Abstract
Despite significant investments in basic science by the US National Institutes of Health, there is a concern that the return on this investment has been limited in terms of clinical utility. In the field of biomarkers, translational research is used to bridge the gap between the results of basic research that identify biomolecules involved in or the consequence of carcinogenesis and their incorporation into medical application. The cultural separation between different scientific disciplines often makes it difficult to establish the multidisciplinary and multi-skilled teams that are necessary for successful translational research. The field of biomarker research requires extensive interactions between academic researchers and industrial developers, and clinicians are needed to help shape the research direction that can only be addressed by multi-disciplinary, multi-institutional approach. In this article, we provide our perspective on the relatively slow pace of cancer biomarker translation, especially those for early detection and screening.
Keywords: Biomarkers, cancer, multidisciplinary, translational research, academic researchers, industrial developers, clinicians
Introduction
Despite significant investments in basic science by the US National Institutes of Health, there is a concern that the return on this investment has been limited in terms of clinical utility. It is frequently stated that translational research is a missing component between basic science and clinical application. Numerous definitions exist for translational research and to make it relevant to this commentary, we define translational research as a discipline that makes the results of basic research applicable to clinical use. The term translational medicine has been applied to a research approach that seeks to move “from bench to bedside” or from laboratory experiments into clinical trials to actual point-of-care patient applications. Outside of the medical domain, the term translational research has been applied more generally where researchers seek to shorten the time-frame of the basic to applied continuum to ‘translate’ fundamental research results into practical applications. In the field of biomarkers, translational research is used to bridge the gap between the results of basic research that identify biomolecules involved in or the consequence of carcinogenesis and their incorporation into medical application. The cultural separation between different scientific disciplines often makes it difficult to establish the multidisciplinary and multi-skilled teams that are necessary for successful translational research. The field of biomarker research requires extensive interactions between academic researchers and industrial developers, and clinicians are needed to help shape the research direction that can only be addressed by a multidisciplinary, multi-institutional approach.
Translational Science and Biomarker Research
In this article, we provide our perspective on the relatively slow pace of cancer biomarker translation, especially those biomarkers for use in early detection and screening. We are members of the US National Cancer Institute’s (NCI) Cancer Biomarkers Research Group and administer NCI’s Early Detection Research Network (EDRN). The mission of this network is to discover and validate biomarkers for the detection of early stage cancers and for risk assessment. We will also discuss how the EDRN is working to accelerate the pace translation of biomarkers from discovery into clinical application.
A cancer biomarker refers to a substance or process that is indicative of the presence of cancer in the body. It might be a molecule secreted by a malignancy itself, or it can be a specific response of the body to the presence of cancer. The National Institutes of Health Biomarkers Definition Working Group1 provided a formalized definition of biomarker as cellular, biochemical, and molecular alterations by which a normal, abnormal, or simply biologic process can be recognized or monitored and are used to objectively measure and evaluate normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention. The majority of cancer biomarkers are measured either in the tumor or in blood. To maximize usefulness and minimize cost for screening or early detection, it is advantageous that a biomarker be measurable in a body fluid that can be obtained using minimally invasive methods, such as blood, urine, sputum or stool. PSA (prostate-specific antigen) is an example of a blood-based protein marker for prostate cancer, methylated vimentin is a stool-based DNA biomarker for colorectal cancer, and DNA FISH assays are urine-based biomarkers for bladder cancer. PAP test is a cell-based biomarker for cervical cancer that has contributed significantly to the 99% reduction in deaths due to cervical cancer in the US. Annual screening using the fecal occult blood test (FOBT) reduces colon cancer mortality by 15 to 33 percent.
Discovering cancer biomarkers is a relatively easy process as judged by the number of papers published every year on this subject. A PubMed search for cancer and biomarkers gives more than 15,000 publications in 2010, and 1100 when limited to cancer biomarkers and early detection. However, translating these discoveries into useful clinical assay is very difficult. To date, less than 30 cancer biomarkers have been approved by the FDA, and most of these are for monitoring response to therapy. A number of explanations have been given for the relatively few cancer biomarkers being moved into clinical use. These include the high performance characteristics needed to a make a biomarker clinically useful, the biology of tumors, a flawed discovery process, a validation process that is cumbersome and expensive, regulatory requirements, and an academic system that does not reward translational research. Issues of credit, publication priority, and patent credit have also slowed progress into validation. Indeed, much of the biomarker research remains “stuck” at the discovery phase, and unfortunately, some investigators appear content to reiterate the discovery process, never proceeding beyond that point.
Successful biomarker research requires a knowledge-driven environment, in which investigators generate, contribute, manage and analyze data from a variety of sources and technology platforms. It is only by integrating these diverse data types that the complex and underlying causes of illness can be revealed, and effective strategies for prevention, early detection and personalized treatments be realized. The translation of biomarker discovery into clinical application is an iterative process with multiple feedback loops, i.e. results from the verification of a biomarkers performance can be used to inform additional discovery efforts. Collaboration, data sharing, data integration and standards are integral. Successful translational research also requires that information and data also flow from hospitals, clinics and participants of studies in an organized and structured format to repositories and research-based facilities and laboratories.
The scale, scope and multi-disciplinary approach of translational research requires a new level of operation’s management capabilities within and across studies, repositories and laboratories. The increased operational requirements of these large studies, with large annotated specimen collections, large complex data sets, and government regulations, necessitate an informatics approach that enables the integration of both operational capabilities and clinical and basic data. Most informatics systems in use today are inadequate to handle the tasks of complicated operations and contextually in data management and analysis.
