Abstract
Alphaviruses, a group of positive-sense RNA viruses, are globally distributed arboviruses capable of causing rash, arthritis, encephalitis, and death in humans. The viral replication machinery consists of four nonstructural proteins (nsP1–4) produced as a single polyprotein. Processing of the polyprotein occurs in a highly regulated manner, with cleavage at the P2/3 junction influencing RNA template use during genome replication. Here, we report the structure of P23 in a precleavage form. The proteins form an extensive interface and nsP3 creates a ring structure that encircles nsP2. The P2/3 cleavage site is located at the base of a narrow cleft and is not readily accessible, suggesting a highly regulated cleavage. The nsP2 protease active site is over 40 Å away from the P2/3 cleavage site, supporting a trans cleavage mechanism. nsP3 contains a previously uncharacterized protein fold with a zinc-coordination site. Known mutations in nsP2 that result in formation of noncytopathic viruses or a temperature sensitive phenotype cluster at the nsP2/nsP3 interface. Structure-based mutations in nsP3 opposite the location of the nsP2 noncytopathic mutations prevent efficient cleavage of P23, affect RNA infectivity, and alter viral RNA production levels, highlighting the importance of the nsP2/nsP3 interaction in pathogenesis. A potential RNA-binding surface, spanning both nsP2 and nsP3, is proposed based on the location of ion-binding sites and adaptive mutations. These results offer unexpected insights into viral protein processing and pathogenesis that may be applicable to other polyprotein-encoding viruses such as HIV, hepatitis C virus (HCV), and Dengue virus.
Alphaviruses are small, enveloped RNA viruses that infect various vertebrates, including humans, horses, birds, and rodents (1). Transmission occurs via an invertebrate vector, usually mosquitoes. In humans, typical alphavirus infection can result in rash, arthritis, encephalitis, and death. Within the past decade, infection by Chikungunya virus (CHIKV), a member of the genus alphavirus, caused massive epidemics in Africa and Southeast Asia, with positive cases reported in Europe and the United States, raising the possibility of an emerging threat (2). Additionally, CHIKV often presents with nonclassic symptoms that go undiagnosed, making global pathogenicity difficult to monitor (3).
The alphavirus genome consists of single-stranded, positive-sense RNA of ∼9–11 kb with a 5′ cap structure and 3′ polyadenosine tail. The genome contains two cistrons. The first, located in the 5′ two-thirds of the genome, encodes the viral replication machinery, termed nonstructural proteins (nsPs), whereas the structural proteins that form virus particles are encoded in the second cistron. The genomic RNA is used as an mRNA for the translation of the nsPs and as a template for the synthesis of complementary negative-sense RNA. The negative-sense strand is a template for subgenomic RNA containing the second cistron and progeny, genomic RNA. Because genomic and subgenomic RNAs are made in vast excess, the negative-sense RNA strand is considered a replication intermediate.
The alphavirus replication machinery is composed of four nonstructural proteins (nsP1 to -4), which are expressed as one of two polyproteins (P123 or P1234). P1234 is expressed as a read-through of an opal termination codon at the end of nsP3. These precursor polyproteins are cleaved by a protease within nsP2 (1, 4). After translation of P1234, cleavage at the P3/4 junction occurs either in cis or trans, followed by the P1/2 junction, which occurs in cis only (4, 5). Both P123+nsP4 and nsP1+P23+nsP4 preferentially synthesize negative strand viral RNA (6, 7). The final cleavage event between P23 produces fully mature nsPs and switches RNA template for synthesis of positive-sense genomic and subgenomic RNAs. The correlation between P23 cleavage and the switch from negative- to positive-sense RNA production is poorly understood.
In mammalian cells, alphavirus infection inhibits host gene expression, causes severe cytopathic effects (CPEs), disrupts the cellular response to IFN, and leads to cell death (8–11). Mature nsP2 contains an amino-terminal RNA helicase domain, a central protease domain that catalyzes all cleavages between the nonstructural proteins, and an inactive RNA methyltransferase-like (MT-like) domain (12). During infection, a portion of nsP2 localizes to the nucleus (13, 14) and plays a role in shutting off host-cell transcription and viral cytopathogenicity (9, 10). Two genetically distinct groups of alphaviruses termed New World and Old World have emerged because geographic isolation (8). In Old World alphaviruses, such as Sindbis virus (SINV) and Semliki Forest virus (SFV), transcriptional shutoff is nsP2-dependent (15). Even in the context of a replicon lacking the structural proteins, the nsPs of SINV and SFV alone can efficiently shut off host transcription and induce severe CPE (16).
Mutations that cause alphaviruses to establish a persistent infection with minimal or no CPE have been identified (16–20). A key feature of these mutants is their inability to inhibit host-cell transcription (10, 15). The best-characterized noncytopathic mutations map to nsP2 proline 726 (P726) within the MT-like domain (10, 16, 17, 21, 22). The P726 mutation has been shown to attenuate the cytotoxic effect caused by the expression of wild-type SINV nsP2 protein alone (10). Noncytopathic mutations of nsP2 P726 also reduce viral RNA replication levels and are unable to shut off host transcription and translation processes, showing the overall importance of nsP2 to viral replication, as well as host-cell interaction (11, 16).
The function(s) of nsP3 in alphavirus replication remains unresolved. Based on sequence conservation, nsP3 is organized into three domains: an amino-terminal macro or X domain (23), a central alphavirus-specific region, and a hypervariable-sequence carboxyl terminus. Macro domains have been shown to bind ADP ribose (24) with certain viral macro domains also retaining ADP ribose-1′′ phosphatase activity (25, 26). Recent work in SFV shows that residues just after the macro domain of nsP3 play a role in positioning of the P23 cleavage site (27). Additionally, the carboxyl-terminal region of nsP3 contains numerous phosphorylation sites targeted by yet unknown cellular kinases (28, 29).
