Abstract
Transcription factor A (TFAM) functions as a DNA packaging factor in mammalian mitochondria. TFAM also binds sequence-specifically to sites immediately upstream of mitochondrial promoters, but there are conflicting data regarding its role as a core component of the mitochondrial transcription machinery. We here demonstrate that TFAM is required for transcription in mitochondrial extracts as well as in a reconstituted in vitro transcription system. The absolute requirement of TFAM can be relaxed by conditions that allow DNA breathing, i.e., low salt concentrations or negatively supercoiled DNA templates. The situation is thus very similar to that described in nuclear RNA polymerase II-dependent transcription, in which the free energy of supercoiling can circumvent the need for a subset of basal transcription factors at specific promoters. In agreement with these observations, we demonstrate that TFAM has the capacity to induce negative supercoils in DNA, and, using the recently developed nucleobase analog FRET-pair tCO–tCnitro, we find that TFAM distorts significantly the DNA structure. Our findings differ from recent observations reporting that TFAM is not a core component of the mitochondrial transcription machinery. Instead, our findings support a model in which TFAM is absolutely required to recruit the transcription machinery during initiation of transcription.
The mtDNA is a double-stranded circular molecule that encodes 22 tRNAs, 2 rRNAs, and 13 subunits of the respiratory chain. Transcription is initiated from two sites, the light- and heavy-strand promoters (LSP and HSP1, respectively), and proceeds to produce near genome-length polycistronic transcripts, which are subsequently processed to generate the individual RNA molecules (1). Transcription from LSP also produces the RNA primers required for initiation of mtDNA replication at the origin of the heavy strand (2–4). In vivo experiments have identified a second transcription initiation site (HSP2) downstream of HSP1, but the sequence requirements of this promoter remain to be defined (5, 6).
In budding yeast, the basic machinery for mtDNA transcription consists only of two factors: the mitochondrial RNA polymerase (Rpo41) and its accessory factor Mtf1, also denoted sc-mtTFB (1). In mammalian cells, it has been reported that mitochondrial transcription also requires the transcription factor A (TFAM), a high-mobility group-box (HMG) protein (7, 8). TFAM plays a role as an mtDNA packaging factor that can bind, wrap, and bend DNA in a non–sequence-specific manner (9–12). TFAM also binds sequence-specifically to sites upstream (−15 to −35) of the HSP1 and LSP transcription start sites (13, 14). The human mitochondrial RNA polymerase (POLRMT) is distantly related to the bacteriophage T7 RNA polymerase and has the capacity to recognize promoter elements (14). In combination, POLRMT, TFAM, and the mammalian Mtf1 homolog TFB2M can initiate transcription from a promoter-containing DNA fragment in vitro (15). TFB2M forms a heterodimeric complex with POLRMT and interacts directly with the priming substrate, indicating that TFB2M acts as a transient component of the catalytic site of the transcription initiation complex (16).
TFAM is required to recruit POLRMT/TFB2M to LSP, and mutations in the TFAM high-affinity binding site abolish promoter-specific transcription (14, 17). The exact distance between the specific TFAM binding site and the LSP transcription start site is important for promoter function (17), and there may be direct physical interactions between the C-terminal domain of TFAM and the other components of the mitochondrial transcription machinery (18). X-ray structural studies have revealed that TFAM bound to LSP induces a dramatic U-turn in DNA and places the C-terminal tail of TFAM next to the transcription start site, where POLRMT and TFB2M are expected to bind (19, 20). Collectively, these observations support a model in which TFAM interacts with the other components of the mitochondrial transcription machinery and recruits them to a precise position at the promoter.
A recent study questioned the importance of TFAM for transcription initiation using a reconstituted in vitro transcription system and suggested that POLRMT and TFB2M could initiate promoter-specific transcription in the absence of TFAM (21). This finding led to the conclusion that mitochondrial transcription is a two-component system also in higher cells, much similar to the situation in yeast. However, this model is not easy to reconcile with findings from other laboratories that have demonstrated that TFAM is an essential mammalian transcription factor both in vivo and in vitro (6, 15, 22). We here investigate the role of TFAM for transcription initiation in a defined in vitro system. We find that the requirement for TFAM can be relaxed if conditions are chosen that allow for promoter breathing, i.e., low salt concentrations or negatively supercoiled DNA templates. The situation is thus similar to what has been described in nuclear transcription where the need for the basal transcription factors TFIIE, TFIIF, and TFIIH can be circumvented by the free energy of supercoiling (23). Our findings reconcile findings in the field and firmly establish TFAM as a basal transcription factor.