Business Model for Translational Research in Biomarkers
Why do “big science” projects that have such great promise often take decades from concept to fruition (if they make it to practice at all)? A likely contributor is that the health care industry, while accounting for more than 13% of the U.S. gross domestic product and growing at triple the rate of inflation, remains a fragmented industry. Perhaps health care can learn from computer electronics, where “big science” is always part of the equation, but so is a business model that drives the translation from “art to part.” Part of the problem is the difference in the business models that drive the respective systems.
Health care organizations have generally grown organically, which typically results in structures that are organized along functional “silos,” areas of expertise where depth of knowledge in one particular area is critical. Although such a horizontal structure fosters excellent solutions for primary scientific problems, it often generates barriers if knowledge must be shared between silos. In contrast, computer chip manufacturers are organized in a more vertical structure. In this structure, formal hand-off procedures have been designed to ensure that discoveries in one aspect of chip design and construction are rapidly and efficiently conveyed to others who require the information. This allows for rapid vetting of ideas, quickly culling out poor concepts, and fostering the rapid acceptance of good concepts.
In a vertical design, there are a number of focused experts within in a single organization, generating economies of scale for the rapid discovery and development, and there is a focus on coordination of multiple entities, using shared resources and emphasizing hand-offs between entities. This is in contrast to a horizontal approach, which may result in rapid discoveries but limited advancement towards a useful deployed product. A horizontal design may also increase duplication and reduce potential synergies across disciplines.
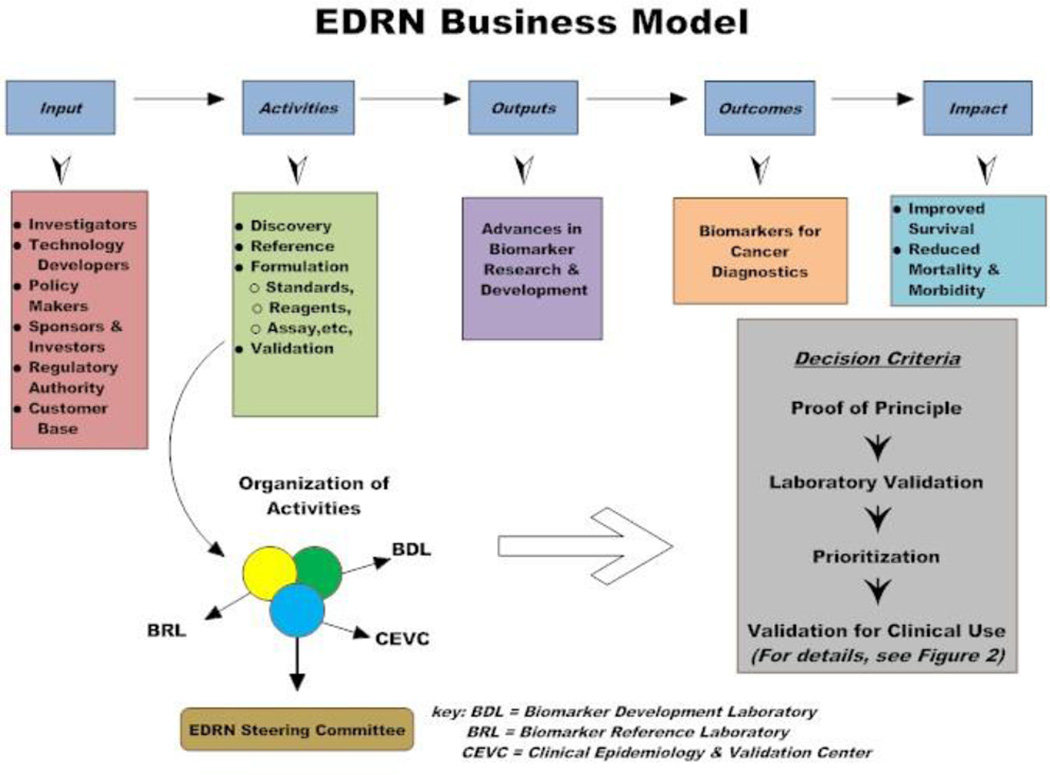
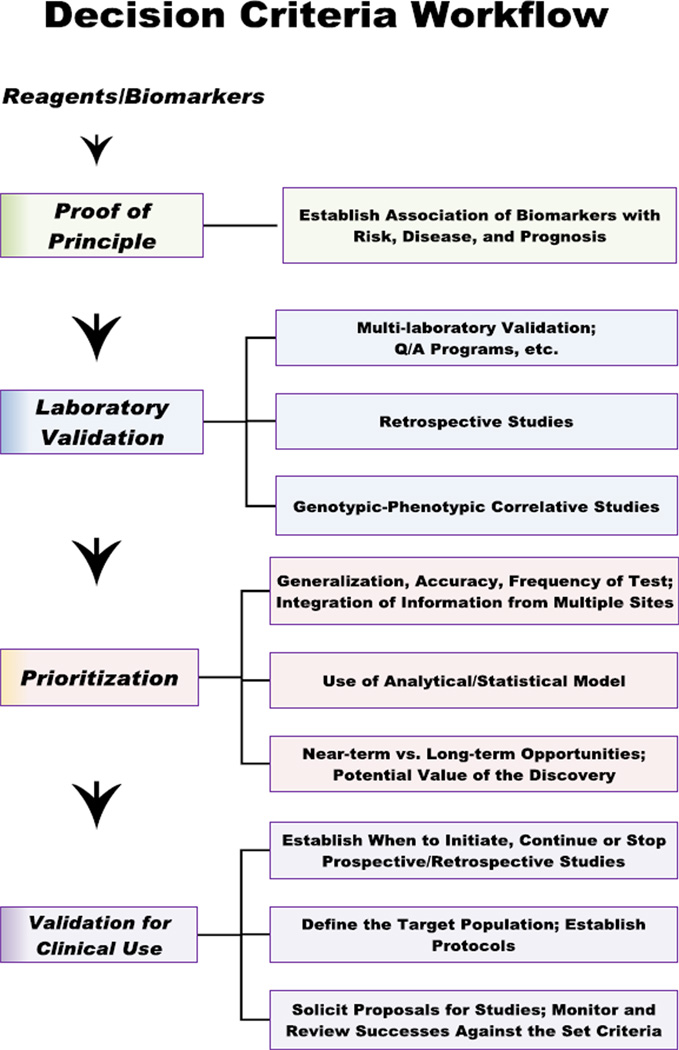
When deciding whether to adopt a horizontal or vertical model, one must consider the influence and interests of constituents that can either drive or hinder the process. NCI’s Early Detection Research Network (EDRN) has developed methods, policies, and procedures for relating with each of the major constituent groups. EDRN promotes a vertical approach for conducting biomarker research, whereby biomarkers are developed, refined and analytically and clinically and validated all within one organization (Figure 1). A critical aspect of the EDRN is its focus on coordinating multiple resources with a goal of minimizing barriers to the rapid and efficient “hand-off” between entities. One method used for achieving this is a structured set of criteria for assessing the roles and clinical significance of each newly discovered biomarker, along with criteria and strategies for judging the use of biomarkers in relationship to one another (Figure 2).
Figure 1.
Vertical approach business model followed by the EDRN (adopted from the Harvard Business Review)
Figure 2.
EDRN decision criteria for judging relationships and strategies in biomarker development and fruition
The Early Detection Research Network (EDRN) as a Model for Biomarker Research
NCI’s EDRN was formed to facilitate the discovery, development, and validation of biomarkers for early cancer detection and risk assessment (http://edrn.nci.nih.gov/). This network includes experts from academia, government, and industry and has the goal of translating research discoveries into substantial benefits that can be implemented in the clinic or the population setting. The EDRN is a vertically integrated network composed of four main components.
