Abstract
RPE65 is a key metalloenzyme responsible for maintaining visual function in vertebrates. Despite extensive research on this membrane-bound retinoid isomerase, fundamental questions regarding its enzymology remain unanswered. Here, we report the crystal structure of RPE65 in a membrane-like environment. These crystals, obtained from enzymatically active, nondelipidated protein, displayed an unusual packing arrangement wherein RPE65 is embedded in a lipid–detergent sheet. Structural differences between delipidated and nondelipidated RPE65 uncovered key residues involved in substrate uptake and processing. Complementary iron K-edge X-ray absorption spectroscopy data established that RPE65 as isolated contained a divalent iron center and demonstrated the presence of a tightly bound ligand consistent with a coordinated carboxylate group. These results support the hypothesis that the Lewis acidity of iron could be used to promote ester dissociation and generation of a carbocation intermediate required for retinoid isomerization.
Keywords: metalloprotein, monotopic membrane protein, extended X-ray absorption fine structure, isomerohydrolase
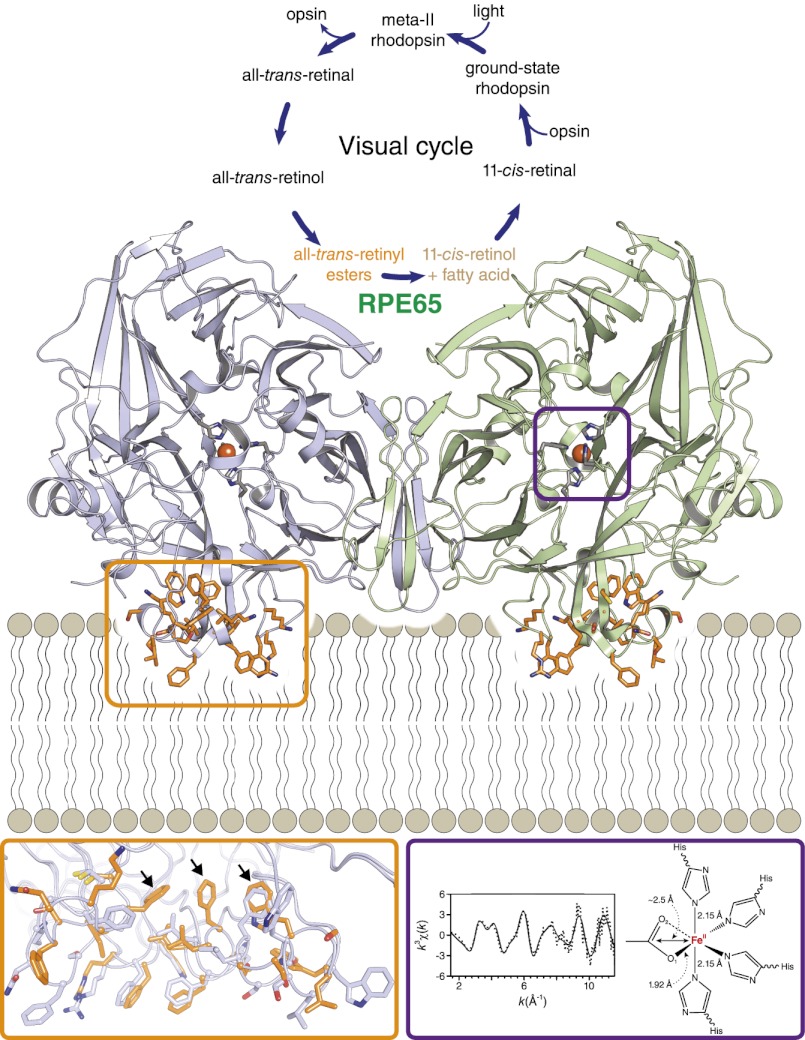
Vision is a complex, multistep process whereby environmental information contained in light rays is converted to chemical and electrical signals in the retina that are relayed to the brain for interpretation. The initial step in this process is mediated by light receptors called rod and cone opsins and their associated G proteins in photoreceptor cells (1). These receptors contain a vitamin A-derived chromophore called 11-cis-retinal that undergoes cis to trans isomerization upon absorption of a photon. Isomerization of the chromophore converts the receptor to a state in which it can activate downstream signaling molecules that alter the electrochemical state of the photoreceptor cell (2, 3). Following photoisomerization, the Schiff base-linked chromophore is hydrolyzed and dissociates from the receptor. Continuous regeneration of 11-cis-retinal, required for maintenance of visual function, is critically dependent on a metabolic pathway known as the visual cycle (4, 5). At the heart of this cycle is the enzymatic conversion of all-trans-retinyl esters to 11-cis-retinol by the retinoid isomerase, RPE65, mutations of which cause the severe childhood blinding disease Leber congenital amaurosis (6, 7).
RPE65 is an iron-dependent, membrane-bound enzyme expressed nearly exclusively in the retinal pigment epithelium (RPE) (8, 9). The iron dependence of RPE65 was anticipated on the basis of its evolutionarily relationship to carotenoid cleavage oxygenases (CCOs). However, CCO activity has not been demonstrated for RPE65, and it is likely that iron serves a different, albeit poorly understood, function in this enzyme. It is now appreciated that phospholipids constitute a second critical factor necessary for RPE65 activity. Purified RPE65 exhibited retinoid isomerase activity only when bound to all-trans-retinyl ester-incorporated membrane vesicles (10). Moreover, destruction of intact microsomal phospholipids by phospholipase A2 greatly reduced the production of 11-cis-retinol by native bovine RPE65 (11). Spectroscopic data also indicated that RPE65 adopts functionally and structurally different conformations when bound to lipid bilayers compared with its detergent-solubilized state, although specific changes could not be ascertained (12).
Previously, we determined the crystal structure of detergent-solubilized RPE65 purified from bovine RPE (13), which revealed a 4-His–ligated iron center contained within seven-bladed β-propeller architecture and a monotopic mode of membrane attachment. The structure featured residual active-site electron density that we speculated might represent a bound fatty acid molecule perhaps derived from a cleaved retinyl ester. However, other possibilities, such as a bound detergent, could not be ruled out. Interestingly, despite the ease with which detergent-solubilized RPE65 crystallizes and the generally well-ordered structure obtained, the protein used for crystallization experiments was catalytically inactive. Historically, the inhibition of RPE retinoid isomerase activity by detergents was the major factor that delayed its link to RPE65, and the mechanism of inhibition remains incompletely understood. In the structure, we observed that residues composing the RPE65 membrane-interaction surface exhibit substantial disorder compared with the rest of the enzyme (11, 13). Owing to the roles of these residues in membrane binding as well as substrate recognition and proper positioning of the substrate in the active site, detergent-induced alterations in their native structure, either by stripping the protein of its native phospholipids or by a direct effect, could inactivate the enzyme.
Here, we present the crystal structure of RPE65 in a membrane-like environment. These crystals, obtained from enzymatically active microsomal extracts, display an unusual “type 1” packing arrangement that is, to our knowledge, unprecedented for monotopic membrane proteins (14). Compared with delipidated RPE65, residues composing the membrane interaction region are more ordered and adopt a substantially different conformation in crystals obtained from catalytically competent, lipid-replete enzyme, indicating a key role for these residues in substrate recognition and uptake. This lipid-embedded structure featured active-site density near the iron cofactor similar in shape to prior structures. To gain further insights into the identity of this cryptic ligand as well as the structure and oxidation state of the iron cofactor we performed an iron K-edge X-ray absorption spectroscopy (XAS) study of detergent-solubilized RPE65. This analysis revealed that the formal oxidation state of iron, as found in the active site, is +2. Extended X-ray absorption fine structure (EXAFS) spectra support a model for the iron center wherein either a carboxylate or a solvent-derived hydroxide group is bound to the iron in its open coordination sites. These results provide key insights into two critical aspects of RPE65 enzymology.