Here, we present the structure of an uncleaved P23 precursor protein spanning the nsP2 protease domain through to the central, zinc-binding domain (ZBD) of nsP3 from SINV (Fig. 1A). The P23 cleavage site is located in a narrow cleft formed between nsP2 and nsP3 that is inaccessible for proteolysis. Surprisingly, all of the previously reported nsP2 noncytopathic mutants lie at the interface between nsP2 and nsP3. Mutagenesis of residues in nsP3 opposite of nsP2 P726 showed inefficient cleavage of P23, delayed onset of pathogenesis and host gene shutoff, and altered viral RNA synthesis. Because P23pro-zbd copurified with large amounts of RNA, we propose a potential RNA-binding surface that extends across both nsP2 and nsP3 and hypothesize that cleaving the P2/3 junction may alter the RNA-binding surface, thereby contributing to template switching by the replication machinery. Our results provide a better understanding of the mechanism of viral polyprotein processing and pathogenesis.
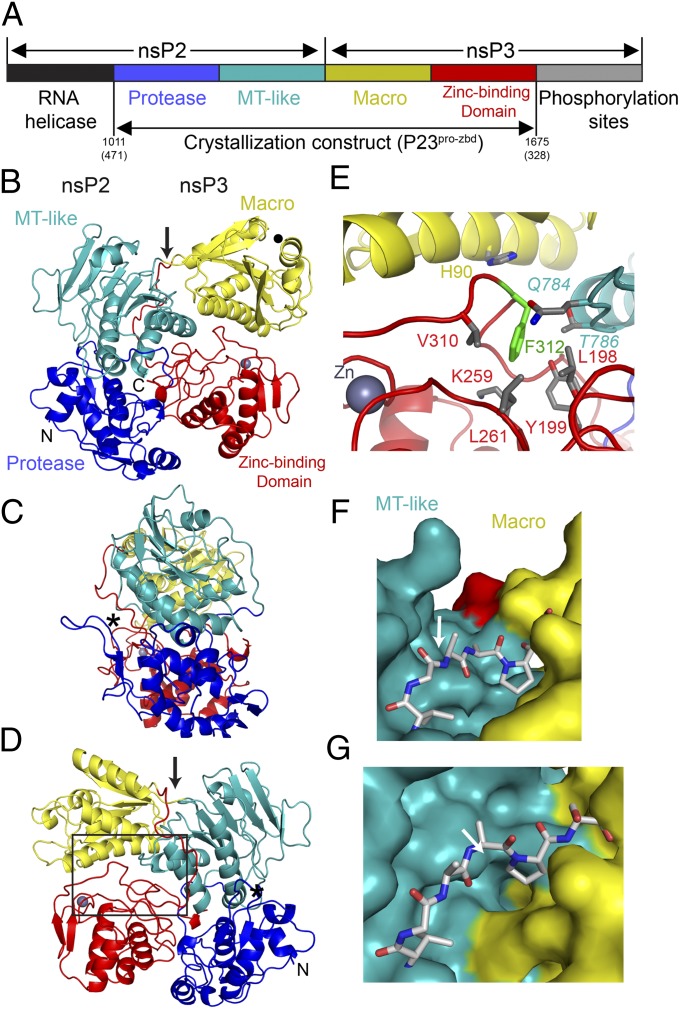
Fig. 1.
Overview of the alphavirus replication machinery and P23pro-zbd structure. (A) Schematic representation of the alphavirus replication machinery with emphasis on domain organization of nsP2 and nsP3. The crystallization construct consists of four domains: protease (blue) and methyltransferase-like (teal) domains of nsP2 and the macro (yellow) and zinc-binding (red) domain of nsP3, encompassing amino acids 1011–1675 of the P1234 polyprotein. Amino acid numbering provided from the amino terminus of nsP2 and nsP3 are also noted in parentheses. (B–D) Ribbon diagram of P23pro-zbd colored according to A. A gray sphere denotes the position of the zinc ion. The P2/3 cleavage site, nsP2 protease active site, and ADP ribose–binding site are labeled with an arrow, asterisk (*), and filled circle, respectively. (C) Ribbon diagram of P23 rotated 90° about a vertical axis from the view in B. (D) Ribbon diagram of P23 rotated 180° about a vertical axis from the view in B. Box shows location of the view in E. (E) Previously described nsP3 temperature-sensitive mutant ts7 (F312S) highlighted in green with the immediately surrounding amino acids located in the macro, ZBD, and MT-like domains. nsP2 residues are shown in italics. (F and G) The P2/3 cleavage site is located at the base of a narrow cleft formed by the MT-like and macro domains. Three amino acids on either side of the scissile bond (arrow) are shown in stick representation. The view in F is similar to that in B, whereas that in G is rotated 90° about a horizontal axis from F, looking directly into the cleft.
Results
Alphavirus P23pro-zbd Structure.
The SINV P23 precursor protein extending from the nsP2 protease domain to the nsP3 central domain was shown to be fully intact by SDS/PAGE and mass spectrometry despite the presence of a competent nsP2 protease active site (Fig. S1 A and B). Sequence alignment of the central domain of nsP3 shows numerous absolutely conserved cysteine residues (Fig. S2), suggesting the presence of a metal ion–binding site. Various purified preparations of the middle region of nsP3 were analyzed by quantitative X-ray fluorescence for common transition metals found in metalloproteins (30, 31) and shown to have one zinc ion per molecule. Crystals of P23pro-zbd diffracted to 2.85-Å resolution and the structure was determined by molecular replacement with structures of nsP2pro (12) and nsP3 macro domain (32) (Table S1). The resulting electron density allowed for an unambiguous trace of the entire polypeptide chain, including the previously uncharacterized nsP3 central region containing the zinc-binding site. The location of the zinc ion was confirmed by anomalous difference maps (Table S1 and see Fig. S4B).
