (2, 3). It is well established that there are species-specific differences in quorum regulons, but there is little information regarding the possibility of intraspecies strain-specific differences. We hypothesized that, particularly for versatile species that occupy diverse niches, there might be a shared core of quorum-controlled genes and, in addition, strain-variable quorum-regulated genes that reflect adaptations to the habitats from which strains are isolated. We tested our hypothesis using isolates of the metabolically versatile γ-Proteobacteria species Pseudomonas aeruginosa.
P. aeruginosa has been isolated from diverse environments. It can be found in soil and water, as a member of the normal microbiota of eukaryotes or as an opportunistic pathogen in a wide range of hosts including plants and humans. Comparative genomic analyses of multiple P. aeruginosa strains have identified core (shared) and accessory (strain-variable) genome sequences (4). Evidence indicates that accessory genes encode functions associated with adaptation and niche diversification (4). P. aeruginosa has a quorum-sensing system comprising two AHL synthases and three receptors. The LasI synthase produces 3OC12-HSL, for which there are two receptors, LasR and QscR. The RhlI synthase produces C4-HSL, for which the receptor is RhlR. There are indications that, although the complete complement of synthase and receptor genes is conserved among strains, there are differences in the quorum-controlled genes (5), and some strains from certain habitats contain LasR mutations (6–8).
Much of the existing data on genes controlled by quorum sensing in P. aeruginosa derive from studies of a single laboratory strain, PAO1 (9-11) an extensively passaged isolate from a wound infection (12). Here we use RNA-seq to identify genes in the quorum regulons of seven P. aeruginosa strains isolated from disparate environments. Specifically we use strain PAO1 as a reference. We generated and annotated draft genome assemblies of the other six isolates. We generated lasI, rhlI quorum-sensing mutants of each isolate and compared the transcriptomes of lasI, rhlI mutants of all seven strains, with and without added AHLs, to each other. As we predicted, there was a set of core quorum-controlled genes in the core genome, and there were elements of the accessory genome that showed quorum-sensing control. There also were genes in the core genome that showed strain-to-strain variation with respect to quorum-sensing control.
Results
Quorum-Sensing Circuit Is Conserved Among Environmental and Clinical P. aeruginosa Isolates.
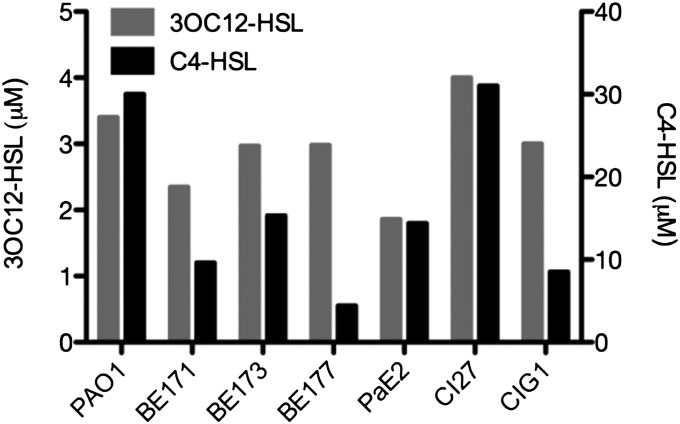
We examined intraspecific diversity in quorum-regulated gene expression by examining seven P. aeruginosa strains, including four environmental isolates, two clinical isolates from chronic cystic fibrosis (CF) lung infections, and the laboratory strain, PAO1 (Table S1). Some information regarding genome content and assembly statistics for the draft genomes is provided in Table 1, and annotations are available at www.ncbi.nlm.nih.gov/genome. The draft genomes show a pangenome for the seven strains consisting of 7,423 genes and a shared core genome of 4,449 genes. To determine if all strains in our panel produced the two P. aeruginosa AHLs, we tested stationary-phase cultures by using bioassays. All strains exhibited generally similar growth rates and produced both 3OC12-HSL and C4-HSL at micromolar levels (Fig. 1). These data indicate that the prototypical P. aeruginosa quorum-sensing circuit is conserved and operational in all examined strains. We generated lasI, rhlI signal-generation mutants (Materials and Methods) which did not produce detectable levels of AHLs.
Table 1.
Content and assembly statistics for the draft genomes and the PAO1 reference genome
| Strain | Source | Size (bp) | Contigs | Coding sequences | Accessory genes | (G+C) content (%) |
| PAO1* | Wound isolate | 6,264,404 | 1 | 5,571 | 1,122 | 66.56 |
| BE171 | Soil | 6,385,231 | 218 | 5,487 | 1,038 | 66.38 |
| BE173 | Air | 7,170,615 | 1138 | 5,828 | 1,379 | 65.69 |
| BE177 | Biofilm | 6,808,690 | 613 | 5,711 | 1,262 | 66.04 |
| PaE2 | Tomato plant | 6,368,819 | 213 | 5,486 | 1,037 | 66.39 |
| CI27 | CF chronic infection | 6,781,513 | 169 | 5,923 | 1,474 | 66.02 |
| CIG1 | CF chronic infection | 6,556,618 | 573 | 5,518 | 1,069 | 65.95 |
*Details for strain PAO1, included for comparison, are from the Pseudomonas genome project (www.pseudomonas.com). G+C, guanine plus adenosine mols percent.
Fig. 1.
AHL concentrations in P. aeruginosa cultures at OD600 3.5. AHLs were not detected in cultures of lasI, rhlI mutants of any isolate.
Identification of Quorum-Sensing–Regulated Genes by RNA-Seq.
As described in the Materials and Methods, RNA-seq libraries were generated by selective cDNA priming with a pool of hexamers, none of which showed a perfect match to any of the P. aeruginosa ribosomal RNAs (rRNAs). This approach enables enrichment of non-rRNA transcripts without a ribosome-depletion step, lowering RNA input requirements and simplifying sample preparation (13, 14). In fact, depending on the isolate examined, the reads mapping to rRNAs ranged from about 30% to about 80%. Overall, the RNA-seq results (Table 2) revealed that at least 89% of the genes for a given strain had their coding sequence covered, indicating that the overall genome coverage afforded by this method was high. Further, sequencing read depth data indicated that the numbers of non-rRNA reads were largely similar for all samples, thus enabling valid comparisons across samples.
Table 2.