Results
Crystallization of Enzymatically Active RPE65 in the Presence of Native Microsomal Phospholipids.
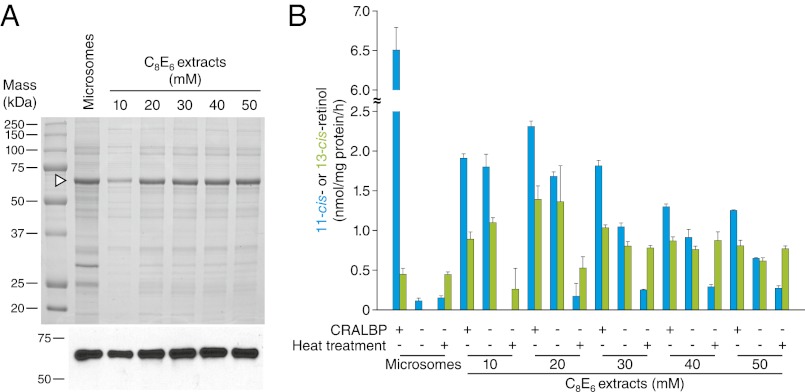
On the basis of reports that RPE65 requires phospholipids for maintenance of its catalytic activity (10, 11), we hypothesized that detergent extracts of RPE microsomes in which RPE65 is solubilized but not separated from the native microsomal phospholipids could maintain retinoid isomerase activity. RPE65 was extracted from microsomes using the nonionic detergent hexaethylene glycol octyl ether (C8E6) and the extracts were assayed for retinoid isomerase activity (Fig. 1 A and B). Analysis of the extract phospholipid composition showed a profile very similar to that of RPE microsomes with phosphatidylethanolamine and phosphatidylcholine constituting the major species (Fig. S1A). Consistent with our hypothesis, we observed substantial production of 11-cis-retinol by the extracts. Maximal activity was observed for the 20-mM C8E6 extracts, corresponding to the minimal concentration needed for complete solubilization of RPE65 (Fig. 1). Production of 11-cis-retinol was about one-third that of untreated microsomes and the activity declined with increasing concentration of C8E6 used for extraction. RPE65 purified from this extract displayed little or no activity when all-trans-retinyl palmitate was provided as a substrate (Fig. S1B). Whereas untreated microsomes almost exclusively produced 11-cis-retinol, the detergent extracts generated substantial quantities of 13-cis-retinol in addition to 11-cis-retinol. Cellular retinaldehyde-binding protein (CRALBP) was conducive but not essential for 11-cis-retinol production by the extracts in contrast to its strict requirement for 11-cis-retinol production by RPE microsomes (15). These results are consistent with data showing that RPE65 is a “leaky” isomerase capable of producing both 11- and 13-cis-retinol (16, 17). Importantly, 11-cis-retinol production was essentially abolished in heat-treated extracts, demonstrating that its production was enzymatic (Fig. 1B).
Fig. 1.
Protein composition and activity of RPE microsomes and detergent extracts. (A) SDS/PAGE analysis of proteins in untreated microsomes and C8E6 extracts. Proteins were visualized by Coomassie Blue staining. An arrowhead shows the position of the RPE65 band. (Lower) An immunoblot analysis of the samples performed using a monoclonal RPE65 antibody (11). (B) Retinoid isomerase activity of untreated microsomes and C8E6 extracts. Blue and green bars represent 11-cis and 13-cis-retinol production, respectively. Error bars are SDs calculated from three independent experiments.
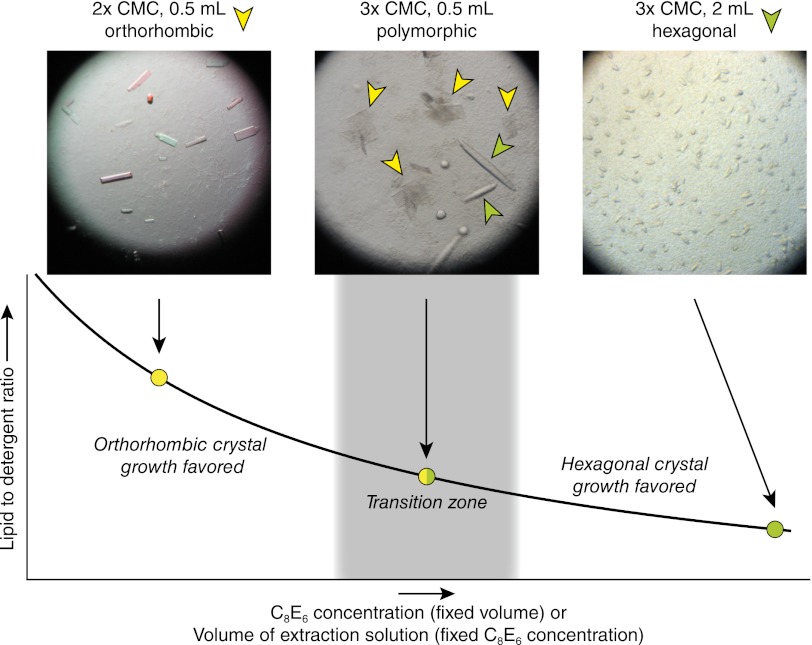
Differential centrifugation and selective detergent extraction gave a preparation that was substantially enriched in RPE65. We therefore initiated experiments to crystallize RPE65 directly from the enzymatically active extracts. Initial trials were conducted with concentrated extracts produced by treatment of 40 mg of RPE microsomes with 1 mL of buffered solution containing 20–30 mM C8E6. Surprisingly, we obtained a unique crystal form under conditions very similar to those previously reported for growth of hexagonal RPE65 crystals (13). These thin, plate-like crystals were extremely fragile and diffracted X-rays in a highly anisotropic manner, thus precluding their use for structure determination (Fig. S2A). Extensive attempts to improve the crystals by changing components of the well solution were unsuccessful, prompting us to consider other parameters affecting crystal growth. By reducing the volume of extraction solution by one-half, thus increasing the lipid/detergent ratio in the preparation, much larger and thicker crystals with markedly improved diffraction properties could be obtained (Fig. 2 and Fig. S2B). SDS/PAGE analysis of the crystals revealed a single protein band at ∼65 kDa (Fig. S2C). Interestingly, reduction of the lipid/detergent ratio by increasing the volume of extraction solution and/or by increasing the detergent concentration led to the growth of the original hexagonal RPE65 crystals and inhibited growth of the alternative crystal form (Fig. 2). This remarkable dependence on the lipid/detergent ratio suggests a critical role for microsomal lipids in the growth of the alternative crystal form.
Fig. 2.
Correlation between lipid/detergent ratio and RPE65 crystal form. At high lipid/detergent ratios corresponding to the minimal amount of detergent required for complete RPE65 extraction, exclusive growth of the orthorhombic crystal form is observed. Low lipid/detergent ratios lead to exclusive growth of the original hexagonal crystal form. Between these two extremes is a transition zone in which growth of both crystal forms can be observed simultaneously.
Unusual Molecular Packing Observed in Orthorhombic RPE65 Crystals.