Biomarker Developmental Laboratories (BDLs) discover, develop and characterize new biomarkers or refine existing biomarkers.
Biomarkers Reference Laboratories (BRLs) serve as Network resources for analytical and clinical validation of biomarkers, including technological development and refinement.
Clinical Validation Centers (CVCs) conduct and support biomarker validation trials, provide high quality biological specimens to other EDRN investigators for use in biomarker discovery, and contribute biospecimens for the formation of EDRN reference sets (common sets of specimens from well characterized and matched cases and controls from specific disease spectra).
Data Management and Coordinating Center (DMCC) provides network coordination, data management and protocol development in support of EDRN biomarker validation trials and conducts theoretical and applied statistical research related to the goals of the EDRN.
This collaborative arrangement allows for the rapid movement of newly discovered or refined biomarkers from the laboratory into clinical validation. When a Biomarker Developmental Laboratory (BDL) has obtained promising data on a biomarker or panel of biomarkers, they work with a Biomarkers Reference Laboratory (BRL) to refine the assay and independently reproduce the results using blinded specimens; these specimens may come from a Clinical Validation Center (CVC). If the assay is reproducible and has sufficient sensitivity and specificity, the BDL works with a CVC, BRL and the Data Management and Coordinating Center (DMCC) to conduct a multisite biomarker validation trial. Investigators outside the EDRN who have promising biomarkers can access the EDRN infrastructure to validate their biomarkers or participate in an ongoing validation trial.
Other goals of the EDRN are to establish scientific criteria to evaluate biomarkers as indicators of early cancer, prognostic factors of pre-invasive cancer, markers of risk, and surrogate endpoints. The EDRN has developed a quality assurance program for biomarker testing and evaluation.
The EDRN follows an adaptive model in which all of the players (stakeholders), investors, policy makers, and technology developers work coordinately towards the goal of developing biomarkers for cancer diagnostics that improve health and decrease mortality. The EDRN has the requisite expertise and resources to create an evidence-based biomarker pipeline. There is no similar collaborative or cooperative group that is capable of conducting research projects from basic discovery to clinical validation. Without the pressure of venture capital, “best science” rather than “best business” practice permits development of novel concepts that otherwise, due to commercial needs, might never reach fruition.
Why are models like the EDRN needed?
There is a perception that translational research receives less support from NIH than does basic research. NCI’s Translational Research Working Group (http://www.cancer.gov/researchandfunding/trwg) reported that in fiscal year 2004, $0.9–1.3 billion of NCI’s $4.4 billion research budget fit the broad inclusion criteria for translational research. Despite this substantial commitment, some investigators believe that translational research is of lower priority than basic or clinical research or that it is somehow less prestigious, i.e., results from translational research are published in lower impact journals. The very nature of translational research is to a certain extent at odds with the current system of academic advancement, which favors the independent investigator. Translational research requires interdisciplinary research teams and the involvement of multiple departments or multiple institutions, and the time required to set up and conduct the studies, especially if they involve a prospective trial, is long with few publication while the study is ongoing.