SINV P23pro-zbd shows a highly compact structure, with the four domains (protease and MT-like from nsP2; macro and ZBD from nsP3) arranged with each domain occupying a vertex of a rectangle (Fig. 1 B–D). The polypeptide chain progresses around the perimeter of the rectangle such that the nsP2 protease and nsP3 ZBD are adjacent to one another. The carboxyl terminus of the P23pro-zbd construct is pointing toward the protease active site (Fig. 1 B–D). A previously described SINV mutant (ts7) defective in RNA synthesis at nonpermissive temperatures contains a F312 to serine mutation in nsP3 (33). This residue is located in the hydrophobic core and surrounded by residues from the macro, ZBD, and MT-like domains (Fig. 1E). Mutation of this residue would likely destabilize the core and disrupt domain interaction, giving rise to the RNA− phenotype at the nonpermissive temperature.
In a previously described nsP2pro structure, the six carboxyl-terminal amino acids were disordered (12). In the P23pro-zbd structure presented here, the entire polypeptide between nsP2 and nsP3 is ordered, permitting modeling of the P2/3 cleavage site. The P2/3 cleavage site is located at the base of a narrow cleft about 11–13 Å wide formed by the MT-like domain of nsP2 and the macro domain of nsP3 (Fig. 1 F and G). The cleavage site is 40 Å away from the nsP2 protease active site, supporting a trans cleavage mechanism (5, 34, 35). Although the 6 aa surrounding the scissile bond are solvent-exposed, attempts to position the protease domain onto the P2/3 cleavage site using the proposed model (36) resulted in numerous steric clashes.
ZBD in nsP3 Is Essential for Viral Replication.
After the P2/3 cleavage site, the polypeptide chain continues into the nsP3 macro domain. The ADP ribose–binding site within the macro domain is solvent-exposed and points away from the other four domains. The polyprotein exits the last helix of the macro domain adjacent to the P2/3 cleavage site and continues into a stretch of ∼40 aa that lacks secondary structure but forms a twist (Fig. S3). The conserved nsP3 central domain contains an antiparallel α-helical bundle, two parallel β-strands, and a previously unknown zinc-coordination site (Fig. S4B). The zinc ion is coordinated by C263, C265, C288, and C306 of nsP3. C263 and C265 are located in the loop between the last two α-helices, whereas C288 and C306 are at the carboxyl termini of the two parallel β-strands (Fig. S3C). Each of the zinc coordinating cysteines is invariant among all alphavirus sequences (Fig. S2). Structural comparison of the nsP3 ZBD against all known folds in the Protein Data Bank using PDBe Fold (37) and Dali servers (38) failed to produce any statistically significant hits, and no metalloprotein folds were identified. The spatial arrangement of secondary structural elements that contribute to nsP3 zinc-binding site, i.e., two cysteines in a loop between alpha helices and the other two cysteines at the end of two parallel β-strands, represents a structural scaffold for zinc-ion coordination (39).
Site-directed mutagenesis was performed to examine the functionality of the four conserved cysteine residues involved in zinc coordination in nsP3 (Fig. S4A). All four SINV mutants (C263A, C265A, C288A, and C306A) failed to produce productive infection in BHK cells, as evaluated by infectious center and plaque assays (Fig. S4C). A conserved cysteine residue in nsP3 (C247) unrelated to the zinc-binding site served as a control and C247A mutant produced infectious particles to near wild-type levels. Zinc-coordination mutants failed to express the nonstructural polyprotein and replicate virus, thereby supporting the structural role of the zinc ion (Fig. S4D).
The coordination site forms a distorted tetragon due to the presence of two highly conserved serine residues (S289 and S290) below the zinc atom (Fig. S2). Mutation of these serine residues did not alter viral replication levels. Furthermore, a double serine to alanine mutant (S289A+S290A) resulted in decreased viral RNA infectivity, but overall viral titers were comparable to wild type (Fig. S4C).
Importance of P23 Interface for Polyprotein Processing and Viral Pathogenesis.
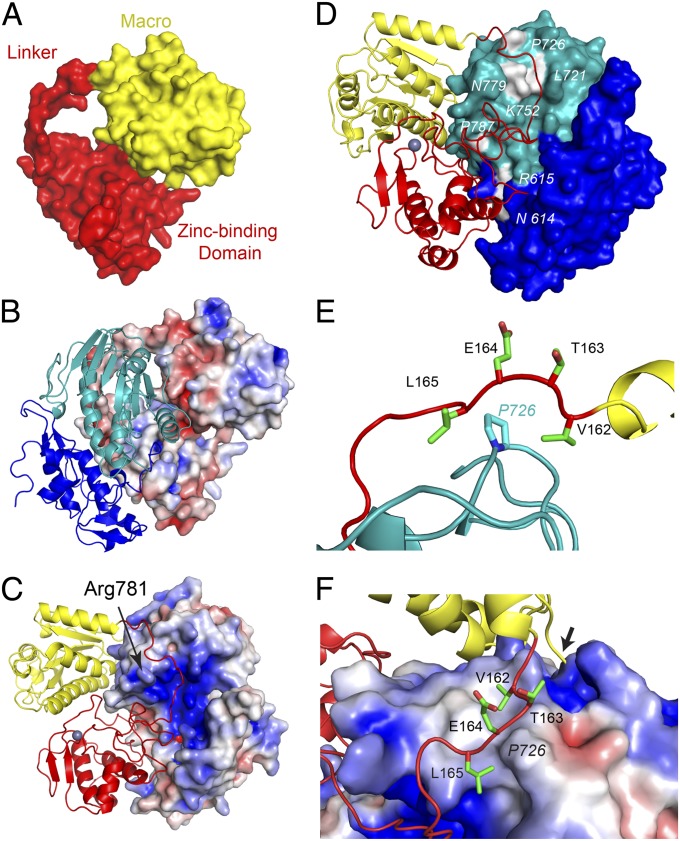
The interface between the macro and ZBD of nsP3 has a buried surface area of 570 Å2, whereas the linker connecting the two domains is extended, making nsP3 into a ring-like structure with an inner diameter of 15–18 Å (Fig. 2A). Together, nsP2 and nsP3 share an extensive interface with 3,000 Å2 of buried surface area. nsP3 encircles the MT-like domain of nsP2, with residue R781 of nsP2 protruding through the opening (Fig. 2C). The interface between nsP2 and nsP3 is charged, with the nsP2 surface being mostly basic, whereas nsP3 is generally acidic (Fig. 2 B and C).