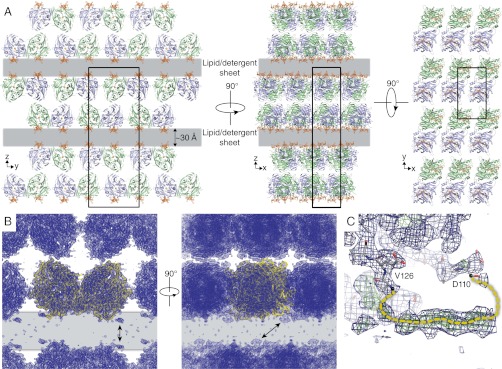
The alternative RPE65 crystals diffracted X-rays anisotropically to Bragg spacings of ∼2.7 Å in the a* and b* directions and ∼3.3 Å along c* and belonged to the orthorhombic space group P212121. The structure was solved by molecular replacement revealing the presence of an RPE65 dimer in the asymmetric unit and refined to an Rfree value of 26.1% against data extending to 3 Å resolution (Table 1, Crystal form A). An extraordinary feature of these crystals is their molecular packing arrangement wherein sheets of RPE65 molecules, arranged back to back, are separated from each other by a gap that is ∼20–30 Å wide and extends in two dimensions throughout the crystal (Fig. 3A). A consequence of this arrangement is that there are no direct protein–protein contacts between sheets. The electron density in the gap is weak and essentially featureless with a single region of relatively strong density corresponding to a loop that was unmodeled in our previous structure (Fig. 3 B and C). Analysis of the diffraction data revealed no evidence of lattice translocation defects or twinning that could cause crystal packing abnormalities (Fig. S3 and SI Materials and Methods). Thus, we could rationalize the gaps in the crystals only by postulating that they are filled with mixed micelles or bilayer-like lipid–detergent sheets. Although disordered and thus essentially invisible in the electron density maps, these assemblies provide a solid support for the sheets of protein molecules in the crystal structure. The arrangement of RPE65 molecules in the unit cell with their membrane-binding surfaces facing the lipid–detergent-filled gap strongly supports this interpretation (Fig. 3A). Moreover, the gap is of the same thickness as the hydrophobic core of a lipid bilayer. This unusual molecular packing arrangement with lipid–detergent sheets forming an integral part of the crystal lattice provides a harmonious explanation for the dependence on the lipid/detergent ratio for crystal growth. Although unusual, a similar crystal packing arrangement has been reported for a polytopic membrane protein (18). These RPE65 crystals are to our knowledge a unique example of a classical type 1 crystal of a monotopic membrane protein (14).
Table 1.
Data collection and refinement statistics
| Dataset name | Crystal form A | Crystal form B Ir-derivative* | Crystal form B native | Crystal form C |
| Data collection† | ||||
| Beamline | X29 | X29 | NE-CAT ID-C | NE-CAT ID-C |
| Wavelength, Å | 1.07500 | 1.09520 | 0.97949 | 0.97950 |
| Space group | P212121 | P6522 | P6522 | P65 |
| Unit cell parameters, Å | a = 55.80 | a = 178.64 | a = 178.50 | a = 175.29 |
| b = 100.23 | ||||
| c = 277.59 | c = 86.47 | c = 86.64 | c = 86.46 | |
| Resolution, Å | 100–3.0 (3.19–3.0) | 50–2.6 (2.75–2.6) | 50–3.15 (3.35–3.15) | 50–2.5 (2.65–2.5) |
| Unique reflections | 31,980 (5,030) | 47,089 (7,080) | 14,356 (2,246) | 52,159 (8,369) |
| Completeness, % | 99.7 (99.0) | 98.7 (91.8) | 99.0 (98.6) | 99.1 (98.8) |
| Multiplicity | 6.8 (6.1) | 10.6 (7.6) | 4.1 (4.1) | 3.7 (3.8) |
| <I/σI> | 13.3 (2.4) | 19.0 (2.2) | 12.1 (2.8) | 10.1 (1.5) |
| RmergeI, %‡ | 13.2 (81.7) | 10.5 (89.8) | 10.8 (49.5) | 11.3 (88.2) |
| RmeasI, %‡ | 14.3 (89.2) | 11 (96.2) | 12.4 (56.8) | 13.2 (103) |
| RpimI, %§ | 5.5 (34.8) | 2.5 (24.3) | 6.0 (27.4) | 6.8 (52.9) |
| Wilson B-factor, Å2 | 54.9 | 55.7 | 54.8 | 49.7 |
| Refinement | ||||
| Resolution, Å | 69.4–3.0 | 48.4–2.6 | 48.4–3.15 | 47.8–2.5 |
| No. reflections | 30,384 | 23,925 | 13,635 | 49,415 |
| Rwork/Rfree, %¶ | 22.4/26.1 | 18.5/22.9 | 21.5/24.7 | 19.3/21.9 |
| No. atoms | ||||
| Protein | 8,338 | 4,159 | 4,213 | 8,286 |
| Water | — | 165 | — | 115 |
| Metal/ion | 2 Fe | 1 Fe, 1 Ir | 1 Fe, 1PO4 | 2 Fe |
| B-factors, Å2 | ||||
| Protein | 75.6 | 49.9 | 63.9 | 45.7 |
| Water | — | 47.1 | — | 36.8 |
| Metal/ion | 60.2 (Fe) | 27.5 (Fe), 68.8 (Ir) | 50.3 (Fe), 68.8 (PO4) | 35.4 (Fe) |
| RMS deviations | ||||
| Bond lengths, Å | 0.006 | 0.01 | 0.005 | 0.006 |
| Bond angles, ° | 1.09 | 1.35 | 1.01 | 1.09 |
| Ramachandran plot║ | ||||
| Favored/outliers, % | 96.5/0 | 95.9/0.39 | 94.2/0 | 98/0 |
| PDB ID code | 4F2Z | 4F3A | 4F30 | 4F3D |
*Bijvoet pairs treated separately.
†Values in parentheses show the highest-resolution shell of data.
‡As defined in XDS.
§As defined in ref. 66.
¶As defined in REFMAC.
║As defined in MolProbity.
Fig. 3.
Molecular packing in orthorhombic RPE65 crystals. (A) Orthogonal views down the crystallographic axes demonstrate well-packed RPE65 sheets separated by 20- to 30-Å gaps (gray layers) that extend in two dimensions and contain lipid–detergent mixed-micelle sheets. The membrane-binding surface of RPE65 (orange) faces the lipid-filled sheet. (B) The final σA-weighted 2Fo − Fc electron density map contoured at the 1σ level (blue mesh) showing clear density for the RPE65 molecules (yellow ribbons) and weak or absent density in the lipid–detergent-filled sheet. Arrows point to regions of relatively strong unmodeled density that correspond to partially ordered residues 111–125 shown in a close-up view in C. The final σA-weighted 2Fo − Fc and Fo − Fc electron density maps contoured at the 1- and 3σ levels are shown in C as blue and green mesh, respectively. Structural figures were prepared with PyMOL (Schrödinger).
Conformational Differences Between Lipid-Embedded and Delipidated RPE65.