Results
TFAM-Depleted Mitochondrial Extracts Cannot Initiate Transcription.
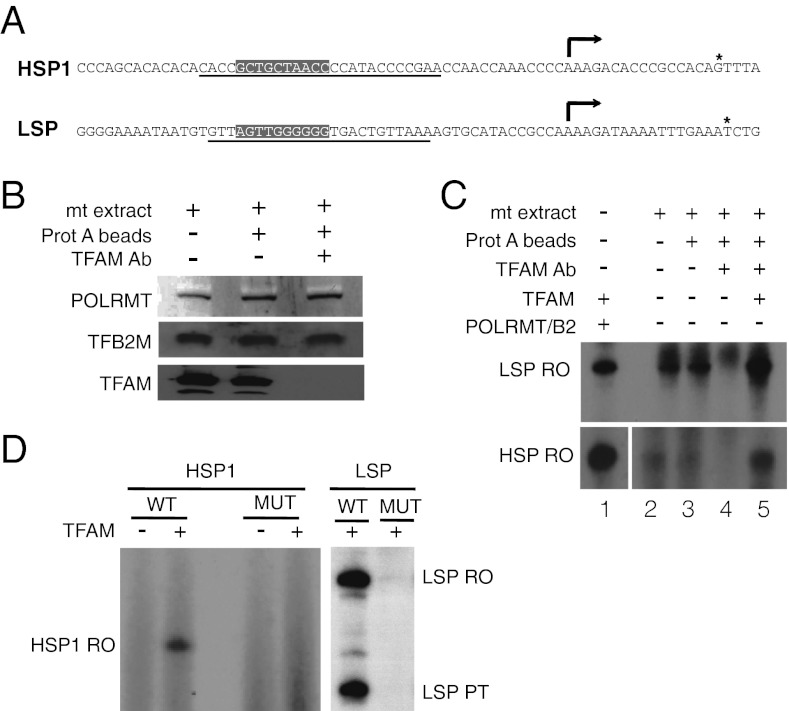
We first investigated if TFAM is required for transcription from the HSP1 and LSP promoters in mitochondrial extracts (Fig. 1A). To this end, we used antibodies to immunodeplete TFAM from transcriptionally active mitochondrial extracts. Immunoblotting analysis demonstrated that TFAM could be efficiently depleted, whereas the levels of POLRMT and TFB2M remained unchanged (Fig. 1B). We monitored run-off transcription using promoter-containing DNA fragments. We could observe transcription initiation from HSP1 and LSP in extracts before and after incubation with protein A beads (Fig. 1C, lanes 2 and 3), whereas loss of TFAM abolished transcription from both promoters (Fig. 1C, lane 4). By adding back recombinant pure TFAM protein to the immunodepleted extract, we could reconstitute mtDNA transcription from both promoters (Fig. 1C, lane 5). We conclude that TFAM is essential for promoter-specific initiation of transcription in mitochondrial extracts.
Fig. 1.
TFAM is required for HSP1 and LSP transcription in mitochondrial extracts. (A) Sequences of the HSP1 and LSP promoters. The TFAM binding sites (13, 14) are underlined. The boxed sequences were mutated to inactivate TFAM-dependent transcription. (B) Immunodepletion of TFAM from mitochondrial extracts. Immunoblotting against the indicated proteins demonstrates that POLRMT and TFB2M levels remained unaffected. (C) Run-off transcription in mitochondrial extracts was monitored using run-off transcription on linearized templates containing HSP1 or LSP. Experiments were performed as described in Materials and Methods. TFAM (1 pmol) was added when indicated. As a control, in vitro transcription was performed with recombinant TFAM, TFB2M, and POLRMT (lane 1). (D) Run-off transcription was monitored on linearized DNA templates containing (WT) or lacking (MUT) the high-affinity TFAM binding site. Experiments were performed as described in Materials and Methods. TFAM (2.5 pmol) was added when indicated. The TFAM binding sites were mutated by changing A to C, C to A, T to G, and G to T at positions −26 to −35 relative to the transcription start site. LSP PT corresponds to a shorter LSP product caused by premature transcription termination at CSBII (3).