Role of industry
Industry plays a unique role in bringing the end-stage products of the Nation’s investment in research through product development and into clinical use. While academic scientists and clinicians are well suited for the discovery, development and initial validation of cancer biomarkers, the final steps of obtaining FDA clearance or CLIA approval requires an industry involvement. It will be the private company that performs the assays on patient specimens or sells the in vitro diagnostic. Thus, it is important for academic investigators to interact with their industrial counterparts, preferably at an early stage of development. The EDRN holds biannual industrial forums that bring together academic investigators and industrial representatives interested in commercializing cancer biomarkers.
Kalorama Information reported that cancer diagnostics sales totaled $4.1 billion in 2004 and projected that the worldwide market for in vitro diagnostic (IVD) tests for cancer by the end of 2012 will be $8 billion, an annual growth rate of nearly 11 percent (http://www.healthimaging.com/index.php?option=com_articles&view=article&id=10094). The worldwide market for cancer IVDs is comparable to that for cardiac and about one third that for infectious diseases.
Choosing the “winners” and eliminating the “losers” among biomarkers
Despite substantial advances in our understanding of the molecular basis of cancer, there continues to be paucity of biomarkers for clinical use. While biomarkers in biological fluids have great potential to help determine the risk of disease or to allow early detection, the rate of success in this area has been disappointing. To increase the likelihood of success, it is imperative for us to learn from those biomarkers that failed to progress or whose performance did not validate when analyzed in independent specimens. There has been some criticism of the experimental approaches with which biomarkers studies have been conducted2. These studies have resulted in the thousands of publications describing promising candidates, but few of these candidates have been pursued to support their clinical utility, and most of those that have been pursued have failed in subsequent validation studies. A case in point is a recent validation study which encompassed 28 promising candidate protein biomarkers for ovarian cancer. When assayed in pre-diagnostic serum specimens from ovarian cancer cases and controls from NCI’s Prostate, Lung, Colorectal, and Ovarian Cancer screening trial, none of the individual markers exhibited performance characteristics that equaled those of CA-125, the current best marker for ovarian cancer3. Neither did their combination into a panel outperform CA-125. This study has been cited as evidence that something is wrong in our approach to biomarkers. Rather than dismissing this as failed experiment, the community should examine why the performances of these biomarkers did not hold up and use this information to guide future discovery.
The lack of clinically useful biomarkers may be partly attributed to inadequate resources and incentives to develop and maintain collaborative teams. As a result, studies are mostly conducted by a single laboratory with limited means to accomplish objectives. Such studies generally consist of comparisons using a limited number of convenience specimens, leading to the discovery of candidate biomarkers, which are not generalizable and need to be further pursued to determine their relevance. In retrospect, most such studies have not been solidly designed to minimize the biological and molecular heterogeneity of cancer. Compounding this problem is a considerable shortage of quality specimens for discovery and validation studies that overcome the biases inherent in retrospective samples. Unfortunately, biomarker discovery is too often performed using poor quality or poorly matched specimens, resulting in the identification of biomarkers that result from differences in specimen handling or storage or differences in the subjects other than cancer.
The size of early stage tumors and preneoplastic lesions and the heterogeneity of cancers contribute to the difficulty of identify biomarkers for early cancer detection or screening. Effective screening and early detection biomarkers should be measurable in bodily fluids that can be obtained using minimally invasive techniques, e.g., blood, urine or stool. This requires that the biomarker be present at a sufficiently high concentration that they can be measured in these fluids. The inherent difficulty is that tumors, especially at early stages, are small and consequently the amount of protein or other type of biomarker shed into the blood is very low, making them difficult to detect. Indeed, the only FDA approved blood based biomarkers for screening are PSA and complex PSA for prostate cancer. However, the use of PSA for screening is a subject of debate, and the U.S. Preventive Services Task Force (USPSTF) recently issued a draft recommendation against using the PSA test to screen healthy men for prostate cancer; primarily because this test, while detecting more cases of prostate cancer, resulted in little or no reduction in prostate cancer-specific mortality and harms associated with subsequent evaluation and treatments. A number of investigators are using more proximal fluids as a source for biomarkers, e.g. urine for prostate cancer, stool for colorectal cancer, and sputum for lung cancer. The FDA approved protein marker NMP22 and FISH assays for chromosomes 3, 7, 9 and 17 are urine based biomarkers for screening for bladder cancer.
As most of the major epithelial cancers, including lung, colorectal, breast, prostate and ovarian, are heterogeneous, it is unlikely that a single biomarker will detect all tumors from a particular organ. Consequently, many investigators are attempting to develop biomarkers panels to increase sensitivity. However, developing biomarker panels requires balancing increased sensitivity with decreasing specificity. Simply adding one biomarker with another may increase the sensitivity of the assay, but it is likely to decrease the specificity (each marker contributes to the number of false positives), and thus one typically has to alter cut off values of the individual biomarkers to maximize performance or develop complex algorithms to interpret the results. The difficulty with simply lowering cut off values of the individual markers in the panel to increase specificity is that this also lowers sensitivity.
It takes a village…
As pointed out by Samir Hanash, an EDRN investigator4, “It will ‘take a village” to implement a paradigm shift in our approach to biomarkers, necessitating a partnership among the various stakeholders, from academia with multi-disciplinary contributions, to philanthropy, government and industry. ․… Initiatives aimed at contributing to a paradigm shift in our approach to the development of biomarkers are burgeoning. They are exemplified by the increased availability of biospecimens through cohort studies with reduced bias. They are also exemplified by consortia to assess and standardize technologies for discovery and assays of biomarker candidates.”