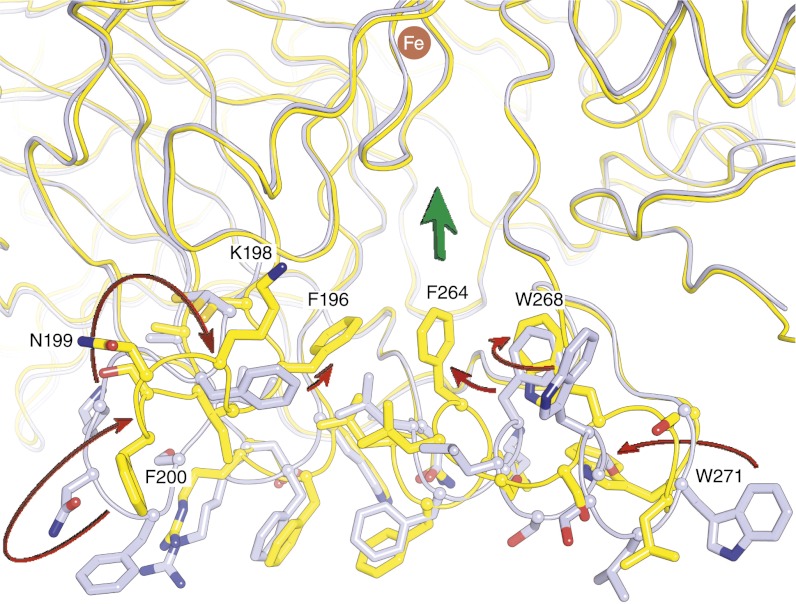
We compared the lipid-embedded RPE65 structure with our previously reported delipidated, enzymatically inactive structure, using error-scaled difference distance matrices (19). Whereas most of the structure is invariant within model errors, there are two regions where large and significant structural differences are observed (Fig. S4A). These regions, consisting of residues 196–202 and 263–271, compose a portion of the RPE65 membrane-binding surface and entrance to the active site. Compared with the delipidated RPE65 structure, residues 196–202 are flipped by ∼90° and adopt a more regular β-sheet structure that packs against blade II of the β-propeller core of the protein with the F196 side chain inserting deeper into the active-site tunnel (Fig. 4 and Movie S1). Additionally, residues 263–271 of α-helix 5 are unwound in the lipid-embedded structure, resulting in loss of their α-helical structure and movement of the F264 and W268 side chains toward the active-site entrance. Notably, these regions exhibit much better order in the lipid-embedded structure as evidenced by lower relative B-factors and substantially clearer electron density despite the fact that the residues lack stabilization by crystal contacts. No specifically bound phospholipid or detergent molecules were located in the structure. The loss of α-helical structure is in good agreement with circular dichroism spectroscopy experiments that detected similar structural differences in membrane-bound vs. detergent-solubilized RPE65 (12).
Fig. 4.
Conformational differences in the membrane-binding surface and active-site entrance observed between lipid-embedded and delipidated RPE65. Arrows pointing from the delipidated structure (gray) to the lipid-embedded structure (yellow) show the general direction of movement between the two structures. Residues F196, F264, and W268 in the lipid-embedded structure form a continuous aromatic surface near the active-site entrance, indicated by the broad green arrow.
The general location of residues 110–126, which were completely disordered and thus not modeled in the previously reported RPE65 structure (13), is apparent in the lipid-embedded structure as a stretch of weak but mostly continuous electron density that runs parallel to the membrane surface (Fig. 3C). This electron density lacked sufficient definition to allow precise model building, but its general location places it in an appropriate position to contribute to RPE65 membrane binding as previously suggested (9).
RPE65 Dimeric Structure Is Preserved in Multiple Crystal Forms.
RPE65 is found in a dimeric assembly with parallel-oriented membrane-interacting surfaces in the orthorhombic RPE65 crystals, which is essentially identical to that observed in the previously reported hexagonal crystal form (Fig. 5). Although the dimer interface is fairly hydrophobic and predicted to be stable in solution, it also features a number of intermolecular hydrogen bonds and salt bridges that could be labile in a high ionic strength environment (13). Support for the stability of the dimeric assembly was provided by additional crystal structures of RPE65 in space group P6522 (Table 1, Crystal form B). One such structure was obtained by soaking a P65 RPE65 crystal in 1 mM IrCl6. Binding of Ir to the RPE65 molecule induced a rearrangement in the crystal such that the dimer axis rotated ∼4° to become aligned with the crystallographic “a” and “b” axes, resulting in elevation of the space group symmetry. Thus, the RPE65 dimer moved as a rigid body in the crystal without undergoing any distortions, thereby providing physical evidence for its stability. A second structure in space group P6522 was obtained in the presence of ∼500 mM ammonium phosphate. Similar to the iridium derivative, the dimeric axis in this crystal is aligned with the crystallographic a and b axes, resulting in a single RPE65 monomer per asymmetric unit. Application of the appropriate symmetry operation generates the preserved RPE65 dimer. These data demonstrate that the dimer is stable even under high ionic strength conditions.
Fig. 5.
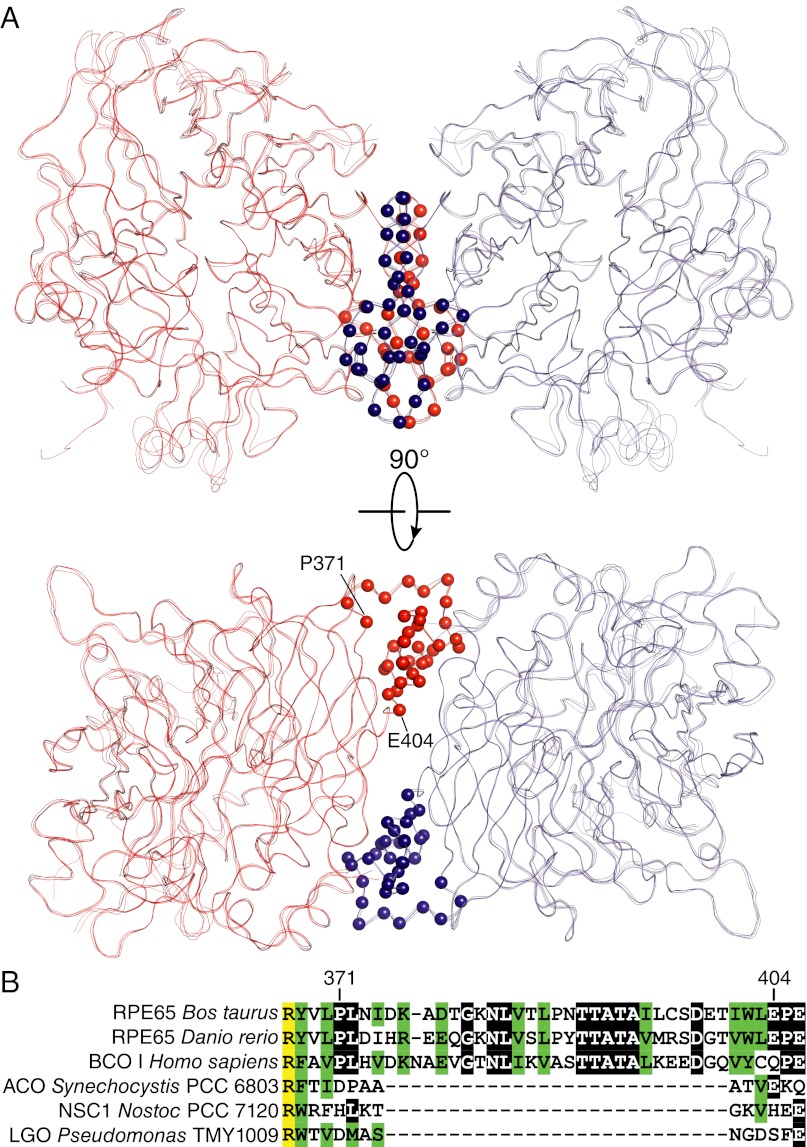
RPE65 dimeric assembly. (A) Superpositioning of the dimeric assembly observed in all crystal forms obtained to date demonstrates its highly preserved nature. The dimer interface is primarily composed of residues 371–404, which are indicated by red and blue spheres for their Cα atoms. (B) Sequence alignment of CCOs shows that the dimer-mediating sequence is present in vertebrate but not bacterial members. Completely identical, blocks of identical and similar residues are shown in yellow, black, and green, respectively. The sequence alignment was generated with the DALI server (65) and AlignX. Abbreviations: ACO, apocarotenoid oxygenase; BCO I, beta-carotene oxygenase I; LGO, lignostilbene-α,β-dioxygenase; NSC1, Nostoc sp. carotenoid oxygenase 1.
A superposition of the RPE65 dimers from the different crystal forms is shown in Fig. 5, demonstrating a near-perfect alignment. The dimer-mediating residues 371–404 constitute a portion of a sequence extension that is found only in vertebrate members of the CCO family. Sequence homology in this region is fairly high among vertebrate proteins, even between those that are evolutionarily well separated.
Cryptic Active-Site Ligand.