It has previously been demonstrated that the high-affinity TFAM binding site upstream of LSP is required for transcription initiation in vitro (14, 17). To investigate the functional importance of the corresponding site upstream of HSP1, we mutated 10 base pairs in the TFAM binding site (Fig. 1A) and investigated the effect on HSP1 activity in a reconstituted in vitro transcription system containing purified TFAM, POLRMT, and TFB2M (Fig. 1D). No transcription could be observed with the mutant template, supporting the conclusion that sequence-specific TFAM binding is required for HSP1 function.
Low-Salt Conditions Relax the Absolute Requirement of TFAM.
The experimental conditions used to demonstrate TFAM-independent initiation of transcription contained low levels of salt (21); the only salt added to the reactions was 5–10 mM MgCl2. We speculated that these conditions could influence TFAM dependency, and to test this we repeated the reported experiments. We monitored run-off transcription on linearized DNA templates containing HSP1, LSP, or both HSP1 and LSP (Fig. 2A). Indeed, in the presence of 5 mM MgCl2 and low levels of NaCl (4 mM), our transcription system generated run-off transcription products in the absence of TFAM (Fig. 2A, lane 1). In agreement with results published by others (21), the highest levels of TFAM-independent transcription were observed at HSP1, whereas relatively low levels of LSP transcription were seen in the absence of TFAM. Interestingly, a gradual increase of NaCl concentrations led to the inhibition of promoter-specific transcription (Fig. 2A, lanes 1–5). The levels of both LSP and HSP1 transcription dropped significantly at 12 mM NaCl, and the reactions were almost completely inhibited in the presence of 24 mM NaCl. When TFAM was added to the reactions, we obtained markedly different results (Fig. 2A, lanes 6–10). Importantly, the transcription reactions gave substantially higher levels of run-off products in the presence of TFAM, even at very low salt concentrations (Fig. 2A, compare lanes 1 and 6). Note that the depicted gel images in Fig. 2A, right hand column, are overexposed on purpose, because we wanted to use the same exposure time for all of the experiments to allow for a direct comparison of the efficiency of promoter-dependent transcription in the presence and absence of TFAM. Furthermore, the TFAM-containing transcription reactions remained unaffected by increasing amounts of NaCl added. The transcription yield was similar at 4 and 48 mM NaCl (Fig. 2A, compare lanes 6 and 10), and we could not observe a clear effect on transcription levels until NaCl concentrations were above 100 mM (Fig. 2B).
Fig. 2.
The requirement of TFAM can be relaxed at low ionic strength. (A) The effect of increasing ionic strength on run-off transcription was monitored on linearized DNA templates containing LSP (Middle), HSP1 (Bottom), or both HSP1 and LSP (Top). Transcription reactions contained the indicated template, POLRMT (400 fmol) and TFB2M (400 fmol). Transcription was monitored in the absence (Left) and presence (Right) of TFAM (2.5 pmol). (B) Promoter-specific transcription is less sensitive to salt concentration than promoter-independent transcription. The effect of ionic strength on HSP1 transcription was monitored in the presence of TFAM as in A. (C) Bacterially expressed POLRMT and TFB2M (indicated with a B) require TFAM for HSP1 transcription at 40 mM NaCl. Identical results were obtained with TFB2M expressed in insect cells (SF9).
DNA is not a static structure. Because the negatively charged phosphates of the DNA backbone repel each other, the double helix may spontaneously denature locally, opening up single-stranded zones even under physiological conditions. Salt shields the negative charges and stabilizes the double helix (24). Therefore, the lower salt concentrations required for TFAM-independent transcription could potentially affect the behavior of the DNA and cause breathing of the DNA template. In support of this notion, we observed that low salt concentrations did not only cause TFAM-independent transcription from promoter sequences, but also high levels of background transcription, suggesting nonspecific initiation of transcription from partially single-stranded regions in the DNA templates (Fig. 2 A and B, black bars). This nonspecific initiation of transcription was completely abolished at higher salt (>70 mM NaCl) without affecting the TFAM-dependent initiation (Fig. 2B, lanes 4–6).
To verify that the requirement of TFAM was not specific to our experimental setup with recombinant proteins expressed in insect cells, we also investigated TFAM dependence using commercially available transcription factors expressed in Escherichia coli, which previously had been used by others to demonstrate TFAM-independent transcription initiation (21). On a linearized template and in the presence of salt (40 mM NaCl in the experiment displayed), we obtained consistent results from the two different transcription systems. In both recombinant systems, transcription was dependent on TFAM (Fig. 2C). From our experiments we could conclude that TFAM is a core component of the mitochondrial transcription machinery, not only in vivo and in mitochondrial extracts, but also in a reconstituted in vitro transcription system.