Guidelines for Developing and Validating Biomarkers
EDRN has developed a five-phase approach for biomarker development that provides a systematic approach to discovery, development and validation and can be used to help identify “winners” or “losers” among biomarkers5. This phased approach has been widely accepted by the biomarker research community.
Phase 1, Discovery - Exploratory studies to identify potentially useful biomarkers
The majority of biomarkers do not progress beyond this phase. Reasons for this include modest differences in the concentration of the biomarker in cases compared to controls and large variability in the levels the biomarker in healthy subjects.
Phase 2, Clinical Assay and Validation – Studies to determine the capacity of biomarkers to distinguish between people with cancer from those without. Specimens used in these studies are from patients with cancer (cases), from healthy subjects (controls), and ideally form patients with confounding conditions, e.g. men benign prostate hyperplasia when evaluating biomarkers for prostate cancer.
Most biomarkers do not progress beyond this phase primarily because the validation study shows that the biomarker does not have sufficient sensitivity and/or specificity to be clinically useful (see below).
Phase 3, Retrospective Longitudinal – Determine how well biomarkers detect preclinical disease by testing the markers against specimens collected longitudinally from research cohorts.
These studies are usually retrospective and use specimens collected from apparently healthy individuals who were monitored for the development of cancer. However, phase 3 studies do not establish the stage of the disease when the biomarker is first detected.
Phase 4, Prospective Screening – Identify the extent and characteristics of disease detected by the test and determine the false referral rate.
Asymptomatic people are screened, and those with a test positive are followed up to determine if they have cancer and if so its stage.
Phase 5, Cancer Control – Evaluate both the role of the biomarkers for detection of cancer and the overall impact of screening on the population through large-scale population studies.
These studies are designed to determine whether the screen results in a reduction in morbidity and mortality.
For a biomarker to have clinical utility (the likelihood that the test will lead to an improved health outcome), it must achieve a certain level of performance in its ability to distinguish patients with cancer from those without, to determine prognosis, or to predict response to treatment. The performance required for a biomarker depends on the cancer and intended use. For example, biomarkers used to detect cancers with high prevalence may be clinically useful with lower specificity than those used to detect cancers with low prevalence. The sensitivity of a biomarker is the proportion of individuals with cancer who test positive for the biomarker, and specificity is the proportion of individuals without cancer who test negative for the biomarker. The lower a biomarker’s sensitivity the more people with cancer will not be detected, and the lower the specificity the more people without cancer will be incorrectly identified as having cancer. Sensitivity is also referred to as the true positive rate (TPR). The false positive rate (FPR) is the probability that a subject without cancer has a positive test result. None of the currently used biomarkers are 100% sensitive and specific. For example, PSA at a cut point of 4 ng/ml has a sensitivity of 70–80% and a specificity of approximately 60%, i.e., 20–30 % of men with cancer will be missed and 40% men without cancer will undergo needless biopsies. Lowering the PSA cut point will increase the sensitivity, resulting in fewer missed cancers but will lower the specificity, resulting in more needless biopsies. Thus, when choosing where to set the cut point, it is necessary to balance the impact of the number of missed cancers with the impact of the number of false positives and the relative harms of each. Does a positive biomarker test result trigger an imaging procedure, a biopsy or surgery? Two other criteria by which to judge a biomarker’s performance are positive predictive value (PPV) and negative predictive value (NPV). PPV is the chance that a person with a positive test has cancer and is very dependent on the prevalence of the cancer. For a given sensitivity and specificity, the higher the prevalence, the higher the PPV. If a cancer’s prevalence is 1% and the biomarker has 100% sensitivity and 95% specificity, 1 out of 6 people with a positive test will actually have cancer (PPV = 0.17). However, even when a biomarker has very high specificity and sensitivity, it may not be useful for screening if the cancer has low prevalence. The prevalence of pancreatic cancer is approximately 1 in 10,000 people at age 50 (0.01%) and 6 in 10,000 at age 70 (0.06%). To achieve 10% PPV (one true positive for every nine false negatives), a biomarker would need to have 100% sensitivity and 99.9% specificity to be used to screen 50 year olds for pancreatic cancer and 100% sensitivity and 99.4% specificity to be used to screen 70 year olds for pancreatic cancer. Similarly a biomarker would need to have 100% sensitivity and 99.6% specificity to be used to screen average-risk population women for ovarian cancer.
EDRN focuses on phases 1 thorough 3. NCI’s Translational Research Working Group has also published a developmental pathway for biospecimen-based assays that overlaps with phases 2 through 4 and is focused on enhancing the efficiency and effectiveness of the process6.
Discovery process
Successful translation of biomarkers into clinical application depends on the biomarkers entering into the validation pipeline. No matter how well thought out and rigorous the translation process, if the biomarkers lack the requisite performance characteristics, they will not be successfully translated into clinical application. Unfortunately, most of the biomarkers reported in the literature have insufficient sensitivity and specificity or lack data using appropriate specimens, e.g. late stage cancers or mismatched cases and controls. Thus, the first step to enhancing the translation of biomarkers is to improve the discovery process. Although a thorough discussion of shortcomings and problems in biomarker discovery and potential solutions are beyond the scope of this review, here we list some of the recurrent problems with study design and biospecimens.