Fig. 2.
nsP2 and nsP3 interface and the location of nsP2 noncytopathic mutants. (A) Solvent-accessible surface of nsP3. (B and C) Surface of nsP3 (B) or nsP2 (C) colored for electrostatic potential at ±5 kT/e; blue (basic), red (acidic), and white (neutral) with ribbon diagram of nsP2 (B) or nsP3 (C). (D) Molecular surface of nsP2 highlighting (white) the location of nsP2 noncytopathic mutations. The numbering corresponds to SINV nsP2 sequence. (E) Location of P726 in nsP2, highlighting interactions with the nsP3 linker. A portion of the last helix in the nsP3 macro domain is shown in yellow. (F) Surface of nsP2 P726 and surrounding area colored for electrostatic potential at ±4 kT/e with nsP3 linker region in stick format. Residues labeled in italics are located in nsP2. Arrow indicates P2/3 cleavage site. Domain coloring in each panel is identical to Fig. 1.
Several groups have identified mutations in the carboxyl-terminal portion of nsP2 that result in noncytopathic virus and persistent infection (16–20). These mutations reduce viral RNA replication levels with minimal effects on host gene expression and viral yields. All of the noncytopathic mutations map to the surface of nsP2 at the nsP3 interface (Fig. 2D). The mutations follow the encircling path made by the macro to ZBD linker. Mutagenesis of P726 in SINV to amino acids with large side chains (Phe, Tyr, Leu, Arg, and Gln) caused a dramatic decrease in RNA replication and were noncytopathic, whereas substitutions with small side chains (Ala or Val) behaved like wild-type virus, with Gly, Ser, and Thr substitutions giving an intermediate effect (16). P726 is located in the loop connecting α9 with β11 and makes direct contact with nsP3 linker just after the macro domain (Fig. 2E). There is a striking, direct correlation between cytopathogenicity and the level of RNA replication with the size of the mutant side chain. Molecular modeling analyses demonstrated that mutating P726 to Phe, Tyr, Leu, Arg, or Gln resulted in numerous steric clashes with nsP3, whereas Ser, Thr, Ala, and Val had fewer conflicts. Moreover, the P726G mutation could destabilize the α9-β11 loop, causing it to interfere with P23 interaction. Attempts to purify P23pro-zbd with a P726S or P726L mutation resulted in poorly soluble and aggregated protein, supporting the importance of this position.
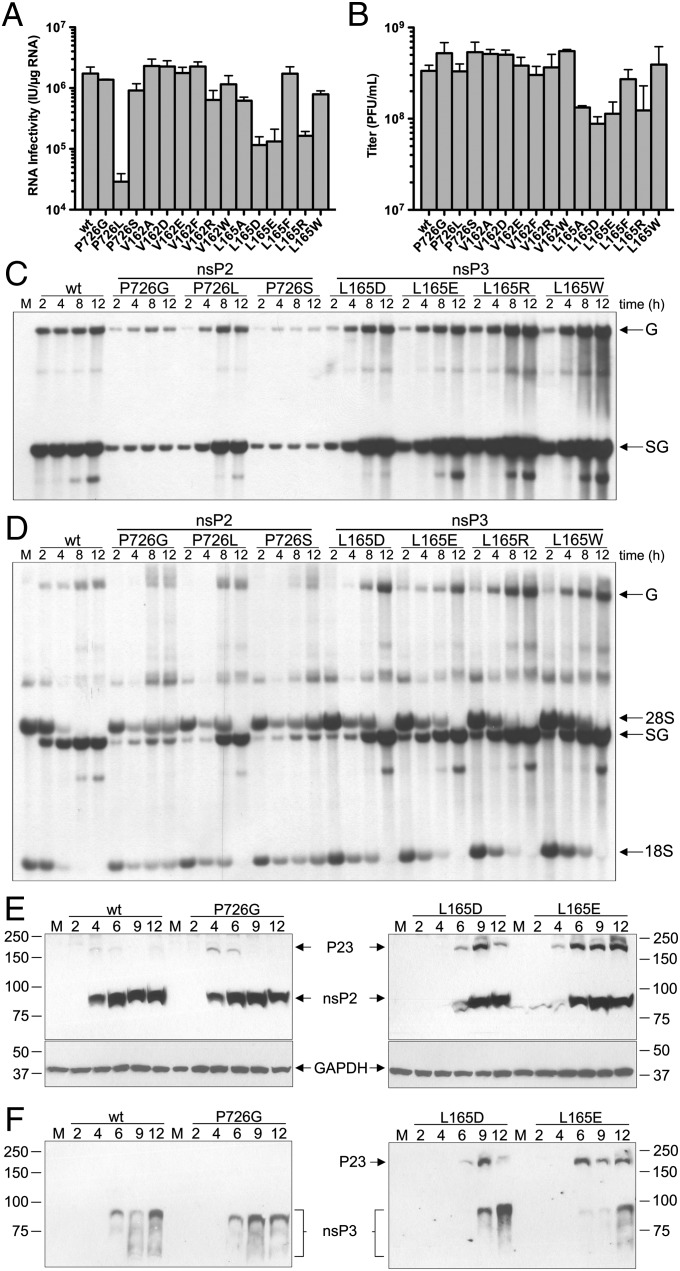
To investigate how destabilizing P23 interface may contribute to changes in viral pathogenesis and replication, site-directed mutagenesis to the corresponding surface of SINV nsP3 in close proximity to nsP2 P726 were created (Fig. 2E). The side chains of V162 and L165 of nsP3 are ∼4.6 and 3.3 Å away from nsP2 P726 side chain, respectively (Fig. 2F). V162 and L165 of nsP3 were mutated to one of the following: Ala, Asp, Glu, Phe, Arg, or Trp. nsP2 P726 to Ser, Gly, or Leu mutants were used for comparison. RNA infectivity measured by infectious center assay on BHK cells were reduced in P726L, L165D, L165E, and L165R mutants compared with the wild-type SINV (Fig. 3A). P726L mutation in nsp2 reduced RNA infectivity to only 2% of wild-type levels, whereas the three nsP3 L165 mutants averaged an RNA infectivity at 8% of wild type. These four mutants also displayed a small size plaque phenotype in infectious center assay (Fig. S5 A–D). In addition, L165A, L165D, L165E, and L165R mutants produced lower viral titers at 24 h after electroporation, only reaching an average of 34% of wild-type levels (Fig. 3B). Of the nsP3 mutants generated, L165D had the lowest titers, only producing 26% compared with wild-type titers. All others, including all three P726 mutants, reached wild-type levels of virus titers. All V162 mutants behaved similarly to wild type in both RNA infectivity and viral titer and were not pursued further. None of the nsP3 mutants displayed a noncytopathic phenotype in the TSG/puromycin N-acetyl-tranferase (PAC) replicon background (16), although onset of CPE in L165 to D, E, and R mutants was delayed.