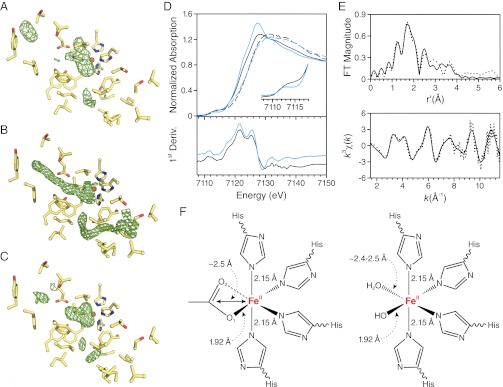
Analysis of residues composing the active-site cavity in the lipid-embedded and delipidated structures did not reveal any significant structural differences consistent with a rigid active-site structure (Fig. S4 A and B). The lipid-embedded structure exhibited residual active-site electron density similar in shape and position to that observed in the original delipidated structure (Fig. 6 A and B) (13). Previously, we speculated that this density could represent a bound fatty acid molecule derived from a cleaved retinyl ester, but other possibilities could not be ruled out. In particular, detergent constitutes an important possibility owing to its inhibitory effects on RPE65 activity. To address this question, we purified and crystallizated RPE65 in the zwitterionic detergent Fos-Choline-10, which is structurally unrelated to the polyoxyethylene detergents. These crystals belonged to space group P65 and were essentially isomorphous to our previously reported crystals. The structure was refined to an Rfree value of 21.9% against data extending to 2.5 Å resolution (Table 1, Crystal form C). As shown in Fig. 6C, the triangular appearance of the residual density in the vicinity of the iron cofactor was unchanged, allowing us to exclude bound detergent as its source.
Fig. 6.
Residual electron density observed in the RPE65 active site and XAS analysis of the RPE65 iron cofactor. (A–C) The final σA-weighted Fo − Fc map in the vicinity of the active-site cavity at the 3σ level. (A) Electron density in the active site of the lipid-embedded RPE65 structure. Despite being weaker, the overall shape and position of the density near the iron center are reminiscent of the density observed in the structure of delipidated RPE65 (PDB ID code 3FSN) shown in B. (C) The residual electron density maintains its shape and position in the crystal structure of RPE65 purified in Fos-Choline-10, confirming that the density does not represent a bound detergent molecule. (D) XANES measurements demonstrate the presence of a ferrous iron cofactor in the RPE65 active site (solid black line). The iron K-edge position at 7,121.5 eV is consistent with divalent iron. H2O2 treatment caused a “blue shift” in the edge position consistent with iron oxidation to the ferric form (dashed black lines). Comparison of the RPE65 iron edge spectrum with that of ACO, an enzyme with a highly similar active-site structure, confirms the assignment of the active-site iron valence state (ferrous ACO, solid blue line; H2O2-treated ACO, dashed blue line). (Inset) An expansion of the pre-edge region is shown for the ferrous forms of RPE65 and ACO. (E) Experimental EXAFS spectra (dotted line) and best-fit curve (solid line) for detergent-solubilized RPE65. The fit shown corresponds to fit 7 in Table S2. (F) Two structural models consistent with the XAS results.
To gain further insights into the identity of the cryptic active-site ligand, we carried out a K-edge XAS study of the iron cofactor. XAS is a sensitive structural technique that can provide information regarding the symmetry and oxidation state of the metal under study as well as accurate metal–ligand bond lengths that can be used to discriminate between different types of bound ligands. The tendency of RPE65 preparations to be contaminated with so-called “junk iron” was a major barrier that had to be overcome before informative XAS data could be obtained. We found that treatment of membrane-bound RPE65 with 10 mM EDTA removed nonspecifically bound junk iron while preserving the active-site iron XAS signal. Because prior studies showing the ferrous iron dependence of RPE65 catalytic activity relied on chelation/add-back experiments, it was important to determine the formal oxidation state of iron as it exists in the RPE65 active site (8).
XAS data were collected on a 0.82-mM (∼50 mg/mL) sample of RPE65 solubilized and purified from RPE microsomes in the presence of C8E6. The X-ray absorption near-edge spectra (XANES) displayed a broad, weak 1s→3d pre-edge feature centered at 7,112.9 eV, which is dipole forbidden in octahedral symmetry and consistent with a five- or distorted six-coordinate iron center (Fig. 6D) (20, 21). Two peaks are observed in the first derivative trace of the absorption edge spectrum, one strong peak at 7,121.5 eV and a weaker peak at 7,125.25 eV (Fig. 6D). The energy of the major peak indicates that most of the iron in the sample is in the ferrous state. To confirm this result, we recorded XANES on a fresh aliquot of concentrated RPE65 treated with a 20-fold molar excess of H2O2, an agent known to abolish RPE65 activity (13). The edge position was “blue-shifted” by ca. 3 eV, consistent with oxidation of iron to the ferric form (Fig. 6D). Interestingly, the oxidized iron underwent rapid photoreduction during subsequent scans. As a final confirmation of the iron oxidation state, we obtained XANES on a 1.85-mM (∼100 mg/mL) sample of Synechocystis apocarotenoid oxygenase (ACO), a soluble CCO with a 4-His–ligated iron center essentially identical to that of RPE65. This preparation displayed high ACO activity, which is known to depend on a ferrous iron cofactor. The Fe(II)-ACO edge shape and position were very similar to that of RPE65 (Fig. 6D). Following treatment of Fe(II)-ACO with a 100-fold molar excess of H2O2, enzymatic activity was abolished and the iron edge position was blue-shifted to a value close to that observed for oxidized RPE65 (Fig. 6D). These data conclusively demonstrate that the iron center of RPE65 is in the divalent state.
EXAFS analysis of detergent-solubilized RPE65 was carried out to help identify nonprotein ligands associated with the RPE65 iron cofactor. Fourier-filtered, first-shell EXAFS data revealed a primary shell consisting of four N/O ligands located ∼2.13 Å from the Fe atom as well as a single short Fe-O/N scatterer at ∼1.9 Å (Table S1). Inclusion of the short scatterer in the simulation was required to obtain a good fit to the experimental data. The difference in bond lengths between these two shells is significantly larger than the experimental resolution of 0.156 Å, thus justifying the two-shell fit. Inclusion of a low Z scatterer (Fe-C/N/O) at a distance of ∼2.5 Å from the iron was required to complete the fit.
To further validate these results, multiple scattering analysis using unfiltered data and a simple starting model based on the crystallographic structure of RPE65 was performed. Inclusion of multiple-scattering paths associated with the Cα and Cβ atoms of coordinated imidazoles conferred a significant improvement in fit quality (Table S2 and Fig. S5) and accurately reproduced the double-hump feature at k ∼ 4 Å−1 in the k3χ(k) EXAFS that is characteristic of coordinated imidazole ligands (Fig. 6E). Variation in the number of imidazoles suggests the presence of three to four bound His ligands, within the constraints of the rigid refinement model that was used, consistent with X-ray crystallography-derived structural data.
Two possible structural models consistent with the EXAFS analysis are shown in Fig. 6F. The first of these consists of a shell of four His ligands with a carboxylate moiety occupying the remaining coordination sites of the iron atom. The short 1.9-Å Fe-O distance assigned to bound carboxylate favors a monodentate or asymmetric bidentate mode of interaction with the iron. In this model, the ∼2.5-Å iron–ligand interaction is attributable to the bridging carbon atom of the carboxylate moiety. This model is quite similar to one we previously proposed on the basis of the RPE65 crystal structure in which the carboxylate moiety of a fatty acid interacts with the iron atom at the open coordination sites (13). The second model features a hydroxide ion and a water molecule in the two open sites at distances of ∼1.92 Å and ∼2.5 Å, respectively, from the iron center. This model is not as well supported by our crystallographic data because discrete peaks attributable to bound solvent molecules are not observed in the electron density maps. Importantly, a complex between iron and molecular oxygen is unlikely on the basis of the observed ferrous oxidation state of the iron as well as the iron–ligand bond lengths derived from the EXAFS analysis (22, 23).