Study design
Biomarker discovery is undertaken without consideration of potential clinical use.
Specimens used come from inappropriate kinds of tissues or stages of cancer. If interested in early detection, perform discovery using early stage cancers or body fluids from patients with early stage disease or if possible, work with preneoplastic lesions or prediagnostic sera/plasma. If interested in detecting aggressive cancer (e.g., prostate cancer) use specimens from patients with aggressive cancer.
Failure to account for confounding conditions. Biomarker levels are altered by factors other than cancer, e.g., PSA is increased in benign prostatic hyperplasia (BPH) as well as in prostate cancer, and CA19-19 is elevated in pancreatitis as well as in pancreatic cancer.
Failure to account for lack of tissue specificity. Other tissues produce the biomarker resulting in high biomarker levels in healthy patients.
Classifier results from overfitting. This occurs in biomarker discovery when models are used that involve a large number of variables (e.g., mass spectrometry peaks) measured on a small number of samples. This can result in a statistical model that describes random error or noise instead of the underlying relationship. Completely independent training and test sets of specimens should be used to rule out the possibility that he observed classification is a result of overfitting.
Biospecimens
Use of poorly matched cases and controls (differences in age, sex, confounding conditions, diet, ethnicity, and exercise) can result in biomarkers that reflect these differences and not the presence of cancer.
Artifacts due to sample collections, processing times, storage can result in biomarkers that reflect these differences and not the presence of cancer.
Cases and controls collected under different conditions – controls collected during a general check up and cases collected immediately prior to surgery. These two very different collection conditions can result in the discovery of biomarkers for stress rather than for disease.
Whenever possible, discovery should be performed using high quality specimens from carefully matched cases and controls that are collected under the same conditions using a standard operating procedure. It has been suggested that discovery should be performed using specimens collected from asymptomatic patients who later go on to develop cancer. Several investigators within the EDRN are basing their discovery efforts on prediagnostic sera or plasma. However, as there are very few collections of prediagnostic sera, it is unclear how widely this approach can be used.
Another approach the EDRN is taking to develop panels of biomarkers or to combining biomarkers developed by different investigators is to support collaborative team projects. EDRN investigators combine resources to evaluate their biomarkers using a common set of well characterized, high quality biospecimens. This allows for a direct comparison of the performance of the individual biomarkers and the possibility of combining them to form a more effective panel.
There a number of reviews and commentaries that discuss in detail problems associated with study design and biospecimen collection7–12. The EDRN brings together investigators from discovery laboratories, clinicians and biostatisticians to discuss issues related to clinical needs and study design and to share resources, such as high quality biospecimens that are collected to address a specific clinical question. An NCI workshop sponsored, Development of Biospecimen Reporting Criteria for Publication, concluded that “human biospecimens are subject to a number of different collection, processing, and storage factors that can significantly alter their molecular composition and consistency” and recommended guidelines for reporting data elements for human biospecimens used in biomedical studies, the Biospecimen Reporting for Improved Study Quality (BRISQ)13. These reporting guidelines are intended to allow other researchers to better evaluate and reproduce experimental results.
Development of the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK)14 was a major recommendation of the National Cancer Institute – European Organization for Research and Treatment of Cancer (NCI-EORTC) First International Meeting on Cancer Diagnostics. These reporting guidelines are intended to address the all too common problem of initial promising reports on prognostic biomarkers not being reproduced in subsequent studies. Guidelines are provided for reporting study design, preplanned hypotheses, patient and specimen characteristics, assay methods, and statistical analysis methods. This information should help other investigators to better understand the reported results. REMARK guidelines did not make specific recommendation on study design or analysis methods. Although these guidelines were developed for prognostic biomarkers, many of the reporting
Validation process
When there is sufficient preliminary data to demonstrate the potential clinical use of a biomarker, the process of translation or validation begins. This process can be divided into two steps; first a prevalidation or verification study to determine if the biomarker is worth pursing, followed by validation trial, the latter often requires a prospective multisite trial. In the past, validation trials have been performed on biomarkers that had not gone through a verification step, frequently resulting in a disappointing or ambiguous outcome. The EDRN requires verification, preferably performed by an independent laboratory, prior to conducting a biomarker validation trial. The process the EDRN follows for verification and validation are described in detail in the following sections.
Verification of a biomarker or panel of markers involves both analytical validation of the assay and an independent evaluation of its clinical performance. Analytical validation or assay validation includes optimization and determination of the accuracy, reproducibility and reliability of the assay for the intended application. As most investigators conducting biomarker discovery have little experience in this area, the involvement of experts in assay development is recommended. Even if the discoverer has the expertise, it is highly recommended that the assay be reproduced in a completely independent laboratory prior to initiating a biomarker validation trial. The EDRN Biomarker Reference Laboratories (BRLs) are experts in assay develop and meet the requirements of the Clinical Laboratory Improvement Amendment (CLIA) for testing patient specimens. These laboratories work with the discoverers to optimize and independently validate the assays. Equally important is the verification of the clinical performance of the biomarker. This can be achieved by performing the assay on a new set of specimens for which the investigators are blinded. It recommended that if possible, the assays be conducted by an independent laboratory, using specimens from an independent source. Within the EDRN the BRLs can act as an independent laboratory to perform the assays and the Clinical Validation Centers (CVCs) can often supply the needed blinded specimens.