Summary of sequencing results
| Strain* | Quorum-sensing condition | Genes with a read (%) | Non-rRNA sequence reads | rRNA sequence reads | Non-rRNA sequence reads (%)† | rRNA sequence reads (%)† |
| PAO1 (1) | Noninduced | 93.0 | 1,228,269 | 3,245,963 | 27 | 73 |
| PAO1 (1) | Induced | 93.3 | 1,283,754 | 4,142,083 | 24 | 76 |
| PAO1 (2) | Noninduced | 95.7 | 1,346,668 | 878,699 | 61 | 39 |
| PAO1 (2) | Induced | 93.8 | 1,148,384 | 987,278 | 54 | 46 |
| BE171 (1) | Noninduced | 97.6 | 2,157,331 | 1,935,792 | 53 | 47 |
| BE171 (1) | Induced | 97.8 | 1,604,273 | 965,877 | 62 | 38 |
| BE171 (2) | Noninduced | 97.6 | 1,755,874 | 1,098,933 | 62 | 38 |
| BE171 (2) | Induced | 97.2 | 1,755,023 | 1,105,781 | 61 | 39 |
| BE173 (1) | Noninduced | 90.6 | 1,293,600 | 1,714,453 | 43 | 57 |
| BE173 (1) | Induced | 89.7 | 1,284,458 | 1,072,559 | 54 | 46 |
| BE173 (2) | Noninduced | 90.7 | 1,237,810 | 5,839,099 | 17 | 83 |
| BE173 (2) | Induced | 90.4 | 1,560,472 | 3,875,074 | 29 | 71 |
| BE177 (1) | Noninduced | 90.6 | 1,560,503 | 2,027,363 | 43 | 57 |
| BE177 (1) | Induced | 90.7 | 1,673,369 | 1,869,622 | 47 | 53 |
| BE177 (2) | Noninduced | 89.7 | 1,616,886 | 1,521,136 | 52 | 48 |
| BE177 (2) | Induced | 90.4 | 1,761,670 | 2,605,704 | 40 | 60 |
| PaE2 (1) | Noninduced | 98.6 | 1,634,679 | 4,469,882 | 27 | 73 |
| PaE2 (1) | Induced | 98.5 | 1,587,419 | 3,766,500 | 30 | 70 |
| PaE2 (2) | Noninduced | 98.3 | 1,572,729 | 2,798,266 | 36 | 64 |
| PaE2 (2) | Induced | 98.7 | 1,707,952 | 3,105,877 | 35 | 65 |
| CI27 (1) | Noninduced | 96.6 | 1,175,828 | 2,752,688 | 30 | 70 |
| CI27 (1) | Induced | 97.1 | 1,499,394 | 2,893,200 | 34 | 66 |
| CI27 (2) | Noninduced | 96.5 | 1,290,217 | 2,016,573 | 39 | 61 |
| CI27 (2) | Induced | 95.8 | 1,291,468 | 1,400,008 | 48 | 52 |
| CIG1 (1) | Noninduced | 95.1 | 2,011,977 | 5,112,862 | 28 | 72 |
| CIG1 (1) | Induced | 95.0 | 1,855,868 | 5,375,882 | 26 | 74 |
| CIG1 (2) | Noninduced | 95.5 | 1,295,516 | 769,182 | 63 | 37 |
| CIG1 (2) | Induced | 96.3 | 1,491,585 | 650,544 | 70 | 30 |
*Replicate numbers are indicated in parentheses.
†Values indicate percent of mapped reads after eliminating tRNA reads.
We identified 161 genes that were AHL-activated in strain PAO1 (Dataset S1) and 15 genes that were AHL-repressed. For this study, we focused only on the AHL-activated genes. We note that quorum-sensing–dependent genes show variable activation at different points in growth (10, 11), and we assessed AHL-dependent gene expression only at one point in growth (OD600 2). To validate our RNA-seq method, we compared our results with previous data generated with a microarray platform. We identified 77 of the 93 genes shown previously to be AHL-induced at OD600 2 (15). This number includes 10 genes that showed AHL induction but with very few reads (<10) in cells grown with AHLs and 0 or 1 reads in samples from cells grown without AHLs. Because of the low expression of these genes, we did not include them in our further analysis of AHL-activated genes. Of the 16 genes identified in the microarray analysis but not in our RNA-seq analysis, nine are in operons that were detected as AHL-induced by RNA-seq. Thus, the RNA-seq and Affymetrix microarray platforms show excellent concordance. Our RNA-seq analysis identified AHL-induced genes not detected in the microarray analysis. The enhanced sensitivity of this technique likely led to the identification of the additional 84 genes.
In strain PAO1 many quorum-controlled genes are activated by AND logic gates. Quorum sensing is required but not sufficient for activation of specific genes (16, 17). Thus, we do not expect our analysis of transcripts in cells from a single point in the growth curve, in a single growth medium, at a single growth temperature to generate a list of all genes activated by quorum sensing. Rather it provides a base of information to allow a test of our hypotheses about strain variability and core and accessory quorum-sensing–controlled genes.
As noted above, we identified 161 quorum-activated genes in PAO1. The number of quorum-activated genes for the remaining strains were 342 for the soil isolate BE171; 301 for the air isolate BE173; 207 for the biofilm isolate BE177; 76 for the tomato plant isolate PaE2; 153 for the CF isolate CI27; and 31 for the CF isolate CIG1. A list of quorum-activated genes for all strains is provided in Dataset S1. Overall, the quorum regulons represent ∼0.5–6.2% of the coding sequences for a given genome. There is no obvious correlation between genome size and the number of quorum-controlled genes detected.
Pairwise Comparisons Between the Quorum Regulon of Strain PAO1 and Other Strains.