Discussion
Mechanism of Enzymatic Inhibition by Detergents and the Role of Lipids in Maintenance of RPE65 Structure.
Inhibition of RPE65 catalytic activity following its purification in detergent-containing solutions has long been a major obstacle in its biochemical and structural characterization. Recently, it has become clear that phospholipid membranes are required for maintenance of RPE65 catalytic activity, suggesting that the inhibitory effects of detergent are a result of protein delipidation. Spectroscopic data have indicated that phospholipids directly influence RPE65 structure.
Several crystallographic methods have been developed to study membrane proteins in lipidic environments including crystallization in lipidic cubic phase (24) and in bicelles (25) as well as 2D electron crystallography (26). Some highly expressed membrane proteins, such as rhodopsin (27), light-harvesting complex-II (28), and the sodium potassium pump (29), have been successfully crystallized directly from solubilized native membranes (30). Owing to its high abundance in bovine RPE microsomes, we applied the latter strategy to RPE65, allowing us to circumvent the above-mentioned problems inherent in using purified enzyme.
In contrast to purified RPE65 samples used for past crystallographic studies, RPE microsomes solubilized with low levels of detergent retained a substantial level of retinoid isomerase activity as well as native phospholipids. Using the enzymatically active preparation, we were able to grow orthorhombic RPE65 crystals that displayed a highly unusual packing arrangement with lipid–detergent sheets forming an integral part of the crystal lattice. Although, to our knowledge, unprecedented for monotopic membrane proteins, a similar crystal packing arrangement has been reported for a polytopic membrane protein (18). The lipid–detergent mixed micelle or bilayer-like structure can be expected to provide a solid support for crystal growth. However, it remains unclear how layers of RPE65 molecules remain in register to ensure long-range translational order. Modification of the lipid–detergent structure by one RPE65 molecule could promote the proper orientation of another RPE65 molecule on the opposite side of the sheet. Additionally, an electrostatic interaction between RPE65 molecules located across from each other might help maintain register in the context of a crystal. Indeed the range of electrostatic effects should be greatly increased in the low dielectric environment of a lipid bilayer. Regardless of the mechanism of crystal growth, our findings suggest the possibility of crystallizing other monotopic membrane proteins in lipid-embedded states, for example through the use of bicelles or lipidic cubic phases.
These unique crystals provide high-resolution insights into the influence of phospholipids on RPE65 structure. We observe that residues composing the membrane-binding surface and active-site entrance adopt a dramatically different and much more ordered conformation in the lipid-embedded structure compared with the delipidated RPE65 structure. The hydrophobicity of the membrane-binding surface is maintained, but the side chains of three aromatic residues are observed to move toward the mouth of the active-site tunnel, thus producing a continuous aromatic surface. We speculate that these aromatic residues could interact favorably with the π-electron–rich polyene chain of a retinyl ester to facilitate its uptake from the membrane. The observed structural changes, including a loss of α-helical secondary structure, are in good agreement with spectroscopic changes observed upon reassociation of detergent-solubilized RPE65 with liposomal membranes (12). In summary, our results demonstrate that phospholipids, in addition to providing a favorable environment for production and delivery of retinyl esters, also directly maintain the structure and catalytic activity of the RPE65 molecule.
RPE65 Dimeric Structure.
The observation of preserved, parallel-oriented RPE65 dimers in multiple crystal forms lends strong support to the idea that RPE65 forms a dimeric complex in vivo. Parallel dimer formation is a common feature of monotopic membrane proteins (31). Acquisition of a dimer-mediating sequence appears to be a distinct, early step in the evolution of many soluble enzymes into monotopic membrane enzymes as first pointed out for fatty acid amide hydrolase (FAAH) (32). CCO family evolution provides a slight variation on this theme with acquisition of a hydrophobic membrane-binding surface preceding the attainment of a dimer-mediating loop. ACO from Synechocystis, an ancient member of the CCO family, is a water-soluble protein as are many other CCOs from lower organisms (33–35). It contains a modest-sized hydrophobic patch that allows it to bind membranes to access its substrates (33, 36) but was not observed to form a dimeric assembly (33). The stronger affinity of RPE65 for membranes seems to be conferred by an expansion of its hydrophobic, membrane-binding surface area together with acquisition of the capacity to dimerize in a parallel fashion.
Proposed Role for the Iron Cofactor in RPE65-Catalyzed Retinoid Isomerization.
Using XAS, we have conclusively demonstrated that the iron cofactor of RPE65 exists predominantly in the divalent oxidation state. Furthermore, our EXAFS and crystallographic data demonstrate that an iron–carboxylate complex is a likely intermediate formed during the isomerization reaction. Multiple studies have indicated that the reaction catalyzed by RPE65 is actually a concerted ester cleavage/double-bond isomerization reaction in which an atypical ester hydrolysis results in the formation of a carbocation intermediate that allows the isomerization step to occur (13, 16, 17, 37–41) (reviewed in ref. 42). Recently, a second retinoid isomerization mechanism involving formation of a radical cation intermediate and subsequent retinyl ester hydrolysis has been proposed (16, 43, 44). The observation of carboxylate binding to the iron cofactor by different experimental methodologies suggests that such an intermediate lies along the isomerization reaction pathway. Examination of proteins containing the 4-His iron coordination motif reveals that carboxylate ligands commonly occupy the two open sites in the iron coordination sphere (45–47). These data support our prior hypothesis that RPE65 uses the Lewis acidity of the iron ion to polarize the ester moiety to facilitate retinyl ester cleavage.
Although iron is more commonly used as a cofactor for redox chemistry (48, 49), its use as a Lewis acid by hydrolases is not particularly unusual. For example, purple acid phosphatases (50), methionyl aminopeptidase (51), peptide deformylase (52), and 2,3-dihydroxyphenylpropionate 1,2-dioxygenase (MhpB) (53) have all been shown to use iron as a Lewis acid to facilitate hydrolysis reactions. The uncharged, 4-His ligand environment of the RPE65 iron cofactor likely strengthens its Lewis acidity. Although ferric iron would generally be a more powerful Lewis acid, divalent iron seems to be used more often in hydrolase enzymes, possibly owing to a better balance of Lewis acid strength and ability to rapidly exchange ligands (54). For RPE65, the choice of ferrous iron as an active-site metal also likely reflects its evolutionary history as a member of the CCO family.
Materials and Methods
Preparation of RPE65-Containing Membranes.
Bovine RPE microsomal membranes were isolated as previously described with the exception that 10 mM EDTA was included in the high-salt wash solution (13). EDTA treatment improved both the Fe-XAS data and RPE65 crystal quality but did not affect active-site iron occupancy or retinoid isomerase activity.
RPE65 Isomerization Assay.
One milligram of RPE microsomes suspended in 12.5 μL of 10 mM Bis-Tris⋅HCl, pH 7.3, containing 1 mM DTT was used for each enzymatic assay. To ensure ample availability of all-trans-retinyl ester substrate, all-trans-retinol (0.01 μmol in 1 μL dimethylformamide) was added to the membranes and the LRAT-catalyzed retinyl esterification reaction was allowed to proceed for 20 min at 22 °C. Microsomes were solubilized with C8E6 (Anatrace) for 15 min and then centrifuged at 166,000 × g for 30 min. Detergent extracts were collected and the untreated membranes were resuspended in 12.5 μL of 10 mM Bis-Tris⋅HCl, pH 7.3, containing 1 mM DTT. Microsomes or extracts were added to a buffer consisting of 50 mM Bis-Tris⋅HCl, pH 7.3, 10 mM ATP, 1 mM DTT, and 1% (wt/vol) BSA. The reaction was initiated by the addition of CRALBP to a final concentration of 12 μM followed by gentle vortexing. The final volume of each reaction was 200 μL. Samples were incubated under dim red light at 30 °C with gentle shaking for 1 h. Control experiments were performed in which either CRALBP was omitted from the reaction mixture or the microsomes or extracts were incubated at 80 °C for 10 min before the assay. Reactions were terminated by addition of 300 μL of 100%(vol/vol) MeOH followed by vigorous vortexing. Retinoids were extracted in 300 μL of 100%(vol/vol) hexane and analyzed by normal-phase HPLC, using an 80%(vol/vol) hexane, 20%(vol/vol) ethyl acetate liquid phase and a Zorbax SIL column (Agilent).