TFAM Is Not Absolutely Required on a Negatively Supercoiled Template.
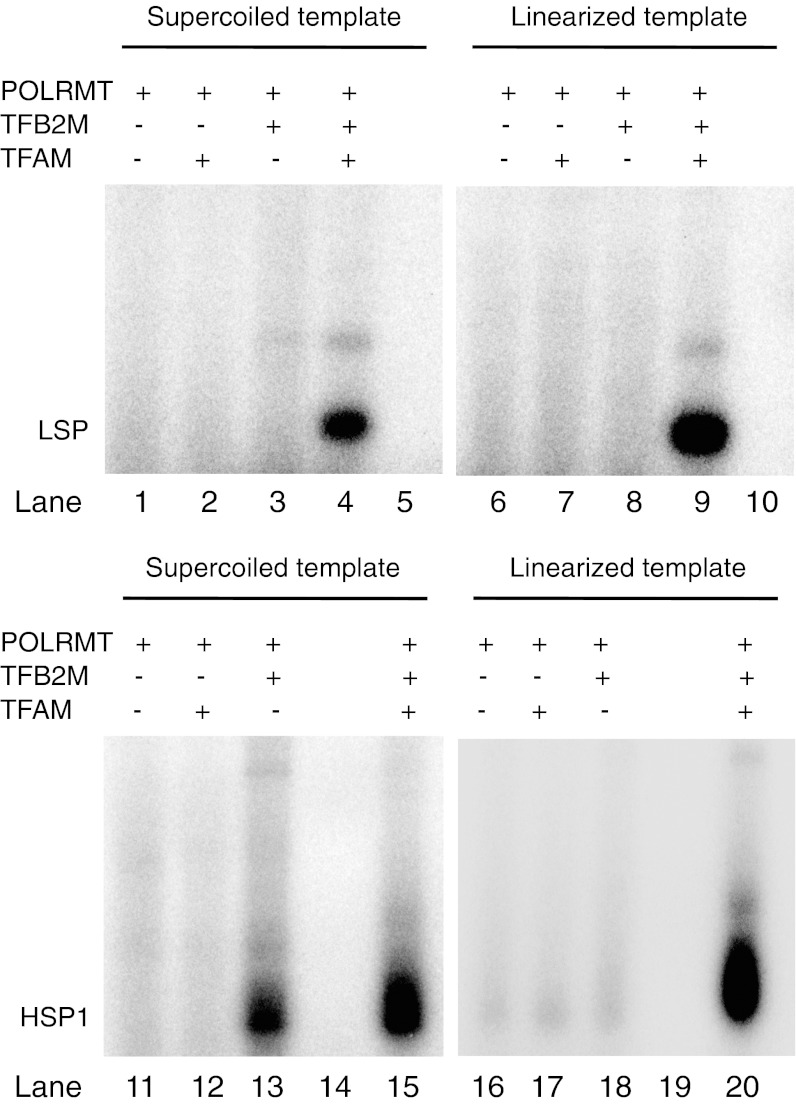
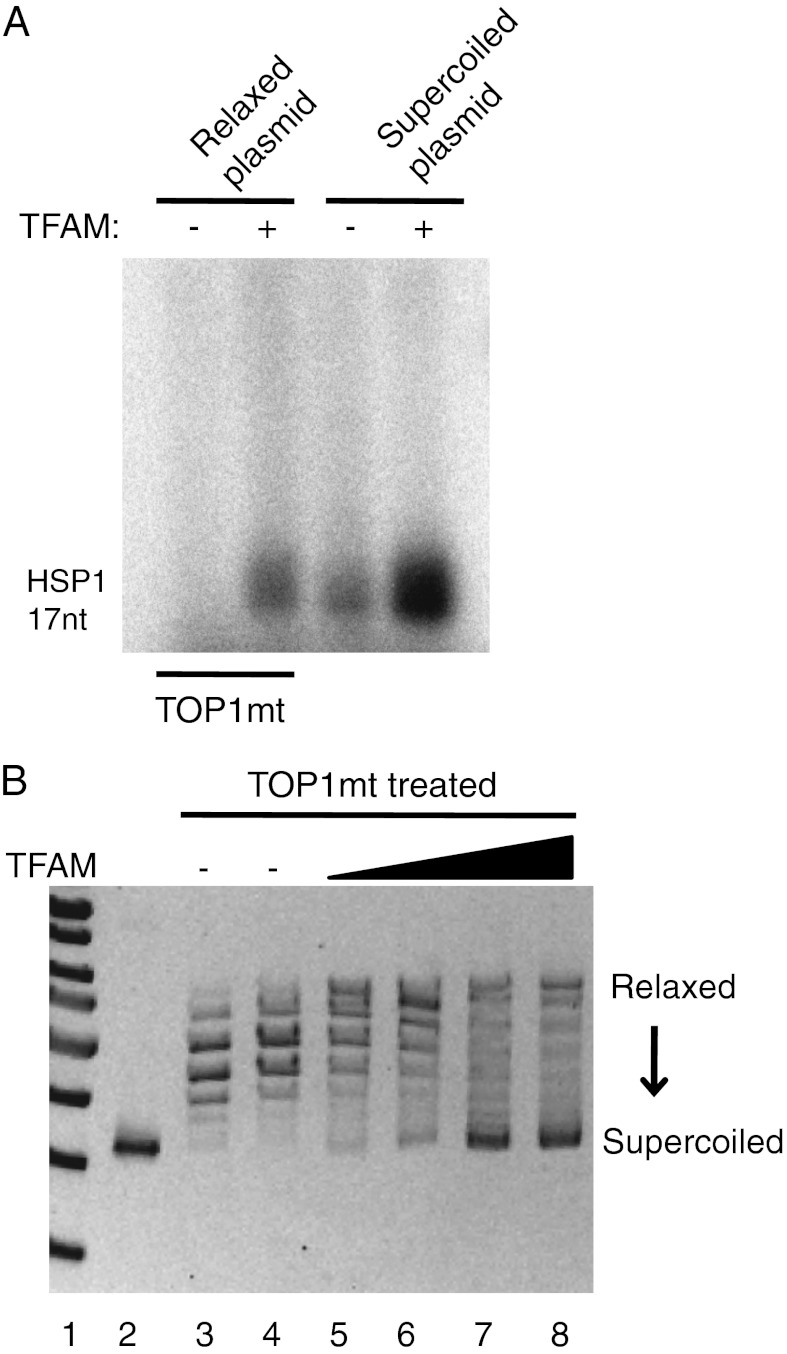
An alternative way to facilitate DNA breathing is negative supercoiling. We therefore tested if negative supercoiling could relax the absolute requirement of TFAM for basal transcription. To obtain transcription products of defined lengths on our circular template, we omitted either CTP, creating an 18-nt–long transcript from LSP, or UTP to generate a 17-nt–long transcript from HSP1 (Fig. 1A; the last transcribed position is indicated with an asterisk). The experiments were performed at 80 mM NaCl, and for comparison we used linearized DNA templates. On a linearized template, TFAM was required at both LSP and HSP1 for transcription initiation. On a negatively supercoiled template, LSP still required TFAM, whereas HSP1 was active also in the absence of TFAM (Fig. 3, Lower, lane 13). Our experiments therefore demonstrated that negative supercoiling could allow HSP1-dependent transcription even in the absence of TFAM. We also monitored transcription initiation on a negatively supercoiled template, before and after addition of mitochondrial topoisomerase I (TOP1mt). TOP1mt relaxed the negative supercoils and restored the strict requirement of TFAM for initiation of transcription at HSP1 (Fig. 4A).
Fig. 3.
Negative supercoiling can relax the requirement of TFAM at HSP1, but not at LSP. TFAM dependence was monitored on negatively supercoiled or linearized templates containing LSP or HSP1. To produce transcripts of a defined length, CTP was omitted from the LSP transcription reaction, generating an 18-nt–long product. Similarly, UTP was omitted from the HSP1 reaction, leading to the formation of a 17-nt transcript. Transcription reactions contained the indicated template, and POLRMT (400 fmol), TFB2M (400 fmol), and TFAM (2.5 pmol) were added when indicated.
Fig. 4.
Topoisomerase I restores the requirement of TFAM for initiation of transcription at HSP1. (A) Transcription reactions were performed as in Fig. 3. When indicated, the template was incubated with TOP1mt (280 fmol) before transcription, as described in Materials and Methods. (B) TFAM induces negative supercoils in plasmid DNA. The input DNA (lane 2) was relaxed by incubation with TOP1mt for 15 min (lane 3) before increasing amounts of TFAM (0, 5.5, 11, 22, or 45 pmol) were added to the DNA and incubated together with TOP1mt for another 20 min (lanes 4–8).