Fig. 3.
In vivo effects of nsP3 linker region mutations. (A and B) Viral RNA infectivity [infectious units (IU)/μg RNA] as measured by infectious center assay (A) and viral titers [plaque-forming units (pfu)/mL] (B) 24 h after electroporation of BHK cells, as determined by plaque assay over two or more independent experiments. Error bars represent the SEM. (C and D) Cells were infected with wild-type or mutant SINV at a multiplicity of infection (MOI) of 10. At indicated time points postinfection, RNAs were labeled for 3 h in the presence (C) and absence (D) of actinomycin D. Total RNAs were harvested and analyzed by denaturing agarose gel electrophoresis. The position of genomic (G), subgenomic (SG), 28S, and 18S RNAs are labeled. M indicates mock-infected cells labeled for 3 h. (E and F) Analysis of P23 cleavage of nsP2 P726 and nsP3 L165 mutants in vivo. Total protein was harvested from wild-type and indicated mutant SINV-infected BHK cells at indicated hours postinfection and analyzed by Western blot using anti-nsP2 (E) and anti-nsP3 (F) antibodies.
Radiolabeling experiments were performed to evaluate both viral and host protein and RNA production over the course of infection and to analyze efficiency of host-cell transcriptional and translational shutoff caused by SINV nsP3 mutations. Wild-type SINV is able to halt host translation completely within 6 h postinfection, whereas P726S and P726G mutants were unable to do so even up to 12 h postinfection, as indicated by persistent actin production (Fig. S6). nsP2 P726L and the four selected nsP3 L165 mutants (Asp, Glu, Arg, Trp) were able to shut off host translation, although at a slightly delayed time postinfection compared with wild type (Fig. S6).
Tritium-labeled uridine was used to track viral and host RNA production over the course of infection. In the presence of actinomycin D, both genomic and subgenomic viral RNA production can be clearly observed in wild-type SINV starting as early as 2 h postinfection and remaining stable until 12 h (Fig. 3C). Levels of subgenomic and genomic RNA in P726G and P726S mutants are reduced compared with wild type. nsP2 P726L and the four nsP3 L165 mutants exhibit a slow initial rate of transcription, especially of genomic RNA; however, these mutants reach wild-type levels by 4 h postinfection. Thereafter, the levels of RNA replication in the L165 mutants continue to increase, and even surpass, wild-type RNA replication, especially subgenomic RNA (Fig. 3C). RNA labeling in the absence of actinomycin D showed complete inhibition of host cellular transcription by 8 h postinfection as demonstrated by decreasing 28S and 18S cellular rRNA (Fig. 3D). P726G and P726S mutants exhibited a lack of host-cell transcriptional shutoff with rRNA persisting up to 12 h postinfection. The nsP2 P726L mutant and the four nsP3 L165 mutants showed a delay in host transcription inhibition but eventually achieved complete shutoff 12 h postinfection (Fig. 3D).
Given that residues R159 and E163 of SFV nsP3 have been shown to affect P2/3 cleavage (27), we assayed the effect of the SINV nsP3 mutations on cleavage activity. nsP3 linker region mutants L165D and L165E revealed drastic inefficiency in P23 cleavage that was not seen in L165W and L165R mutants (Fig. 3 E and F and Fig. S7). The presence of uncleaved P23 in the nsP3 L165D and L165E mutants started 6 h postinfection and persisted up to 12 h, whereas P726 mutants, as well as nsP3 L165R and nsP3 L165W, showed substantially lower amounts of uncleaved P23. Western blots probed with anti-nsP3 antibodies were used to confirm the presence of uncleaved P23 (Fig. 3F). nsP3 appears as a smear, indicative of alternate phosphorylation states and possibly other posttranslational modifications (40). The nsP2 P726 mutants may also accumulate precleavage forms but at very low concentrations that do not build up over time, as in the case of the nsP3 linker mutants.