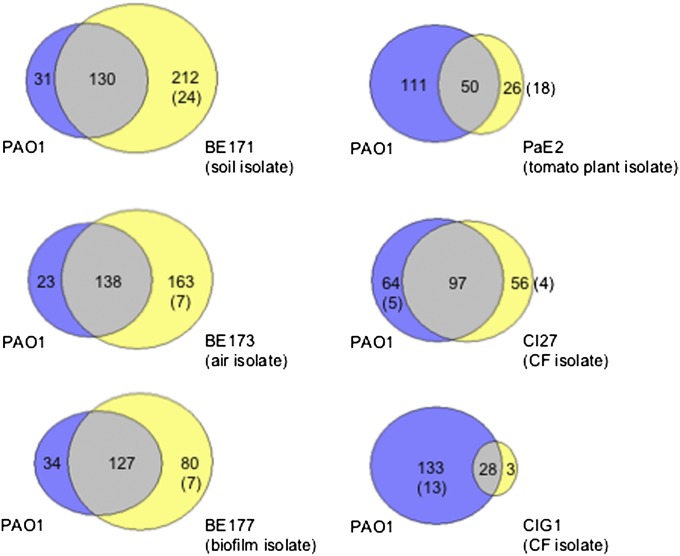
To get a sense of the variation in quorum-activated genes from strain to strain and to lend some validity to conclusions that previously have been drawn about the role of quorum sensing in the regulation of cooperative activities in P. aeruginosa using strain PAO1, we first performed pairwise comparisons of the P. aeruginosa PAO1 quorum regulon with the quorum regulons of the other strains. The largest set of quorum-activated genes (342 genes in BE171) was more than twice the size of the PAO1 set (161 genes), suggesting that P. aeruginosa employs quorum sensing to regulate many more traits than previously identified. With the exception of CIG1, which has a relatively small quorum-controlled regulon of 31 genes, there were pronounced overlaps between the strain PAO1 regulon and the other regulons (Fig. 2). Thus, we consider the P. aeruginosa PAO1 quorum regulon to be reasonably representative of the species. Well-documented examples of some quorum-controlled genes in PAO1, such as lasB, the elastase gene, the apr operon for alkaline protease, rsaL, encoding a quorum-sensing modulator protein, and cbpD encoding a putative chitin-binding protein, were activated in all strains. Genes within the shared subset exhibited variation at levels of both quorum control and transcript abundance. For example, both lasB and cbpD showed a lower quorum response in PaE2 than in PAO1, but expression levels of both genes under non–quorum-sensing conditions were higher in PaE2. This pattern also was noted for lasR and rhlR in PaE2; both genes showed a small quorum response (1.8-fold for lasR and 2.07-fold for rhlR), and neither reached our threshold for differentially regulated genes. lasR induction in strains BE177 (2.84-fold) and CI27 (2.89-fold) was just under the threefold threshold. The PAO1 quorum regulon showed the most overlap (more than 110 genes; about 79%) with the environmental isolates BE171, BE173, and BE177. There are many genes that show quorum control in strains BE171 and BE173 but not in PAO1. Among the 212-gene BE171-specific subset were genes encoding the σ factor AlgU, alginate regulatory protein AlgP, and the alginate and motility regulator AmrZ. Several iron-responsive genes such as PA1363 (encoding an extracytoplasmic-function σ-70 factor), fpvR (encoding an anti-σ factor), and pvdL and tonB1 were among the 163 genes in the BE173-specific quorum regulon. A surprising finding was the strain-specific regulation of several genes coding for extracellular products. For example, phenazine biosynthesis and rhamnolipid biosynthesis genes were quorum-activated in PAO1 but not in two other isolates, PaE2 and CIG1.
Fig. 2.
Venn diagram showing the relationship between quorum-sensing–controlled genes in P. aeruginosa PAO1 and the environmental and CF isolates. Areas within the Venn diagram are drawn approximately to scale, and the number of genes in each is indicated. The number of genes absent from the other genome is shown in parentheses.
Variations in quorum regulons may be caused by differences in gene content between strains or by differences in gene expression. In P. aeruginosa, both determinants appear to dictate strain-specific quorum regulation. Although the entire set of quorum-controlled PAO1 genes was present in all environmental isolates, some genes were absent in one or both CF isolates. The hcnABC operon for hydrogen cyanide production was among the five genes in the pan quorum-controlled set absent in the CF infection isolate CI27. Likewise, 13 genes were absent in the other CF isolate CIG1. PA1874, which codes for a hypothetical protein, was absent in both CI27 and CIG1. Conversely, we identified 48 strain-variable genes that were absent in PAO1 but were present and quorum-activated in one or more of the remaining isolates; these can be considered as belonging to the P. aeruginosa accessory genome.
Differential regulation of genes present in both genomes in pairwise comparisons may be the result of sequence divergence in cis-regulatory regions. Previous work indicates that some genes are indirectly quorum-sensing–activated by other determinants including regulators, which are themselves quorum controlled (16, 17). Thus, differential regulation may be affected by variations in the activity or presence of transcriptional regulators or two-component signal transduction proteins. We examined the defined or putative promoter regions of a few genes that showed significant strain-specific variations in quorum response relative to PAO1. These included PA3724 (lasB) in CIG1 and BE173, PA0143 (nuh) in PaE2, and PA2570 (lecA) in BE173 and CIG1. In all cases examined, either the upstream regions were identical or sequence differences could not account for the observed differential regulation. An example of the latter case is the region upstream of lasB. This gene shows a quorum response of about 130-fold in strain PAO1, a lower response of about threefold in CIG1, and a higher response, about 276-fold in BE173. Thus, there was no correlation between the intensity of the response and levels of the generated AHLs (Fig. 1). Although the promoter region of lasB in both CIG1 and BE173 differed from PAO1, the mismatches in the low-responsive CIG1 promoter were identical to those in the high-responsive BE173 promoter. Collectively, these data suggest that differential regulation for some genes is indirect and may be caused by variations in quorum-controlled or other regulatory factors. For all strains except CIG1, the strain-specific subset of the quorum regulons included transcriptional regulators and/or two component regulatory systems.
Correlations in Quorum Regulons of the Environmental Isolates.
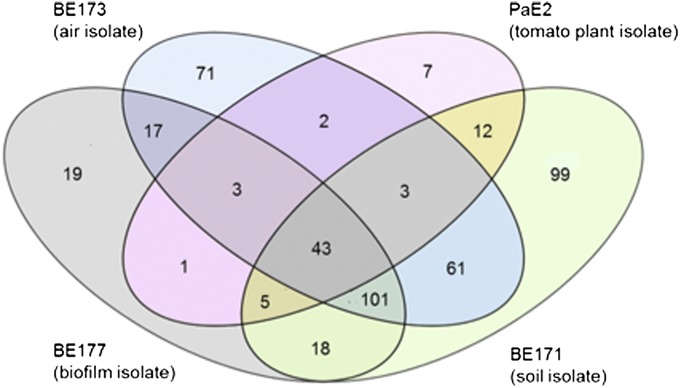
A comparison of the four environmental isolates revealed a set of 43 shared quorum-activated genes (Fig. 3). This subset included genes encoding the production of a number of extracellular factors such as the LasA and LasB proteases, ClpP2 protease, alkaline protease, hydrogen cyanide, and the antibiotic methoxyvinylglycine. The soil isolate BE171, the air isolate BE173, and the biofilm isolate BE177 shared a larger set of 101 genes. Among this set were genes in the flp-tad-rcp locus, which is required for Flp pilus assembly and bacterial adherence, lecA, which codes for a lectin, and the bphO-bphP genes encoding a heme oxygenase and a phytochrome. Also included in this set were 57 genes coding for hypothetical proteins of unknown function including three probable transcription factors. In addition to the shared genes, quorum control of a large subset of 99 genes was unique to BE171, and quorum control of a subset of 71 genes was unique to BE173.