TFAM Structurally Alters Promoter DNA.
The observation that negative supercoiling can relax the absolute requirement of TFAM was interesting because Abf2, the TFAM homolog in budding yeast, has the capacity to introduce negative supercoils into DNA (25). We could demonstrate a similar activity for TFAM (Fig. 4B), suggesting that this could be an essential aspect of the molecular mode by which TFAM activates promoters. To further verify that TFAM induces structural changes in DNA, and to investigate if these alterations could be directed specifically to a region around the transcription start site, we used FRET experiments (SI Text). We incorporated the fluorescent cytosine analog tCO (26, 27) at several positions close to the HSP1 transcription initiation site and also synthesized the corresponding complementary oligonucleotides, including some sequences in which one cytosine was replaced by the nonemissive cytosine analog tCnitro (28), which functions as a FRET acceptor for tCO (Table S1 and Fig. S1). Our data confirmed that the binding of TFAM induces significant structural changes in the template that could be consistent with DNA breathing (Table S2). We also investigated if a mutation in the TFAM binding site that inactivated HSP1 transcription in vitro (Fig. 1C) could affect TFAM-dependent structural changes around the transcription start site. Our data revealed similar changes in FRET efficiency with the WT and mutant template (Tables S1 and S2, duplex DMUT–AMUT). Therefore, the observed effect of TFAM on DNA structure close to the transcription start site appears to be independent of the protein’s sequence-specific binding activity, but rather a consequence of TFAM’s ability to bind and bend DNA in a sequence-independent fashion. In support of this notion, mutations in the TFAM binding site that inactivated HSP1 promoter activity did not significantly affect TFAM binding to a promoter-containing DNA fragment in a gel retardation experiment (Fig. S2). Therefore, even if TFAM requirements may be relaxed by conditions that promote DNA breathing, the role of TFAM under physiological conditions cannot simply be reduced to structural changes in promoter sequences, but most likely also involves complex interactions with the other components of the transcription machinery.
Discussion
In nuclear transcription, RNA polymerase II requires a set of basal transcription factors to initiate promoter-specific transcription in vitro. Among these factors, TFIIH, which harbors a helicase activity, helps to unwind template DNA and provides a single-stranded DNA bubble for the transcription machinery. Even if TFIIH is required for basal transcription in vivo, the requirement of this factor may be relaxed in vitro. Using negatively supercoiled template, it is possible to reconstitute transcription from some promoters in the absence of TFIIH, e.g., at the Ig heavy-chain gene promoter. The free energy of supercoiling circumvents the requirement of TFIIH by promoting the formation of an open complex (23).
In a similar fashion, conditions that stimulate DNA breathing, i.e., low salt or negative supercoiling, help to relax the absolute requirement of TFAM for transcription initiation at mitochondrial promoters. POLRMT has an intrinsic capacity to recognize specific DNA sequence elements at the promoter (14), which may explain why DNA breathing helps the enzyme to initiate transcription in a sequence-specific manner. In addition, it is clear from results presented here and elsewhere that low salt and negative supercoiling lead to unspecific initiation of transcription at many other sites on a DNA template (Fig. 2A) (29). Recent structural studies of TFAM in complex with a LSP-containing promoter fragment have revealed that TFAM binding forces promoter DNA to undergo a sharp U-turn. The two HMG-box domains of the protein wedge into the DNA minor groove and thus cause two kinks on one face of the DNA. TFAM arranged in a similar fashion is supposed to cover the entire mitochondrial genome, thus compacting and protecting the mtDNA molecule (19, 20). In support of the structural findings, FRET analysis using end-labeled DNA fragments have demonstrated that TFAM binding causes DNA bending, and that this effect is stronger on promoter DNA than on a nonspecific DNA fragment (20, 30). FRET analysis have also demonstrated that the C-terminal tail of TFAM, which is required for efficient initiation of transcription, confers increased affinity and bending to a LSP promoter fragment, further demonstrating a link between DNA structure and promoter-dependent transcription (30). Interestingly, mutations in TFAM, which cause a defect in DNA bending, also impair promoter-specific transcription in vitro. The effect is particularly pronounced at the LSP promoter, whereas HSP1 is less affected (20). Our data also demonstrate that the structural requirements may differ between LSP and HSP1, because negative supercoiling only activates TFAM-independent transcription from HSP1, whereas LSP remains inert. In a related way, low-salt conditions have a stronger stimulatory effect on HSP1 compared with LSP.