Discussion
The structure of alphavirus precursor polyprotein P23pro-zbd presented here reveals the arrangement of four domains (protease, MT-like, macro, and ZBD) in a precleavage form. Based on the structure of P23pro-zbd, several inferences can be made about the mechanism of alphavirus polyprotein cleavage. First, the carboxyl terminus of the P23pro-zbd structure is pointing toward the nsP2 protease active site about 40–45 Å away. It is possible that the large, flexible carboxyl region of nsP3 could span the distance, placing the P3/4 junction in the protease active site for cis cleavage (34). Second, the 40 Å distance between the P2/3 cleavage site and the nsP2 protease active site supports the current trans model of cleavage (5, 34, 35). Third, the inaccessibility of the P23 cleavage site itself indicates that access is tightly regulated. It is possible the activator sequence present in the extreme amino terminus of nsP2, which becomes exposed after cleavage at the P1/2 site, may be responsible for this regulation (5).
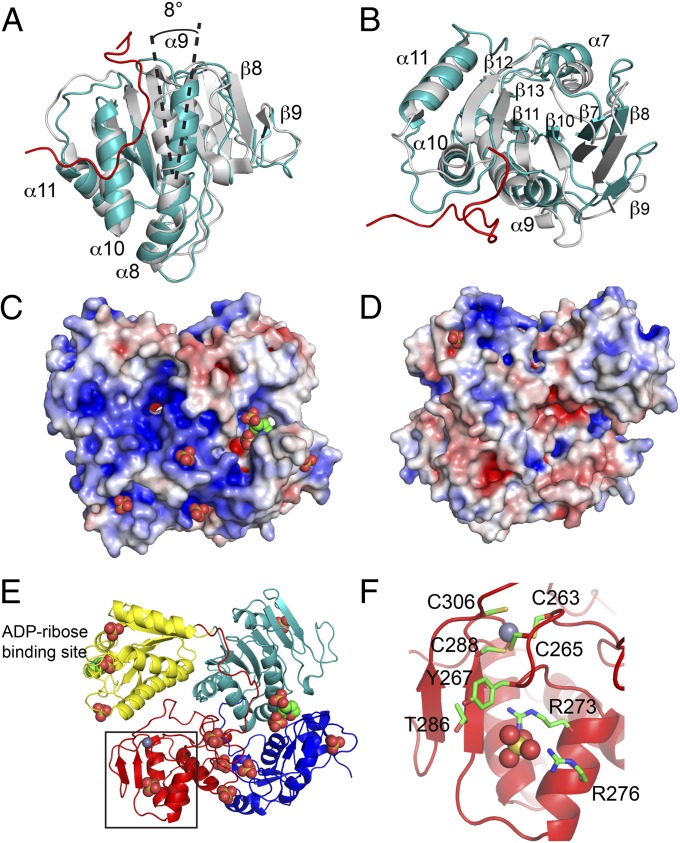
During the course of alphavirus infection, a portion of nsP2 is localized to the nucleus, whereas nsP3 remains cytoplasmic (13, 14, 41). At the moment, it is unclear how nsP2 would dissociate postcleavage from the extensive, encircling grasp of nsP3. Comparison of the domains in P23pro-zbd with the isolated structures of nsP2pro (12) and nsP3 macro domain (32) demonstrated that the protease and macro domains were highly similar with a root mean square deviation (rmsd) of <0.6 Å, whereas the MT-like domains have an rmsd of 1.3 Å for similar α carbon. A superposition of the nsP2 structures shows the discrepancy between nsP2pro and P23pro-zbd is limited to specific regions within the MT-like domain (Fig. 4 A and B). α-Helices α8, α10, and α11, as well as β-strands β11, β12, and β13, between the two structures superimpose well with an rmsd of <1 Å, although the β13-α10 loop moves 5 Å because of interactions with the nsP3 linker. In addition, the nsP3 linker interacts with α9, causing the helix to rotate 8° with the carboxyl-terminal end of the helix moving 3.8 Å to form a single continuous helix with α8. This movement of the α9 helix causes the β-sheet to become more concave. The distortion of the MT-like domain in the P23pro-zbd becomes more pronounced the further the secondary structures are from α9, with α7 and β9 moving about 4 Å from their corresponding positions in nsP2pro. Perhaps movement of the MT-like domain facilitates separation of nsP2 and nsP3 postcleavage, which is triggered by binding viral proteins or RNA to the concave side of the nsP2 MT-like β-sheet.
Fig. 4.
(A and B) Superposition of the MT-like domains of P23pro-zbd (teal) and nsP2pro (gray) (12). The nsP3 linker (for simplicity only residues V162-W178 are shown) connecting the macro domain and ZBD is shown in red. The view in B is rotated 90° about a horizontal axis from A. (C and D) Potential RNA-binding surface of P23pro-zbd. The location of sulfate and MES ligands are represented by spheres. Surface of P23pro-zbd is colored for electrostatic potential at ±5 kT/e. D is a 180° rotation about the vertical axis from the view in panel C. (E) Ribbon diagram of P23pro-zbd in the identical orientation as in C. (F) ZBD of nsP3 showing the four zinc-coordinating cysteines, T286, a sulfate ion represented by spheres bound to Y267, R273, and R276, and surrounding residues. The close-up view corresponds to the box in E.
Previously described noncytopathic mutations at the surface of nsP2 contacting the nsP3 linker region appear by molecular modeling to destabilize P23 interaction, decreasing efficiency of the RNA replication complex during virus infection. Altering the corresponding nsP3 residue (L165), which contacts nsP2 P726, did not produce virus with the same noncytopathic phenotype. In contrast to the nsP2 P726G and P726S mutants, nsP3 L165 mutants had enhanced RNA replication levels, indicating the apparent stabilization of the replication complex. Despite the large, bulky nature of a tryptophan substitution at L165, the virus remained similar in phenotype to wild type. The L165D and L165E mutants, however, exhibited the most extreme phenotype in terms of reduced RNA infectivity and inefficient P23 cleavage. The surface of nsP2 surrounding P726 is both basic and hydrophobic (Fig. 2F), suggesting that the P23 interface may be stabilized by the introduced acidic amino acid at L165 through interaction with the surface of nsP2. This stabilization could cause the interface to become “locked in,” preventing the flexibility needed for conformational change and, thus, disallowing nsP2 access to the P2/3 cleavage site.