Fig. 3.
Venn diagram showing the relationship between the quorum regulons of the four environmental isolates. The number of genes in each is indicated.
Perhaps the most interesting relationship emerged between the plant isolate PaE2 and the soil isolate BE171. The quorum regulons of these isolates showed the most extensive overlap in pairwise comparisons. Sixty-three of the 76 quorum-activated genes in PaE2 were shared with BE171. An interesting finding was that, unlike other subsets of overlapping genes among the environmental isolates, the overlap between these two strains included genes belonging to the accessory genome. Based on orthologs in the P. aeruginosa LESB58 genome, these 11 genes were annotated as part of an 18-gene pyoluteorin biosynthesis (plt) operon. Closer inspection of the genomes revealed that this entire 31,613-bp locus was conserved in both PaE2 and BE171. Although the remaining seven genes all exhibited some level of quorum activation, the response was less than threefold in one or both strains. Thus, six genes that satisfied our filtering criteria sorted to the PaE2-unique quorum-controlled subset (Fig. 3). As previously documented for LESB58 (18), the plt gene cluster was found at the same chromosomal location adjacent to PA2593 in both PaE2 and BE171.
Correlation Between the Quorum Regulons of the Clinical Isolates.
The two clinical isolates were from two different patients with chronic CF lung infections. The CI27 quorum regulon was much larger (153 genes) than that of CIG1 (31 genes). A comparison of the two regulons identified an overlapping set of 25 genes. If signal synthase levels in the wild types of both strains are similar, it is interesting that almost all shared genes showed a significantly lower quorum response in CIG1. Of the set of 128 genes quorum-controlled in CI27 but not in CIG1, six were absent from the CIG1 genome. These include PA2300, which is annotated as a chitinase, PA2566, encoding a hypothetical protein, and genes belonging to the amb operon for methoxyvinylglycine biosynthesis. A small set of six genes was quorum-controlled in CIG1 but not in CI27. Although LasR sorted to the CIG1-unique subset of six genes, it showed a 2.89-fold induction in CI27 (just below the threefold threshold filter used in this study). The remaining five genes unique to CIG1 included the first two genes of the hcnABC operon. However, hcnC is absent in CIG1, and the entire hcnABC operon is absent in CI27. Thus, regardless of the quorum activation of hcnA and hcnB in CIG1, both strains should have a hydrogen cyanide-negative phenotype. Collectively, these observations indicate that the only quorum-activated genes in CIG1 not shared with CI27 belong to the P. aeruginosa quinolone signal (PQS) biosynthesis operon (pqsA, pqsD, and pqsE).
A small number of quorum-activated genes in CI27 were in the accessory genome. An ortholog for one of these genes, encoding a hypothetical protein, also was present in the accessory genome of CIG1, but it was not quorum-responsive. There were no quorum-controlled genes in the CIG1 accessory genome.
One might expect that genes encoding quorum-induced virulence determinants should show a robust response in both CF isolates, but this was not the case. The pattern of expression for these genes appears to be more complicated and dictated variously by differences in gene expression and the absence of genes. For example, lasB, which codes for elastase, showed a 59-fold response in CI27 but only a 3.3-fold response in CIG1. Likewise, the apr genes encoding alkaline protease showed a greater response in CI27 than in CIG1, and genes specifying pyocyanin biosynthesis were quorum-induced only in CI27. As discussed above, a noteworthy finding was that both strains had genomic deletions that should render them incapable of producing hydrogen cyanide. It is possible that conserved selective pressures contributed to the deletion of these genes within the environment of the CF lung.
Core Quorum-Controlled Regulon.
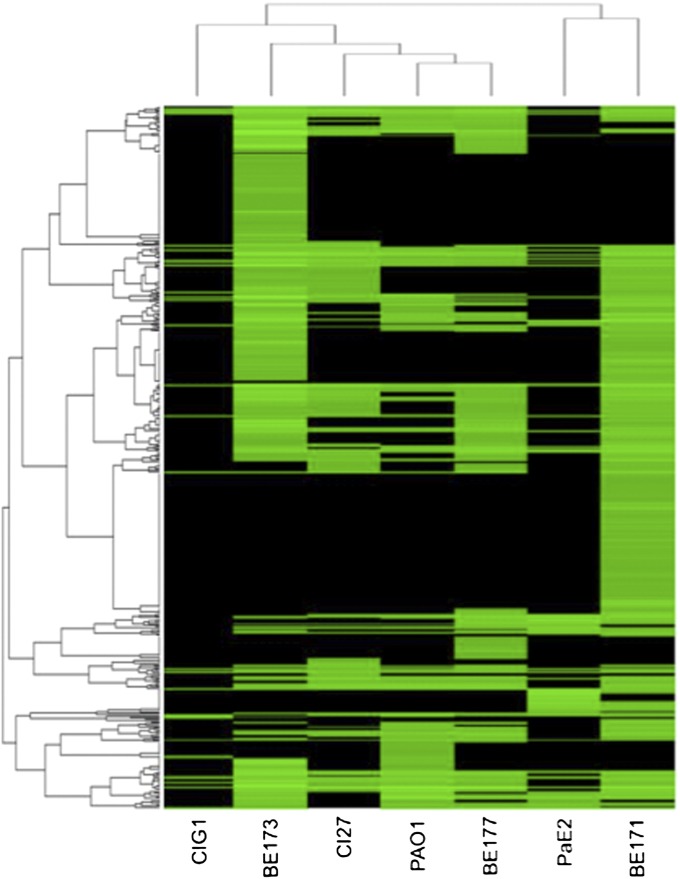
All 43 quorum-controlled genes shared by the four environmental isolates were in the core genome. We sought to determine if this set might represent a P. aeruginosa core quorum-controlled regulon. A comparison revealed that all but two of the 43 genes also were quorum-controlled in PAO1. One of these two genes, pheC (PA3475), is in fact downstream of rhlI and part of the chromosomal deletion in our lasI, rhlI mutant PAO-MW1 (19, 20). Thus, it appears that many P. aeruginosa strains maintain quorum control of this set of about 41 or 42 genes (Table 3) even in the absence of natural selective pressures. Is this set robust to the inclusion of the two CF isolates? A comparison of the set of 41 genes with each CF isolate revealed an overlap of 30 genes in CI27 and an even smaller overlap of 17 genes in CIG1. Of the core genes that were quorum-controlled in PAO1 and the environmental isolates but not in the CF isolates, three were absent in CI27, and seven were absent in CIG1. We view the 41 or 42 quorum-controlled genes shared by strain PAO1 and the environmental isolates as a core. The CF isolates show degraded quorum-sensing regulons. This observation is of interest because it is consistent with previous findings that P. aeruginosa quorum-sensing mutants are abundant in the lungs of some chronically infected CF patients (6, 7). Hierarchical clustering analysis (Fig. 4) supports the view that the quorum-sensing regulon of the tomato plant isolate PaE2 is most closely related to that of the soil isolate BE171, and the quorum-sensing regulon of the CF isolate C1G1 shows the least relatedness to the rest.