POLRMT is unable to melt promoter DNA in the absence of TFB2M (16). A molecular explanation for this observation was recently offered with the solution of the POLRMT X-ray structure (31). In POLRMT, the two regions that melt DNA in phage RNA polymerases (the fingers domain and the intercalating hairpin) are repositioned, which leads to a clash between the intercalating hairpin and the template strand of DNA and causes the fingers domain to block single-stranded DNA from the active site. Based on these findings, it was suggested that TFB2M may help to reposition the fingers domain and the intercalating hairpin so that they could function in promoter melting. It is intriguing to speculate that promoter melting is a rate-limiting step in transcription initiation and that TFAM may stimulate this process further, by inducing negative supercoils in promoter DNA. In addition, TFAM could also stimulate inefficient promoter melting, by recruiting the transcription machinery via direct protein–protein interactions that positions POLRMT and TFB2M in direct proximity of the transcription start site. TFAM has been shown to interact directly with TFB2M and its paralogue, TFB1M (32). The functional importance of this finding is still unclear, because TFB1M is not involved in mitochondrial transcription, but rather plays a role in ribosomal biogenesis (33). However, even if further investigations are required, the close proximity between TFAM and the transcription start site makes direct physical interactions with TFB2M and/or POLRMT very likely. Such interactions may strongly stimulate POLRMT recruitment to the promoter region and help to overcome inefficient promoter melting at physiological salt conditions.
Based on findings reported here, we conclude that TFAM indeed is a core component of the mitochondrial transcription machinery. We also demonstrate that even very small changes in salt concentration may have a strong effect on in vitro transcription reactions, which may complicate interpretations of experimental results. As a rule, in vitro findings should correlate with findings made in vivo or in cellular extracts. The effect of salt should always be considered, e.g., when new regulators of mitochondrial transcription are being identified by in vitro transcription assays, and in vivo verification of novel transcriptional mechanisms is essential.
Materials and Methods
Expression and Purification of Recombinant Proteins.
POLRMT (amino acids 42–1230, N-terminal 10× His-tag), TFAM (amino acids 43–246, N-terminal 6× His-tag), and TFB2M (amino acids 1–396, C-terminal 6× His-tag) were expressed from recombinant baculoviruses in insect cells and purified as described previously (15). POLRMT (amino acids 42–1230), TFAM (amino acids 43–246), and TFB2M (amino acids 31–396) expressed in E. coli were obtained from Enzymax LLC and used as described in Shutt et al. (21). Recombinant Autographa californica nuclear polyhedrosis virus expressing TOP1mt (amino acids 1–601, C-terminal 6× His-tag) was constructed using the BacPAK system, following the manufacturer’s instructions (Clontech). TOP1mt was expressed in insect cells and purified as described in SI Text.
FRET Measurements.
The DNA sequences listed in Table S1 were provided by ATDBio Ltd. The cytosine analogs tCO and tCnitro incorporated in these sequences were purchased from Glen Research Corp. Detailed protocols for DNA duplex formation and FRET measurements can be found in SI Text.
In Vitro Transcription.
We used DNA fragments corresponding to base pairs 1−741 (LSP and HSP1), 1−477 (LSP), or 499−741 (HSP1) of human mtDNA cloned into pUC18 for analysis of promoter-specific transcription in run-off assays as previously described (15). For detailed information, please see SI Text. In HSP1 and LSP transcription reactions using circular DNA templates, radioactive UTP was replaced with 0.2 μM [α-32P]CTP (3,000 Ci/mmol) and 0.2 μM [α-32P]ATP (3,000 Ci/mmol), respectively. When indicated, the HSP1 template was preincubated with 280 fmol of TOP1mt for 15 min at 30 °C before in vitro transcription was initiated.
Topoisomerase Assay.