To help expedite the verification process, the EDRN has established reference sets for many of the major epithelial cancers, including breast, lung, colon, prostate, pancreas and liver. These sets of specimens are from well characterized and matched cases and controls. The specimens are collected using standard operating procedures, and common data elements are collected on all patients. For example, the EDRN colon cancer reference set consists of multiple replicate aliquots of sera, plasma, and urine from 50 subjects with colorectal adenocarcinoma, 50 subjects with adenomas confirmed by pathology, and 50 subjects with normal colons after colonoscopy. Any investigator with promising a biomarker or panel of biomarkers can apply to the EDRN to receive these reference sets to verify the clinical performance of their markers on blinded specimens (http://edrn.nci.nih.gov/resources/sample-reference-sets). These reference sets also allow for the comparison of individual candidate biomarkers from different investigators on the same samples and for the complementary or additive performance of these biomarkers to be assessed. The results of biomarkers or panel of biomarkers in the colon cancer reference set are shown in Table 1.
Table 1.
Verification of colon cancer biomarkers using EDRN colon cancer reference set.
| Biomarker | Sensitivity | Specificity |
|---|---|---|
| Galectin-3 ligand | 72% | 80% |
| K-ras in urine | 77% | 65% |
| K-ras on FOBT card | 14% | 65% |
| GOS | 77% | 49% |
| GOS +FOBT | 27% | 95% |
| Proteomics-A | 78% | 88% |
| Proteomics-B | 70% | 66% |
| Proteomics-SELDI-TOF | 19% | 98% |
| Proteomics-MALDI-TOF | 63% | 48% |
| p53 | 40% | 70% |
| CEA | 40% | 70% |
| Topoisomerase II | 35% | 70% |
| Cathepsin D | 50% | 70% |
| Cyclin B | 40% | 70% |
| IGF Binding Protein 2 | 35% | 70% |
| TRAILR2 | 10% | 96% |
| CIN248 | 12% | 92% |
| P108 | 27% | 94% |
| TRAILR2 & CIN248 & P108 | 29% | 60% |
| FOBT TRAILR2 & CIN248 & P108 | 42% | 97% |
Results from the prevalidation/verification studies should be used to determine whether the biomarker or panel of biomarkers has sufficient performance to warrant a full validation trial to determine clinical validity – does the biomarker accurately and reliably distinguish those with cancer from those without. Most biomarkers evaluated in a prevalidation study do not progress to a validation trial, primarily due to lack of performance when assayed using independently collected specimens that represent the broad spectrum of the disease and its confounding conditions (e.g., benign prostatic hyperplasia for prostate cancer).
Only after the assay has been analytically validated and the clinical performance of the biomarker been verified using an independent set of blinded specimens should a biomarker validation trial be undertaken. This trial may require a prospective specimen collection, or if appropriate specimens exist, it can be retrospective. Whether the trial is retrospective or prospective, the study must be well designed, conducted following a standard operating procedure, and sufficiently powered.
Investigators EDRN at the DMCC have developed a study design for phase 2/3 biomarker validation trials13. They have termed this the PRoBE design for Prospectivespecimen- collection-Retrospective-Blinded-Evaluation. A key feature of this design is the avoidance of bias by collecting all specimens prior to diagnosis, such that cases and controls are enrolled under the same conditions and all specimens collected and processed identically. This design can require large numbers of subjects as the incidence of most cancers are low – most of those enrolled will not have cancer. The design has four main components;
- Clinical context and population
- Define clinical context in which the biomarker will be used
- Define the clinically relevant outcome
- Biomarker performance criteria
- Define what the biomarker will do
- Define discriminatory accuracy – minimally acceptable performance
- Compare with existing biomarker if one exits or determine if it is additive
- Biomarker test
- Biomarker or panel of biomarkers are fixed based on discovery and verification studies
- Define protocol for specimen collection, processing and storage
- Define assay and fix procedure
- Assay performed blinded to outcome
- Study size
- Sample size based on minimally acceptable performance values and the performance values obtained from discovery and verification studies
These four components of the PRoBE design are described in detail in the paper by Pepe and co-workers15, and the EDRN has begun using this as the basis for the design of its biomarker validation trials and reference set collections.
When the EDRN conducts a prospective biomarker validation trial, it collects multiple aliquots of sera and plasma and if applicable urine from each subject. In some studies, lymphocyte DNA is also collected. For example, the EDRN conducted a phase 2 biomarker validation for the early diagnosis of hepatocellular carcinoma (HCC); the biomarkers tested were alpha-fetoprotein (AFP), the currently used biomarker, AFP-L3%, a specific glycoform of AFP, and des-gamma carboxyprothrombin (DCP)16. A total of 424 cirrhotic controls and 422 cases of HCC (208 early stage cancers) were enrolled. Serum and plasma were collected and stored from all patients, and genomic DNA was collected from the majority of the patients. After completion of the trial, the samples were divided into two sets; a prevalidation reference set composed of 50 early stage cases and 50 cirrhotic controls, and a validation set composed of 372 cases, 158 of which are early stage, and 374 cirrhotic controls. These specimens are available to any investigator with promising biomarkers. The reference set is provided to investigators who have candidate biomarkers with preliminary data indicating that the markers perform as well as or complement AFP. If the biomarker performs as well as or complements AFP, AFP-L3%, or DCP in the reference set, the validation set is made available to the investigator. The investigators are blinded as to whether the specimens are case or control, and the EDRN Data Management and Coordinating Center performs the data analysis. The reference set has been used to test candidate biomarkers from six different investigators, and three of these investigators have been approved for access to the larger validation set. This two-step procedure allows the efficient testing of potential biomarkers on a smaller prevalidation reference set to determine performance characteristics before access to the larger validation set.