Table 3.
Core quorum-controlled genes
| ORF no.* | Gene name | Description |
| PA0122 | rahU | RahU |
| PA0852 | cbpD | Chitin-binding protein CbpD precursor |
| PA1131 | Probable major facilitator superfamily (MFS) transporter | |
| PA1216 | Hypothetical protein | |
| PA1221 | Hypothetical protein | |
| PA1245–49 | aprX-F, aprA | Alkaline protease biosynthesis gene cluster |
| PA1431 | rsaL | Regulatory protein RsaL |
| PA1869 | Probable acyl carrier protein | |
| PA1871 | lasA | LasA protease precursor |
| PA2193–95 | hcnABC | Hydrogen cyanide synthase operon |
| PA2302–05 | ambBCDE | l-2-amino-4-methoxy-trans-3-butenoic acid biosynthesis gene cluster |
| PA2330 | Hypothetical protein | |
| PA2331 | Hypothetical protein | |
| PA2591 | Probable transcriptional regulator | |
| PA2592 | Probable periplasmic spermidine/putrescine-binding protein | |
| PA2939 | Probable aminopeptidase | |
| PA3326 | clpP2 | ClpP2 |
| PA3327 | Probable nonribosomal peptide synthetase | |
| PA3329 | Hypothetical protein | |
| PA3332 | Conserved hypothetical protein | |
| PA3475 | pheC | Cyclohexadienyl dehydratase precursor |
| PA3476 | rhlI | Autoinducer synthesis protein RhlI |
| PA3535 | Probable serine protease | |
| PA3724 | lasB | Elastase LasB |
| PA3904 | Hypothetical protein | |
| PA3907 | Hypothetical protein | |
| PA4128 | Conserved hypothetical protein | |
| PA4129 | Hypothetical protein | |
| PA4130 | Probable sulfite or nitrite reductase | |
| PA4131 | Probable iron-sulfur protein | |
| PA4132 | Conserved hypothetical protein | |
| PA4134 | Hypothetical protein | |
| PA4677 | Hypothetical protein |
The set of 42 genes activated by quorum sensing in all but the CF isolates.
*PA ORF number from the Pseudomonas genome project. (www.pseudomonas.com).
Fig. 4.
Relative expression profiles of quorum-activated genes. Fold changes for each gene are depicted in the heat map. The genes are displayed in the order of the hierarchical clustering of their fold changes according to the Spearman correlation coefficient. Quorum-activated genes are depicted in green, and genes that are absent or do not satisfy our filtering criteria are depicted in black.
Overall, an integrated examination of the quorum regulons of the seven strains revealed a pan quorum-controlled set of genes (Dataset S1) consisting of a small set of shared genes, the core quorum-controlled genes (Table 3), and a larger set of strain-variable genes, including genes on the accessory rather than the core genome (Table 4).
Table 4.
Quorum-controlled accessory genes in the pangenome
| ORF no.* | Description | Strain |
| pdPaBE171_012269 | cbb3-type cytochrome c oxidase subunit I | BE171 |
| pdPaBE171_024629 | Transport protein HasD | BE171 |
| pdPaBE171_029070 | Hypothetical protein | BE171 |
| pdPaBE171_031333 | Hypothetical protein | BE171 |
| pdPaBE171_031592 | Hypothetical protein | BE171 |
| pdPaBE171_048695 | Hypothetical protein | BE171 |
| pdPaBE171_048713 | Hypothetical protein | BE171 |
| pdPaBE171_048737 | Hypothetical protein | BE171 |
| pdPaBE171_048744 | Hypothetical protein | BE171 |
| pdPaBE171_070311 | Hypothetical protein | BE171 |
| pdPaBE171_070323 | Hypothetical protein | BE171 |
| pdPaBE171_073685 | Hypothetical protein | BE171 |
| pdPaBE171_079999 | Hypothetical protein | BE171 |
| pdPaBE173_012396 | Hypothetical protein | BE173 |
| pdPaBE173_012892 | Hypothetical protein | BE173 |
| pdPaBE173_031292 | Hypothetical protein | BE173 |
| pdPaBE173_039028 | Protein of unknown function DUF932 | BE173 |
| pdPaBE173_054647 | Hypothetical protein | BE173 |
| pdPaBE173_054656; pdPac1_27_048360, | Hypothetical protein | BE173, CI27 |
| pdPaBE173_090230 | Hypothetical protein | BE173 |
| pdPaBE177_030508 | Hypothetical protein | BE177 |
| pdPaBE177_044115 | Hypothetical protein | BE177 |
| pdPaBE177_046639 | Helicase domain-containing protein | BE177 |
| pdPaBE177_048658 | Hypothetical protein | BE177 |
| pdPaBE177_048697 | Hypothetical protein | BE177 |
| pdPaBE177_049646 | Hypothetical protein | BE177 |
| pdPaBE177_086637 | Secreted protein Hcp | BE177 |
| pdPac1_27_001579 | Putative nucleoside-binding outer membrane protein | CI27 |
| pdPac1_27_015496 | Hypothetical protein | CI27 |
| pdPac1_27_022579 | Hypothetical protein | CI27 |
| pdPaPaE2_010531 | Periplasmic spermidine/putrescine-binding protein | PaE2 |
| pdPaBE171_042124; pdPaPaE2_033353 | Putative alkylhalidase PltM | BE171, PaE2 |
| pdPaBE171_042148; pdPaPaE2_033377 | PltR | BE171, PaE2 |
| pdPaBE171_042170; pdPaPaE2_033399 | PltL | BE171, PaE2 |
| pdPaBE171_042175; pdPaPaE2_033404 | Putative halogenase, PltA | PaE2 |
| pdPaPaE2_033422 | Polyketide synthase type I, PltB | PaE2 |
| pdPaPaE2_033510 | Polyketide synthase type I, PltC | PaE2 |
| pdPaPaE2_033585 | Putative halogenase, PltD | PaE2 |
| pdPaPaE2_033609 | Putative acyl-CoA dehydrogenase, PltE | PaE2 |
| pdPaPaE2_033619 | Putative acyl-CoA synthetase, PltF | PaE2 |
| pdPaPaE2_033649 | Putative thioesterase, PltG | PaE2 |
| pdPaBE171_042439; pdPaPaE2_033667 | PltZ | BE171, PaE2 |
| pdPaBE171_042449; pdPaPaE2_033677 | Membrane fusion protein, PltH | BE171, PaE2 |
| pdPaBE171_042458; pdPaPaE2_033686 | ATP-binding protein, PltI | BE171, PaE2 |
| pdPaBE171_042485; pdPaPaE2_033714 | Inner membrane permease protein, PltJ | BE171, PaE2 |
| pdPaBE171_042500; pdPaPaE2_033730 | Inner membrane permease protein, PltK | BE171, PaE2 |
| pdPaBE171_042511; pdPaPaE2_033741 | Outer membrane channel protein, PltN | BE171, PaE2 |
| pdPaBE171_042538; pdPaPaE2_033767 | Putative transmembrane protein, PltO | BE171, PaE2 |
*ORF number from draft genomes in the PGAT database (http://tools.nwrce.org/pgat/). ORF numbers for both homologs are listed for ORFs that were quorum-activated in two strains.