The buildup of uncleaved P23 is evidence that the nsP3 linker region, which encircles nsP2, may function in positioning and recognition of the P2/3 cleavage site. Given the nsP3 linker residues and nsP2 P726 are within reasonable distance to the P2/3 cleavage site (21 and 17 Å from Cα of L165 or P726 to the cleavage site, respectively), it is plausible that mutation of the nsP3 linker region shifts local structural features, altering recognition of the cleavage site (Fig. 2F). A previously described E163R mutation in SFV nsP3 that altered P2/3 cleavage is completely solvent-exposed (Fig. 2E) (27). These results may indicate that the nsP3 linker residues surrounding nsP2 P726 are recognized by the nsP2 protease or another factor necessary for P2/3 cleavage.
Cleavage between P2/3 dictates template use by the alphavirus replication machinery. During purification, P23pro-zbd bound extensive amounts of bacterial RNA, which could only be removed by high concentrations of salt (Fig. S1 A and C). The electrostatic potential of the P23pro-zbd structure identified a large basic surface that centers on the zinc-binding site of nsP3 and extends to nsP2 (Fig. 4C). Many zinc metalloproteins are involved in nucleic acid–binding and gene regulation (42); therefore, the location of a basic patch in close proximity to the zinc-binding site suggests a potential role in RNA binding. Several molecules of sulfate and 2-(N-morpholino)ethane sulfonic acid (MES), used in crystallization, were bound to P23pro-zbd along the basic surface, which may indicate potential binding sites for the phosphate groups of nucleic acids (Fig. 4 C–E). In fact, a molecule of MES occupies the ADP ribose–binding site in each of the three macro domains in P23pro-zbd in the asymmetric unit, as well as in the macroH2A1.1 structure (Fig. 4E) (43). The alphavirus genome contains conserved sequence elements (CSEs) that serve as promoters for production of positive- and negative-sense genomic RNA, as well as subgenomic RNA. Mutations in a 51-nt-long CSE in the nsP1-coding region of SINV disrupted the RNA structure without affecting the coding sequence and demonstrated reduced replication in mosquito cells but not in mammalian cells (44). Adaptive mutations (E118K of nsP2 and T286I of nsP3) were shown to restore replication of the mutated CSE. T286 is 2 aa before one of the zinc-coordinating cysteine residues (C288) (Fig. 4F) and is located on the basic surface of the ZBD. T286 is 6.1 Å away from a sulfate ion that is coordinated by three highly conserved amino acids in nsP3: Y267, R273, and R276 (Fig. 4F). Taken together, these data strongly implicate this surface as a potential RNA-binding site.
Many important human viral pathogens encode a polyprotein that is cleaved by viral or cellular proteases. Given the wide prevalence of regulated polyprotein processing seen in a number of viral families, there is lack of structural information regarding the precursor forms. Currently, alphavirus P23pro-zbd and poliovirus 3CD (45) are the only structures of polyprotein precursor from a viral replication machinery. Both alphavirus P23pro-zbd and poliovirus 3CD structures contain the protease domain responsible for cleaving the precursor, which is thought to proceed by a trans mechanism. However, 3CD has no intramolecular contacts and a solvent accessible cleavage site that is regulated by intermolecular contacts, whereas P23pro-zbd has an extensive interface and an inaccessible cleavage site. The results presented here further expand our current understanding of viral polyprotein processing, replication, and cytopathogenesis, which may be applicable to other positive-sense RNA viruses.
Materials and Methods
Complete materials and methods are provided in SI Materials and Methods.
Structure Determination.
P23pro-zbd of the SINV strain Toto1101 was expressed in Escherichia coli as a fusion with GST. The protein was purified by sequential chromatography over GST affinity, hydroxyapatite, and heparin columns. Crystals of P23pro-zbd were grown by vapor diffusion against 2 M ammonium sulfate and 0.1 M MES (pH 6.5). Crystals were transferred to a cryoprotectant solution containing 2 M ammonium sulfate, 0.1 M MES (pH 6.5), 35% (vol/vol) xylitol, and 5% (vol/vol) trimethylamine N-oxide and then flash-cooled in liquid nitrogen. Diffraction data were collected at the zinc, K absorption edge (1.2824 Å) to 2.85 Å resolution at beam line X25 at Brookhaven National Synchrotron Light Source (Table S1). Phases were determined by molecular replacement using the nsP2pro (12) and nsP3 macro structures (32). The final model contains 3 molecules of P23pro-zbd, 3 zinc ions, 31 sulfate ions, and 5 molecules of MES. Atomic coordinates have been deposited in the Protein Data Bank under PDB ID code 4GUA.
Virological Studies.
Infectious center and plaque assays were performed similarly to those described in ref. 46. Protein and RNA radiolabeling were performed as described in ref. 9. Harvested protein from BHK-J cells at time points postinfection were used in Western blotting experiments, and membranes were probed with anti-nsP2, anti-nsP3, and anti-GAPDH antibodies.
Supplementary Material
Acknowledgments
We thank J. Bonanno for assistance with the data processing; C. Rice and M. MacDonald for providing BHK-J cells, Sindbis virus DNA, and nsP3 antibody; J. Millonig and P. Matteson for the GAPDH antibody; and A. Basant, J. Chiu, F. Jiang, A. Khan, C. Rice, A. Stock, V. Stollar, and J. Whidby for insightful discussions. S.A.Y. was supported by the NIH-NIAID training Grant 2T32AI007403-18. This study was supported by National Institutes of Health Grants AI080659 (to J.M.) and DK083356 and AI070101 (to A.G.), New Jersey Commission on Cancer Research Grant 10-1962-CCR-EO (to J.M.), and Yerkes Research Center Base Grant RR-00165 (to A.G). We wish to express our profound gratitude to the late Dr. Aaron Shatkin for his unwavering support, sage advice, and insightful discussions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4GUA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210418109/-/DCSupplemental.