In 2011, FDA cleared ROMA Algorithm (Fujirebio), which uses the CA-125 and HE4 blood markers, to determine the likelihood that an ovarian pelvic mass is malignant. Stephen Skates and his colleagues of EDRN helped validate CA-125 and HE4 in preclinical samples received from NIC’s Prostate, Lung, Colon, and Ovarian (PLCO) Screening Trial and develop an algorithm for measuring the these two markers.17 PLCO data and biospecimens are available to all qualified researchers though a peer review process (http://prevention.cancer.gov/plco/biospecimens).
Conclusions
The translation of biomarkers into clinical use is a long and difficult process and the vast majority of biomarkers will never make it beyond verification as they lack sufficient sensitivity and specificity to be clinically useful. Successful translation requires not only close collaboration between investigators involved in discovery research, clinicians, assay developers and statisticians, but the investigators have to willing to have their assays and biomarkers verified by an independent laboratory. Some investigators have expressed reluctance to “hand-over” their biomarkers or to have them independently verified, asking “what’s in it for me.” The EDRN is designed to foster collaboration amongst investigators with different expertise and to allow for the biomarker discoverer to actively participate in the validation process.
EDRN investigators Zen Zhang and Daniel Chan18 have described lessons learned during the six years it took from biomarker discovery to FDA approval for the in vitro diagnostic OVA1. As they noted, “From discovery of biomarkers to their use for a specific clinical indication, it requires the resolution of many interwoven issues and knowledge and expertise from diverse areas.”
Investigators work with the EDRN because of its vision for translational research. Such a vision, while challenging to implement, allows investigators access to expertise, technologies, and resources that would not otherwise be available to them. Investigators have built strong collaborative ties with other first-rate laboratories across the country that has enabled implementation of a translational research paradigm. The EDRN system and resources attracts outstanding investigators who make their scientific resources available to other network investigators.
The EDRN is a model for translational research that links strong basic scientists with translational clinical investigators in formal ways. While sometimes challenging, the resources and collaborative environment have made a major difference in the quality and innovation in biomarker research. The biomarker community foresees extension of the EDRN model to other aspects of biomarker application for cancer prognosis and therapeutic monitoring.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have read the journal's policy on disclosure of potential conflicts of interest and have no potential conflicts to declare.
References
- 1.Biomarkers Definitions Working Group Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2.Diamandis EP. Cancer biomarkers: can we turn recent failures into success? Natl Cancer Inst. 2010;102:1462–1467. doi: 10.1093/jnci/djq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu CS, Pinsky PF, Cramer DW, et al. A framework for evaluating biomarkers for early detection: validation of biomarker panels for ovarian cancer. Cancer Prev Res. 2011;4:375–383. doi: 10.1158/1940-6207.CAPR-10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanash SM. Why have protein biomarkers not reached the clinic? Genome Med. 2011;3:66–67. doi: 10.1186/gm282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava S, Gray JW, Reid BJ, Grad O, Greenwood A, Hawk ET. Translational Research Working Group developmental pathway for biospecimen-based assessment modalities. Clin Cancer Res. 2008;14:5672–5677. doi: 10.1158/1078-0432.CCR-08-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker SG, Kramer BS, McIntosh M, Patterson BH, Shyr Y, Skates S. Evaluating markers for the early detection of cancer: overview of study designs and methods. Clin Trials. 2006;3:43–56. doi: 10.1191/1740774506cn130oa. [DOI] [PubMed] [Google Scholar]
- 8.Ransohoff DF. How to improve reliability and efficiency of research about molecular markers: roles of phases, guidelines, and study design. J Clin Epidemiol. 2007;60:1205–1219. doi: 10.1016/j.jclinepi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Brenner DE, Normolle DP. Biomarkers for cancer risk, early detection, and prognosis: the validation conundrum. Cancer Epidemiol Biomarkers Prev. 2007;16:1918–1920. doi: 10.1158/1055-9965.EPI-07-2619. [DOI] [PubMed] [Google Scholar]
- 10.Baker SG. Improving the biomarker pipeline to develop and evaluate cancer screening tests. J Natl Cancer Inst. 2009;101:1116–1119. doi: 10.1093/jnci/djp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn BK, Wagner PD, Anderson D, Greenwald P. Molecular markers for early detection. Semin Oncol. 2010;37:224–242. doi: 10.1053/j.seminoncol.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Ransohoff DF, Gourlay ML. Sources of bias in specimens for research about molecular markers for cancer. J Clin Oncol. 2010;28:698–704. doi: 10.1200/JCO.2009.25.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore HM, Kelly AB, Jewell SD, et al. Biospecimen Reporting for Improved Study Quality (BRISQ) J Proteome Res. 2011;10:3429–3438. doi: 10.1021/pr200021n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 15.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrero JA, Feng Z, Wang Y, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;37:110–118. doi: 10.1053/j.gastro.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore RG, Miller MC, Disilvestro P, et al. Evaluation of the diagnostic accuracy of the risk of ovarian malignancy algorithm in women with a pelvic mass. Obstet Gynecol. 2011;118:280–288. doi: 10.1097/AOG.0b013e318224fce2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;9:2995–2999. doi: 10.1158/1055-9965.EPI-10-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]