RPE65 Crystallization.
All crystallization trials were carried out with the hanging-drop vapor-diffusion method. One microliter of concentrated protein sample was added to 1 μL of well solution at room temperature and the drop was allowed to equilibrate against 0.5 mL of well solution at 8 °C. Details of the crystallization procedures can be found in SI Materials and Methods.
Diffraction Data Collection and Reduction, Structure Solution, and Refinement.
Crystals were flash cooled in liquid nitrogen before X-ray exposure. Datasets were collected at the National Synchrotron Light Source (NSLS) X29 beamline or at the Northeastern Collaborative Access Team (NE-CAT) 24-ID-C beamline. Data were indexed, integrated, and scaled with XDS (55). Intensities were converted to amplitudes using CTRUNCATE (56, 57). The structures were solved by molecular replacement using Phaser (58) or from single-wavelength anomalous scattering data using SHELXC/D and Phaser (59, 60) and refined using REFMAC (61) and Coot (62). Details of the phasing and refinement can be found in SI Materials and Methods. The stereochemical quality of the models was assessed with the MolProbity server (63). All structural models exhibited Molprobity scores in at least the 99th percentile for structures of comparable resolution. A summary of the crystallographic data and refinement statistics is provided in Table 1.
Preparation of RPE65 and ACO for XAS Studies.
RPE65 was purified from washed RPE microsomal membranes obtained from ∼700 bovine eyes as previously described (13), concentrated to ∼50 mg protein/mL (0.82 mM), and flash frozen in liquid nitrogen. Addition of glassing agent was unnecessary owing to the presence of C8E6 in the sample. This sample is referred to as RPE65red. A 30-μL portion of the RPE65 sample was treated with a 20-fold molar excess of H2O2 to generate RPE65ox. The ACO coding region (GI:81671293) was PCR amplified from Synechocystis sp. strain PCC 6803 genomic DNA (ATCC) and cloned into a pET3a expression vector. The coding region was sequenced and found to be identical to the published sequence. ACO was expressed in Escherichia coli and the protein was purified by (NH4)2SO4 fractionation and gel filtration chromatography. ACO activity was assayed as previously described (35). The enzymatically active ACO samples (ACOred) used for XAS studies were concentrated to 70–100 mg/mL. One concentrated sample was treated with a 100-fold molar excess of H2O2 to generate an oxidized ACO preparation (ACOox). Samples were transferred to Kapton-wrapped XAS sample cells and frozen in liquid nitrogen before data collection.
XAS Data Collection and Analysis.
Fe K-edge XAS data were collected on beamline X3B of the NSLS over an energy range of 6,930–7,810 eV (k = 13.5 Å−1). Internal energy calibration was provided by simultaneous measurement of spectra from an iron foil. The energy of the first inflection point of the foil K-edge was assigned as 7,112.0 eV. The data were analyzed with EXAFSPAK and Athena. Theoretical models used for curve fitting were generated with FEFF 8.40 (64). Details of the data collection and analysis can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mr. David Peck for assistance with collection of bovine RPE; Dr. Jennifer Bohon (National Synchrotron Light Source) for performing initial XAS experiments; Dr. Marcin Golczak (Case Western Reserve University) for help with TLC; Dr. Georg E. Schulz (University of Freiburg) for helpful discussions concerning membrane protein crystal packing; and Dr. Leslie T. Webster, Jr. (Case Western Reserve University) for valuable comments on the manuscript. This research was supported in part by National Institutes of Health (NIH) Grants EY009339, EY020551, and P30-EY011373. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, supported by Grants 5P41RR015301-10 and 8P41GM103403-10, as well as at the National Synchrotron Light Source on the X3B and X29 beamlines, which are operated by the Case Center for Synchrotron Biosciences, supported by NIH National Institute of Biomedical Imaging and Bioengineering Grant P30-EB-009998. K.P. is John H. Hord Professor of Pharmacology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4F2Z, 4F3A, 4F30, and 4F3D).
See Author Summary on page 16410 (volume 109, number 41).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212025109/-/DCSupplemental.
References
- 1.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 3.Salom D, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Lintig J, Kiser PD, Golczak M, Palczewski K. The biochemical and structural basis for trans-to-cis isomerization of retinoids in the chemistry of vision. Trends Biochem Sci. 2010;35:400–410. doi: 10.1016/j.tibs.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wald G. The molecular basis of visual excitation. Nature. 1968;219:800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 6.Gu SM, et al. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat Genet. 1997;17:194–197. doi: 10.1038/ng1097-194. [DOI] [PubMed] [Google Scholar]
- 7.Marlhens F, et al. Mutations in RPE65 cause Leber’s congenital amaurosis. Nat Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 8.Moiseyev G, et al. RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. J Biol Chem. 2006;281:2835–2840. doi: 10.1074/jbc.M508903200. [DOI] [PubMed] [Google Scholar]
- 9.Hamel CP, et al. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J Biol Chem. 1993;268:15751–15757. [PubMed] [Google Scholar]
- 10.Nikolaeva O, Takahashi Y, Moiseyev G, Ma JX. Purified RPE65 shows isomerohydrolase activity after reassociation with a phospholipid membrane. FEBS J. 2009;276:3020–3030. doi: 10.1111/j.1742-4658.2009.07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golczak M, Kiser PD, Lodowski DT, Maeda A, Palczewski K. Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J Biol Chem. 2010;285:9667–9682. doi: 10.1074/jbc.M109.063941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolaeva O, Moiseyev G, Rodgers KK, Ma JX. Binding to lipid membrane induces conformational changes in RPE65: Implications for its isomerohydrolase activity. Biochem J. 2011;436:591–597. doi: 10.1042/BJ20110091. [DOI] [PubMed] [Google Scholar]
- 13.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc Natl Acad Sci USA. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostermeier C, Michel H. Crystallization of membrane proteins. Curr Opin Struct Biol. 1997;7:697–701. doi: 10.1016/s0959-440x(97)80080-2. [DOI] [PubMed] [Google Scholar]
- 15.Stecher H, Gelb MH, Saari JC, Palczewski K. Preferential release of 11-cis-retinol from retinal pigment epithelial cells in the presence of cellular retinaldehyde-binding protein. J Biol Chem. 1999;274:8577–8585. doi: 10.1074/jbc.274.13.8577. [DOI] [PubMed] [Google Scholar]
- 16.Redmond TM, Poliakov E, Kuo S, Chander P, Gentleman S. RPE65, visual cycle retinol isomerase, is not inherently 11-cis-specific: Support for a carbocation mechanism of retinol isomerization. J Biol Chem. 2010;285:1919–1927. doi: 10.1074/jbc.M109.027458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBee JK, et al. Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: Dependence on the specificity of retinoid-binding proteins. Biochemistry. 2000;39:11370–11380. doi: 10.1021/bi001061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz O, Vinothkumar KR, Goswami P, Kühlbrandt W. Structure of the monomeric outer-membrane porin OmpG in the open and closed conformation. EMBO J. 2006;25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider TR. Objective comparison of protein structures: Error-scaled difference distance matrices. Acta Crystallogr D Biol Crystallogr. 2000;56:714–721. doi: 10.1107/s0907444900003723. [DOI] [PubMed] [Google Scholar]