Our study adds substantially to the list of P. aeruginosa quorum-controlled genes, but it does not extend the range of functional categories to which P. aeruginosa quorum-controlled genes have been assigned previously. Thus, we did not identify annotated genes encoding factors involved in the following categories: (i) cell division; (ii) chaperones and heat shock proteins; (iii) chemotaxis; (iv) DNA replication, recombination, modification and repair; (v) phage-, transposon-, or plasmid-related; or (vi) RNA processing and degradation.
Discussion
In most cases what we know about genes regulated by AHL quorum sensing in a given species comes from studies on a single strain. In the case of V. fischeri, quorum-sensing control of luminescence shows conservation, but genomic sequencing revealed that an additional set of about 10 genes regulated by quorum sensing in a squid light organ isolate (21) is not present in the genome of a fish isolate. This limited information provided impetus for our investigation of quorum-controlled genes in P. aeruginosa, which is known to be metabolically flexible and to thrive in diverse habitats (22). Therefore, we examined quorum-sensing regulons of multiple P. aeruginosa isolates from different free-living and host-associated habitats. We used high-throughput DNA sequencing to create draft genomes of six P. aeruginosa isolates, generated quorum-sensing signal-generation mutants of each isolate, and compared transcriptomes of mutants grown with and without added signals (a phenotypic complementation). Our analysis provides a snapshot of the quorum-controlled regulons at one point in growth. This approach certainly has limitations and cannot provide an exhaustive census of quorum-controlled genes. Nevertheless, it supports a view that P. aeruginosa genes controlled by quorum sensing are a reflection of the habitat from which a strain was isolated.
We found that identical quorum-sensing circuits regulate sets of genes that partially overlap in different strains. We show that there are quorum-controlled genes on the P. aeruginosa core genome and on accessory elements of the pan-genome. There also is a core of quorum-controlled genes, and our limited analysis indicates this core set of about 42 loci degenerated during evolution in the specialized environment imposed by chronic colonization of the CF lung. Although deeper analyses are warranted, our studies indicate that the accessory components of the quorum regulon reflect ecologic differences in the habitats from which isolates were obtained.
The presence of several genes that code for secreted products among the shared or core subset for all strains is consistent with the view that P. aeruginosa quorum sensing functions to coordinate the production of public goods, an argument for the idea that quorum sensing coordinates cooperativity (23). Our examination of the strain-variable subsets of quorum-controlled genes was revealing. Several isolates controlled accessory genes (plt genes, for example) by quorum sensing. With the exception of the CF isolate CIG1, the quorum-controlled regulons included several known and putative transcriptional regulators and two-component systems as accessory elements. These findings suggest the existence of feed-forward systems and allude to the possibility of strain-specific integration of quorum sensing with other environmental cues affecting transcription. Also included in the strain-variable subset of quorum-controlled elements were genes for general metabolic functions, for example, PA2144, which codes for glycogen phosphorylase, and PA3183, which codes for glucose-6-phosphate dehydrogenase. One can imagine circumstances in which quorum-regulated alterations in metabolic versatility have implications for survival in different nutritionally restricted environments (24). Given the role of quorum sensing in coordinating the production of extracellular products, it is of interest that some isolates had decoupled production of specific extracellular factors from quorum sensing (e.g., the phenazine biosynthesis operon in CIG1 and genes for rhamnolipid synthesis in PaE2). The selective advantage afforded by the exclusion of these genes from quorum regulation is unclear.
With the environmental isolates, we found evidence that P. aeruginosa can adjust its cooperation strategies via modifications of the quorum-sensing regulon. Two strains, PaE2 and BE171, isolated from geographically separate but ecologically related environments (tomato plant and soil) shared an identical pyoluteorin-coding region (plt) that was not present in other strains. Pyoluteorin is a polyketide with antifungal and antibacterial activity. This antimicrobial polyketide suppresses plant diseases (25–27). The plt gene cluster is found in plant-associated pseudomonads (e.g., P. fluorescens Pf-5 and CHAO) and contributes to the ecological fitness of these pseudomonads in the rhizosphere (27). The plt operon has been identified in a few P. aeruginosa isolates, namely, PACS171b, PACS88, and LESB58 (18, 28). LESB58, the earliest archived P. aeruginosa isolate from the Liverpool CF epidemic, carries the plt gene cluster on a genomic island, suggesting that it was acquired through horizontal transfer (18). Curiously, in LESB58 there is a frameshift mutation caused by a deletion in the pltB gene, and the operon is nonfunctional. This finding suggests that the selective pressures that led to the acquisition of this operon were lifted within infected patients. Interestingly, as in the case of PACS171b, PACS88, and LESB58, the plt gene cluster in the two environmental strains in this study is located at the same place in the chromosome, downstream of PA2593. This example of rapid adaptation illustrates the dexterity with which P. aeruginosa both uses and evolves its quorum-sensing system.