References
- 1.Griffin DE. Alphaviruses. In: Knipe DM, editor. Fields Virology. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1023–1067. [Google Scholar]
- 2.Thiboutot MM, et al. Chikungunya: A potentially emerging epidemic? PLoS Negl Trop Dis. 2010;4:e623. doi: 10.1371/journal.pntd.0000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nkoghe D, et al. Clinical forms of chikungunya in Gabon, 2010. PLoS Negl Trop Dis. 2012;6:e1517. doi: 10.1371/journal.pntd.0001517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot RJ, Hardy WR, Shirako Y, Strauss JH. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasiljeva L, et al. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem. 2003;278:41636–41645. doi: 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 6.Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: Uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemm JA, Rümenapf T, Strauss EG, Strauss JH, Rice CM. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss JH, Strauss EG. The alphaviruses: Gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79:9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garmashova N, Gorchakov R, Frolova E, Frolov I. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J Virol. 2006;80:5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolova EI, et al. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J Virol. 2002;76:11254–11264. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo AT, White MA, Watowich SJ. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure. 2006;14:1449–1458. doi: 10.1016/j.str.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Rikkonen M, Peränen J, Kääriäinen L. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology. 1992;189:462–473. doi: 10.1016/0042-6822(92)90570-f. [DOI] [PubMed] [Google Scholar]
- 14.Rikkonen M, Peränen J, Kääriäinen L. Nuclear targeting of Semliki Forest virus nsP2. Arch Virol Suppl. 1994;9:369–377. doi: 10.1007/978-3-7091-9326-6_37. [DOI] [PubMed] [Google Scholar]
- 15.Garmashova N, et al. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol. 2007;81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frolov I, et al. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73:3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dryga SA, Dryga OA, Schlesinger S. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: The importance of a mutation in the nsP2 gene. Virology. 1997;228:74–83. doi: 10.1006/viro.1996.8364. [DOI] [PubMed] [Google Scholar]
- 18.Mayuri, Geders TW, Smith JL, Kuhn RJ. Role for conserved residues of sindbis virus nonstructural protein 2 methyltransferase-like domain in regulation of minus-strand synthesis and development of cytopathic infection. J Virol. 2008;82:7284–7297. doi: 10.1128/JVI.00224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perri S, et al. Replicon vectors derived from Sindbis virus and Semliki forest virus that establish persistent replication in host cells. J Virol. 2000;74:9802–9807. doi: 10.1128/jvi.74.20.9802-9807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrakova O, et al. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss B, Rosenthal R, Schlesinger S. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J Virol. 1980;33:463–474. doi: 10.1128/jvi.33.1.463-474.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frolov I, Schlesinger S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin EV, et al. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karras GI, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saikatendu KS, et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuvonen M, Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lulla A, Lulla V, Merits A. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J Virol. 2012;86:553–565. doi: 10.1128/JVI.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vihinen H, Ahola T, Tuittila M, Merits A, Kääriäinen L. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J Biol Chem. 2001;276:5745–5752. doi: 10.1074/jbc.M006077200. [DOI] [PubMed] [Google Scholar]
- 29.Vihinen H, Saarinen J. Phosphorylation site analysis of Semliki forest virus nonstructural protein 3. J Biol Chem. 2000;275:27775–27783. doi: 10.1074/jbc.M002195200. [DOI] [PubMed] [Google Scholar]
- 30.Chance MR, et al. High-throughput computational and experimental techniques in structural genomics. Genome Res. 2004;14(10B):2145–2154. doi: 10.1101/gr.2537904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W, et al. Metalloproteomics: High-throughput structural and functional annotation of proteins in structural genomics. Structure. 2005;13:1473–1486. doi: 10.1016/j.str.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Malet H, et al. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hahn YS, Strauss EG, Strauss JH. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: Assignment of complementation groups A, B, and G to nonstructural proteins. J Virol. 1989;63:3142–3150. doi: 10.1128/jvi.63.7.3142-3150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirako Y, Strauss JH. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology. 1990;177:54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- 35.Hardy WR, Strauss JH. Processing the nonstructural polyproteins of sindbis virus: Nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo AT, Malmstrom RD, White MA, Watowich SJ. Structural basis for substrate specificity of alphavirus nsP2 proteases. J Mol Graph Model. 2010;29:46–53. doi: 10.1016/j.jmgm.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 38.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 39.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lastarza MW, Grakoui A, Rice CM. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: Effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology. 1994;202:224–232. doi: 10.1006/viro.1994.1338. [DOI] [PubMed] [Google Scholar]
- 41.Peränen J, Rikkonen M, Liljeström P, Kääriäinen L. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J Virol. 1990;64:1888–1896. doi: 10.1128/jvi.64.5.1888-1896.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg JM. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990;265:6513–6516. [PubMed] [Google Scholar]
- 43.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 44.Fayzulin R, Frolov I. Changes of the secondary structure of the 5′ end of the Sindbis virus genome inhibit virus growth in mosquito cells and lead to accumulation of adaptive mutations. J Virol. 2004;78:4953–4964. doi: 10.1128/JVI.78.10.4953-4964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcotte LL, et al. Crystal structure of poliovirus 3CD protein: Virally encoded protease and precursor to the RNA-dependent RNA polymerase. J Virol. 2007;81:3583–3596. doi: 10.1128/JVI.02306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemm JA, Durbin RK, Stollar V, Rice CM. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol. 1990;64:3001–3011. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.