Individual reaction mixtures (9 μL) contained supercoiled pUC19 (0.2 μg), 10 mM Tris⋅HCl (pH 8.0), 1 mM DTT, 100 μg/mL BSA, 10 mM MgCl2, and 10% (vol/vol) glycerol. The supercoiled plasmid template was relaxed by incubation in the presence of 280 fmol TOP1mt for 15 min at 30 °C. When indicated, TFAM (0, 5.5, 11, 22, or 45 pmol) was added to the reaction, and the reaction was further incubated for 20 min at 30 °C before being stopped by the addition of 2 μL of stop solution [90 mM EDTA (pH 8.0), 6% (wt/vol) SDS, 30% (wt/vol) glycerol, 0.25% (wt/vol) bromophenol, 0.25% (wt/vol) xylene cyanol]. The samples were loaded on a 1% (wt/vol) native agarose gel in 1× Tris⋅acetate- EDTA buffer, and the gel was run for 1.5 h at 100 V before stained with ethidium bromide.
TFAM Depletion from Mitochondrial Extracts.
Protein A Sepharose beads (50 μL; GE Healthcare) were equilibrated with mitochondrial lysis buffer [10% (vol/vol) glycerol, 25 mM Hepes (pH 7.6), 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, 500 mM KCl, 0.5% (vol/vol) Tween 20] and incubated with or without 30 μL TFAM antibody (raised by Agrisera AB) in a 130-μL volume reaction. After a 6-h rotation at 4 °C, the beads were washed 3× with mitochondrial lysis buffer. The 80-μL mitochondrial extracts prepared as described (34) were added to the beads and incubated overnight at 4 °C with rotation. After centrifugation at 800 × g for 1 min at 4 °C, TFAM-depleted mitochondrial extracts were ready for use in transcription assays.
Supplementary Material
Acknowledgments
This study was supported by Swedish Research Council Grants 2010-2766 (to N.-G.L.), 2009-4848 (to C.M.G.), 2008-4990 (to L.M.W.), and 2010-3545 (to M.F.); Swedish Strategic Foundation Grant FFL-3; European Research Council Starting Grant 261248 and Advanced Grant 268897; the Swedish Cancer Foundation; the Olle Engkvist Byggmästare Foundation (L.M.W.); and the Swedish Society for Medical Research (to S.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119738109/-/DCSupplemental.
References
- 1.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 2.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pham XH, et al. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J Biol Chem. 2006;281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 4.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc Natl Acad Sci USA. 2010;107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litonin D, et al. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 9.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 10.Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parisi MA, Xu B, Clayton DA. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol Cell Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufman BA, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 14.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 16.Sologub M, Litonin D, Anikin M, Mustaev A, Temiakov D. TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell. 2009;139:934–944. doi: 10.1016/j.cell.2009.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim Biophys Acta. 1995;1271:127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- 18.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 19.Rubio-Cosials A, et al. Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat Struct Mol Biol. 2011;18:1281–1289. doi: 10.1038/nsmb.2160. [DOI] [PubMed] [Google Scholar]
- 20.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat Struct Mol Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanki T, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 24.Metzler R, Ambjornsson T. Dynamic approach to DNA breathing. J Biol Phys. 2005;31:339–350. doi: 10.1007/s10867-005-2410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diffley JF, Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J Biol Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- 26.Börjesson K, et al. Nucleic acid base analog FRET-pair facilitating detailed structural measurements in nucleic acid containing systems. J Am Chem Soc. 2009;131:4288–4293. doi: 10.1021/ja806944w. [DOI] [PubMed] [Google Scholar]
- 27.Sandin P, et al. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Res. 2008;36:157–167. doi: 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preus S, Börjesson K, Kilså K, Albinsson B, Wilhelmsson LM. Characterization of nucleobase analogue FRET acceptor tCnitro. J Phys Chem B. 2010;114:1050–1056. doi: 10.1021/jp909471b. [DOI] [PubMed] [Google Scholar]
- 29.Fukuoh A, et al. DNA conformation-dependent activities of human mitochondrial RNA polymerase. Genes Cells. 2009;14:1029–1042. doi: 10.1111/j.1365-2443.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 30.Malarkey CS, Bestwick M, Kuhlwilm JE, Shadel GS, Churchill ME. Transcriptional activation by mitochondrial transcription factor A involves preferential distortion of promoter DNA. Nucleic Acids Res. 2012;40:614–624. doi: 10.1093/nar/gkr787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringel R, et al. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 32.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metodiev MD, et al. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Fernández-Silva P, Micol V, Attardi G. Mitochondrial DNA transcription initiation and termination using mitochondrial lysates from cultured human cells. Methods Enzymol. 1996;264:129–139. doi: 10.1016/s0076-6879(96)64014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.