P. aeruginosa infections typically are acquired from environmental reservoirs (29). Overall, the quorum-controlled regulon was somewhat more conserved among the environmental strains and strain PAO1 than it was between strain PAO1 and the CF isolates. It is apparent from our analysis that the CF isolate CIG1 has diverged significantly from the other strains, at least with respect to quorum sensing. Differences include its small quorum regulon, the tempered response for most genes that remain quorum-controlled, and deletions in a number of genes that are quorum-controlled in one or more of the other strains. We also found examples of strain-variable quorum regulation in the CF isolate CI27 that were associated with gene deletions. These findings are consistent with the genetic variations associated with P. aeruginosa adaptation during chronic CF infections (6, 30). Because of the small sample size of two CF isolates, our results must be viewed with caution and must be considered as suggestive. The results lead us to imagine that quorum control of certain genes (such as the hcn genes for hydrogen cyanide production) confers a fitness advantage in the environment or early during infection but not during chronic CF lung infection. Traits that otherwise are beneficial may be unnecessary or even detrimental in the context of a long-term infection and therefore may be uncoupled from quorum sensing during adaptation. A finding that argues in favor of this hypothesis is that hcnA and hcnB, the two genes of the hcnABC operon that were not deleted in CIG1, were in fact activated by quorum sensing. We find it interesting that LasR quorum-sensing mutations accumulate in P. aeruginosa during long-term CF lung colonization. Uncoupling of certain genes from quorum-control represents a different molecular solution to decreasing or eliminating expression of quorum-controlled genes.
The findings that both CF isolates in our panel harbored deletions in the hcn operon and that many CF isolates have lasR mutations also are of clinical significance. There has been recent interest in using the cyanogenic properties of P. aeruginosa to develop a rapid method for its detection in CF patients (31, 32). Although further work with a larger collection of strains is required, the reliability of this approach is brought into question by our findings.
This study extends the list of genes reported to be quorum-controlled in P. aeruginosa and demonstrates that quorum control of gene expression has a strain-variable component. We expect that an extension of this analysis to a larger collection of strains not only will identify additional strain-variable quorum-responsive genes but also will allow a correlation of the variations with ecological origins.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Bacterial strains and plasmids used are listed in Table S1. For plasmid and strain constructions, bacteria were grown in LB broth, supplemented with antibiotics when appropriate at the following concentrations (per mL): 10 μg of gentamicin, 10 μg of tetracycline, and 100 μg of ampicillin for Escherichia coli and 100 μg of gentamicin, 100 μg of tetracycline, and 150 μg of carbenicillin for P. aeruginosa. For transcript profiling, midlogarithmic-phase cells were used to inoculate LB broth buffered with 50 mM 3-(N-morpholino) propanesulfonic acid (MOPS) (pH 7.0). The optical densities (OD600) at inoculation were 0.05. When appropriate, synthetic AHLs were added to the medium at final concentrations of 2 μM for 3OC12-HSL and 10 μM for C4-HSL before inoculation.
Chromosomal deletions were constructed using either pEXG2- or pEX18Tc-derived plasmids (33, 34). PCR-amplified DNA fragments flanking lasI and rhlI from P. aeruginosa PAO1 were cloned into pEXG2 and pEX18Tc, resulting in a deletion of codons 31–191 in lasI and codons 34–184 in rhlI. The resulting plasmids were used to construct lasI, rhlI mutants using standard methods. Candidate mutants were screened by PCR and by demonstrating that mutants did not make detectable 3OC12 and C4-HSL in stationary-phase culture extracts. The lasI, rhlI mutants had no discernible growth defects compared with their respective parents.
Measurement of AHLs.
Cells were grown to an OD600 of 3.5 in LB broth buffered with 50 mM MOPS (pH 7.0). Concentrations of 3OC12-HSL and C4-HSL were measured with bioassays as described previously (35, 36).
Sequencing, Assembly, and Annotation of P. aeruginosa Strains.
DNA Sequencing for the six previously unsequenced isolates was done with the Illumina Genome Analyzer according to the manufacturer's instructions (Illumina). A random fragment library was constructed by using a custom paired-end protocol. The genomes were assembled from 76-bp paired-end reads using Velvet (37). Genome annotation was predicted by a software annotation pipeline associated with the Prokaryotic Genome Analysis Tool (PGAT) (38). Manual annotation of the genome assemblies also was performed using PGAT and the Pseudomonas genome database (www.pseudomonas.com) (39). The first version draft assemblies and annotation of the six genomes sequenced in this study have been deposited at DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank (www.ncbi.nlm.nih.gov/genome). PGAT also was used to determine the presence and absence of genes and for interstrain comparisons of promoter sequences (http://tools.nwrce.org/pgat/).
RNA Isolation.
Approximately 1 × 109 cells at OD600 2 were mixed with RNA Protect Bacteria reagent (Qiagen) and stored at −80 °C. Thawed cells were resuspended in QIAzol reagent and disrupted by bead beating. RNA was purified using a miRNeasy minikit (Qiagen) according to the manufacturer’s instructions. RNA was treated with Turbo DNase (Ambion) and purified using a RNeasy MinElute cleanup kit (Qiagen). Two biological replicates were processed for each condition (without and with added AHLs).
RNA-Seq Library Construction and Sequencing.
P. aeruginosa-specific selective primers were based on the genome sequences of seven P. aeruginosa strains (PAO1, PA14, PA7, LESB58, PACS2, C3719, and 2192). We used a pool of 1,507 selective hexamers with no perfect match to any rRNA genes. An additional set of 200 hexamers, responsible for the majority of rRNA-priming events in test libraries, was removed, leaving a final set of 1,307 selective hexamers. The hexamer sequences of the final primer pool, the forward and reverse adaptor sequences, and the PCR primer sequences are given in Table S2. An in silico assessment of selective hexamer-binding sites in P. aeruginosa PAO1 mRNA showed that there was an average of one binding site for every five bases of potential template sequence. A detailed description of the multiplexed library generation protocol is provided in SI Materials and Methods. Briefly, first- and second-strand syntheses were carried out by using the pool of selective primers and RNA template to create double-stranded cDNA. Subsequent adaptor ligation and PCR-amplification steps were used to generate DNA libraries with sample-specific 3-bp barcode tags. Eight uniquely barcoded DNA libraries were sequenced per lane as 36-mers on an Illumina Genome Analyzer II flowcell using the standard Illumina protocol at the University of Washington Genome Center.