- 20.Westre TE, et al. A multiplet analysis of Fe K-edge 1s->3d pre-edge features of iron complexes. J Am Chem Soc. 1997;119:6297–6314. [Google Scholar]
- 21.Randall CR, et al. X-ray-absorption pre-edge studies of high-spin iron(II) complexes. Inorg Chem. 1995;34:1036–1039. [Google Scholar]
- 22.Emerson JP, Farquhar ER, Que L., Jr Structural “snapshots” along reaction pathways of non-heme iron enzymes. Angew Chem Int Ed Engl. 2007;46:8553–8556. doi: 10.1002/anie.200703057. [DOI] [PubMed] [Google Scholar]
- 23.Carrondo MA, Bento I, Matias PM, Lindley PF. Crystallographic evidence for dioxygen interactions with iron proteins. J Biol Inorg Chem. 2007;12:429–442. doi: 10.1007/s00775-007-0213-2. [DOI] [PubMed] [Google Scholar]
- 24.Cherezov V. Lipidic cubic phase technologies for membrane protein structural studies. Curr Opin Struct Biol. 2011;21:559–566. doi: 10.1016/j.sbi.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faham S, Bowie JU. Bicelle crystallization: A new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 26.Raunser S, Walz T. Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu Rev Biophys. 2009;38:89–105. doi: 10.1146/annurev.biophys.050708.133649. [DOI] [PubMed] [Google Scholar]
- 27.Okada T, et al. X-Ray diffraction analysis of three-dimensional crystals of bovine rhodopsin obtained from mixed micelles. J Struct Biol. 2000;130:73–80. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 28.Standfuss J, Terwisscha van Scheltinga AC, Lamborghini M, Kühlbrandt W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 2005;24:919–928. doi: 10.1038/sj.emboj.7600585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 30.Gourdon P, et al. HiLiDe-systematic approach to membrane protein crystallization in lipid and detergent. Cryst Growth Des. 2011;11:2098–2106. [Google Scholar]
- 31.Bracey MH, Cravatt BF, Stevens RC. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567:159–165. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 32.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 33.Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 34.Marasco EK, Vay K, Schmidt-Dannert C. Identification of carotenoid cleavage dioxygenases from Nostoc sp. PCC 7120 with different cleavage activities. J Biol Chem. 2006;281:31583–31593. doi: 10.1074/jbc.M606299200. [DOI] [PubMed] [Google Scholar]
- 35.Ruch S, Beyer P, Ernst H, Al-Babili S. Retinal biosynthesis in Eubacteria: In vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Mol Microbiol. 2005;55:1015–1024. doi: 10.1111/j.1365-2958.2004.04460.x. [DOI] [PubMed] [Google Scholar]
- 36.Scherzinger D, Ruch S, Kloer DP, Wilde A, Al-Babili S. Retinal is formed from apo-carotenoids in Nostoc sp. PCC7120: In vitro characterization of an apo-carotenoid oxygenase. Biochem J. 2006;398:361–369. doi: 10.1042/BJ20060592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barry RJ, Cañada FJ, Rando RR. Solubilization and partial purification of retinyl ester synthetase and retinoid isomerase from bovine ocular pigment epithelium. J Biol Chem. 1989;264:9231–9238. [PubMed] [Google Scholar]
- 38.Bernstein PS, Rando RR. In vivo isomerization of all-trans- to 11-cis-retinoids in the eye occurs at the alcohol oxidation state. Biochemistry. 1986;25:6473–6478. doi: 10.1021/bi00369a020. [DOI] [PubMed] [Google Scholar]
- 39.Jang GF, McBee JK, Alekseev AM, Haeseleer F, Palczewski K. Stereoisomeric specificity of the retinoid cycle in the vertebrate retina. J Biol Chem. 2000;275:28128–28138. doi: 10.1074/jbc.M004488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law WC, Rando RR. Stereochemical inversion at C-15 accompanies the enzymatic isomerization of all-trans- to 11-cis-retinoids. Biochemistry. 1988;27:4147–4152. doi: 10.1021/bi00411a037. [DOI] [PubMed] [Google Scholar]
- 41.Golczak M, Kuksa V, Maeda T, Moise AR, Palczewski K. Positively charged retinoids are potent and selective inhibitors of the trans-cis isomerization in the retinoid (visual) cycle. Proc Natl Acad Sci USA. 2005;102:8162–8167. doi: 10.1073/pnas.0503318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiser PD, Palczewski K. Membrane-binding and enzymatic properties of RPE65. Prog Retin Eye Res. 2010;29:428–442. doi: 10.1016/j.preteyeres.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandal MN, et al. Alpha-phenyl-N-tert-butylnitrone (PBN) prevents light-induced degeneration of the retina by inhibiting RPE65 protein isomerohydrolase activity. J Biol Chem. 2011;286:32491–32501. doi: 10.1074/jbc.M111.255877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chander P, Gentleman S, Poliakov E, Redmond TM. Aromatic residues in the substrate cleft of RPE65 govern retinol isomerization and modulate its progression. J Biol Chem. 2012;287:30552–30559. doi: 10.1074/jbc.M112.364596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lancaster CR, Michel H. Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid. J Mol Biol. 1999;286:883–898. doi: 10.1006/jmbi.1998.2532. [DOI] [PubMed] [Google Scholar]
- 46.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 47.Gillmor SA, Villaseñor A, Fletterick R, Sigal E, Browner MF. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nat Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 48.Que L, Jr, Ho RY. Dioxygen activation by enzymes with mononuclear non-heme iron active sites. Chem Rev. 1996;96:2607–2624. doi: 10.1021/cr960039f. [DOI] [PubMed] [Google Scholar]
- 49.Holm RH, Kennepohl P, Solomon EI. Structural and functional aspects of metal sites in biology. Chem Rev. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 50.Davis JC, Averill BA. Evidence for a spin-coupled binuclear iron unit at the active site of the purple acid phosphatase from beef spleen. Proc Natl Acad Sci USA. 1982;79:4623–4627. doi: 10.1073/pnas.79.15.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’souza VM, Holz RC. The methionyl aminopeptidase from Escherichia coli can function as an iron(II) enzyme. Biochemistry. 1999;38:11079–11085. doi: 10.1021/bi990872h. [DOI] [PubMed] [Google Scholar]
- 52.Rajagopalan PTR, Yu XC, Pei DH. Peptide deformylase: A new type of mononuclear iron protein. J Am Chem Soc. 1997;119:12418–12419. [Google Scholar]
- 53.Sanvoisin J, Langley GJ, Bugg TDH. Mechanism of extradiol catechol dioxygenases - Evidence for a lactone intermediate in the 2,3-dihydroxyphenylpropionate 1,2-dioxygenase reaction. J Am Chem Soc. 1995;117:7836–7837. [Google Scholar]
- 54.Merkx M, Averill BA. Probing the role of the trivalent metal in phosphate ester hydrolysis: Preparation and characterization of purple acid phosphatases containing (AlZnII)-Zn-III and (InZnII)-Zn-III active sites, including the first example of an active aluminum enzyme. J Am Chem Soc. 1999;121:6683–6689. [Google Scholar]
- 55.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.French S, Wilson K. Treatment of negative intensity observations. Acta Crystallogr A. 1978;34:517–525. [Google Scholar]
- 57.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Read RJ, McCoy AJ. Using SAD data in Phaser. Acta Crystallogr D Biol Crystallogr. 2011;67:338–344. doi: 10.1107/S0907444910051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Real-space multiple-scattering calculation and interpretation of x-ray-absorption near-edge structure. Phys Rev B. 1998;58:7565. [Google Scholar]
- 65.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Cryst. 1997;30:203–205. [Google Scholar]