Sequence Mapping and Analysis.
Raw sequencing reads (36 nucleotides in length) were first sorted on the basis of their barcodes. For PAO1 samples, reads were mapped to the PAO1 genome (sequence downloaded from www.pseudomonas.com) and analyzed using Avadis NGS software (version 1.2.3, Build 149378; Strand Scientific Intelligence, Inc.). For all other P. aeruginosa samples, reads were mapped to the corresponding strain’s draft genome using Burrows–Wheeler alignment (40) and were analyzed using custom Python scripts (http://nwrce.org/nwrce-org/resources/web-resources/software-utilities). For each sample, a set of input reads for comparison analyses was generated in two steps. First, we derived a set of uniquely mapped reads, defined as reads that mapped to the non-rRNA regions of the genome with two or fewer mismatches. Next, we filtered this set to eliminate reads that mapped to tRNA genes. For comparisons, we used sample pairs with generally similar numbers of mRNA sequencing reads in the two conditions (without and with added AHLs). Sequence-read mapping and genome coverage information are summarized in Table 2.
Identification of Differentially Expressed Genes.
For all samples, the number of raw reads mapping to each gene was normalized based on the total number of input reads (non-rRNA and non-tRNA reads) for that sample. This normalization procedure allowed comparison of gene-expression patterns across strains, within and between experiments. Reads that partially overlapped a gene contributed to its total raw read value. The raw read value for each gene was incremented by 1 before normalization to avoid errors caused by instances of division by 0. The fold-change values for each gene were computed by first calculating the geometric mean across replicate samples and then calculating ratios across conditions. We chose geometric averaging to dampen the effects of possible outlying values in transcript abundances between replicates on fold-change estimates. Next, we applied filtering criteria designed to allow a more robust estimation of the quorum-activated component of the quorum regulon. Only genes that had an average of >10 reads in the two replicates for the plus AHL condition were considered for further analyses, and genes with at least a threefold change between conditions were considered differentially expressed. Avadis NGS was used to compare differentially expressed genes between strains and for visualization of reads mapped to the PAO1 genome.
Supplementary Material
Acknowledgments
We thank Chris Armour for invaluable technical assistance and discussions, the staff at the University of Washington Genome Center for their contributions, and Matthew Radey for help with data analysis. This work was supported by US Public Health Service (USPHS) Grants GM-59026 (to E.P.G.) and GM-56665 (to C.S.H.) and by resources from the Microbiology and Genomics Cores of USPHS Grant P30DK089507. B.S.K. and S.H.C. were supported by the Korea Research Foundation Grant KRF-2009-013-F00014 (Republic of Korea). We also acknowledge the services and use of software developed by the Data Integration Core of the Northwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research funded by the National Institutes of Health, National Institute of Allergy and Infectious Diseases Grant U54 AI057141.
Footnotes
The authors declare no conflict of interest.
Data deposition: The draft genome assemblies and annotations have been deposited in the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank database (accession nos. AKZD00000000, AKZE00000000, AKZF00000000,AKZG00000000, AKZH00000000, and AKBD00000000).
See Author Summary on page 16426 (volume 109, number 41).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214128109/-/DCSupplemental.
References
- 1.Lerat E, Moran NA. The evolutionary history of quorum-sensing systems in bacteria. Mol Biol Evol. 2004;21:903–913. doi: 10.1093/molbev/msh097. [DOI] [PubMed] [Google Scholar]
- 2.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol. 2003;50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 3.Kuo A, Blough NV, Dunlap PV. Multiple N-acyl-L-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J Bacteriol. 1994;176:7558–7565. doi: 10.1128/jb.176.24.7558-7565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathee K, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci USA. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosson P, et al. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bacteriol. 2002;184:3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Argenio DA, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J Bacteriol. 2003;185:7222–7230. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hentzer M, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holloway BW. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol. 1955;13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 13.Armour CD, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6:647–649. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- 14.Hirakawa H, et al. Activity of the Rhodopseudomonas palustris p-coumaroyl-homoserine lactone-responsive transcription factor RpaR. J Bacteriol. 2011;193:2598–2607. doi: 10.1128/JB.01479-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müh U, et al. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006;50:3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73:1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuster M, Greenberg EP. A network of networks: Quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 18.Winstanley C, et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2009;19:12–23. doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brint JM, Ohman DE. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antunes LC, et al. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol. 2007;189:8387–8391. doi: 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiehlmann L, et al. Population structure of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2007;104:8101–8106. doi: 10.1073/pnas.0609213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hense BA, et al. Does efficiency sensing unify diffusion and quorum sensing? Nat Rev Microbiol. 2007;5:230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- 24.Studer SV, Mandel MJ, Ruby EG. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J Bacteriol. 2008;190:5915–5923. doi: 10.1128/JB.00148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howell C, Stipanovic R. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology. 1979;69(5):480–482. [Google Scholar]
- 26.Howell C, Stipanovic R. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology. 1980;70(8):712–715. [Google Scholar]
- 27.Haas D, Keel C. Regulation of antibiotic production in root-colonizing Peudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol. 2003;41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- 28.Hayden HS, et al. Large-insert genome analysis technology detects structural variation in Pseudomonas aeruginosa clinical strains from cystic fibrosis patients. Genomics. 2008;91:530–537. doi: 10.1016/j.ygeno.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Römling U, Wingender J, Müller H, Tümmler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst RK, et al. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ Microbiol. 2003;5:1341–1349. doi: 10.1111/j.1462-2920.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 31.Enderby B, Smith D, Carroll W, Lenney W. Hydrogen cyanide as a biomarker for Pseudomonas aeruginosa in the breath of children with cystic fibrosis. Pediatr Pulmonol. 2009;44:142–147. doi: 10.1002/ppul.20963. [DOI] [PubMed] [Google Scholar]
- 32.Gilchrist FJ, et al. Variation in hydrogen cyanide production between different strains of Pseudomonas aeruginosa. Eur Respir J. 2011;38:409–414. doi: 10.1183/09031936.00166510. [DOI] [PubMed] [Google Scholar]
- 33.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 35.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brittnacher MJ, et al. PGAT: A multistrain analysis resource for microbial genomes. Bioinformatics. 2011;27:2429–2430. doi: 10.1093/bioinformatics/btr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winsor GL, et al. Pseudomonas Genome Database: Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39(Database issue):D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]