Abstract
Growth rate regulation in bacteria has been an important issue in bacterial physiology for the past 50 years. This review, using Escherichia coli as a paradigm, summarizes the mechanisms for the regulation of rRNA synthesis in the context of systems biology, particularly, in the context of genome-wide competition for limited RNA polymerase (RNAP) in the cell under different growth conditions including nutrient starvation. The specific location of the seven rrn operons in the chromosome and the unique properties of the rrn promoters contribute to growth rate regulation. The length of the rrn transcripts, coupled with gene dosage effects, influence the distribution of RNAP on the chromosome in response to growth rate. Regulation of rRNA synthesis depends on multiple factors that affect the structure of the nucleoid and the allocation of RNAP for global gene expression. The magic spot ppGpp, which acts with DksA synergistically, is a key effector in both the growth rate regulation and the stringent response induced by nutrient starvation, mainly because the ppGpp level changes in response to environmental cues. It regulates rRNA synthesis via a cascade of events including both transcription initiation and elongation, and can be explained by an RNAP redistribution (allocation) model.
Keywords: growth rate regulation, rRNA synthesis, RNA polymerase distribution, transcription factories, nucleolus-like structure, ppGpp
Introduction
More than half a century ago two accompanying papers by Maaloe and his colleagues were the first to describe the growth rate regulation in bacteria, an important issue in bacterial physiology (Kjeldgaard et al., 1958; Schaechter et al., 1958). They reported that during balanced growth, cell mass and RNA level can be described as an exponential function of the growth rate afforded by the various media. Further, during transitions between different growth rates, either by nutrient up-shift from a minimal medium to rich broth or, conversely, by nutrient down-shift, RNA synthesis is the first to respond to the change in medium, either accelerating or decreasing rapidly to the rate characteristic of the new condition. Changes in the relative rates of synthesis of other macromolecules, including DNA and protein, are delayed relative to that of RNA synthesis during periods of changing growth rates (Neidhardt & Fraenkel, 1961; Maaloe & Kjeldgaard, 1966). rRNA, along with tRNA (collectively they are called stable RNA), comprises the majority of total RNA in bacterial cells. Because most of the stable RNA is rRNA, for simplicity, total RNA reflects rRNA in the cell. In Escherichia coli, the number of ribosomes in the cell is proportional to the growth rate in order to meet the demand for protein synthesis (Nomura et al., 1984; Bremer & Dennis, 1996; Keener & Nomura, 1996). However, it is the synthesis of rRNA, but not of ribosomal protein (r-protein), that is sensitive to the growth rate (Gausing, 1977).
In the past 50 years, many researchers have studied growth rate regulation in bacteria, with a focus on the regulation of rRNA synthesis. The main challenge has been to understand how rRNA synthesis is regulated during two extreme growth conditions (analogous to two opposite ends of the spectrum): in rapidly growing cells, rRNA is actively synthesized and commandeers the majority of RNA polymerase (RNAP) in the cell, whereas rRNA synthesis is suppressed upon nutrient starvation, which induces the stringent response (Cashel et al., 1996). In the latter case, because Cashel & Gallant (1969) discovered >40 years ago that two alarmone molecules or magic spots, pppGpp and ppGpp (hereafter collectively named ppGpp) are rapidly accumulated immediately following an amino acid starvation, accompanied by concomitant inhibition of almost total RNA synthesis in the cell, the question has also become how rRNA synthesis is stringently controlled by ppGpp. Subsequent works by many groups showed that both the stringent control and the growth rate regulation are mediated by ppGpp, indicating an interconnection of the two regulatory mechanisms for rRNA synthesis. Relatively recently, Gourse and colleagues discovered that DksA acts with ppGpp synergistically in the regulation of rRNA synthesis (Paul et al., 2004a).
The extensive literature on the subject, although at times controversial and contentious, demonstrates the significance of the research and reflects a long intellectual journey in seeking to solve the puzzle. Many reviews on various aspects of the subject have been written (Nomura et al., 1984; Gourse et al., 1996; Wagner, 2002; Dennis et al., 2004; Paul et al., 2004b; Gralla, 2005). Recognizing that different mechanisms of growth rate regulation have evolved in different bacteria, however, this review intends to summarize mechanisms for the regulation of rRNA in E. coli with systems biology perspectives. The authors hope this review will facilitate discussions in the continuation of the journey toward understanding this important issue.
Organization of rRNA operon and bacterial growth
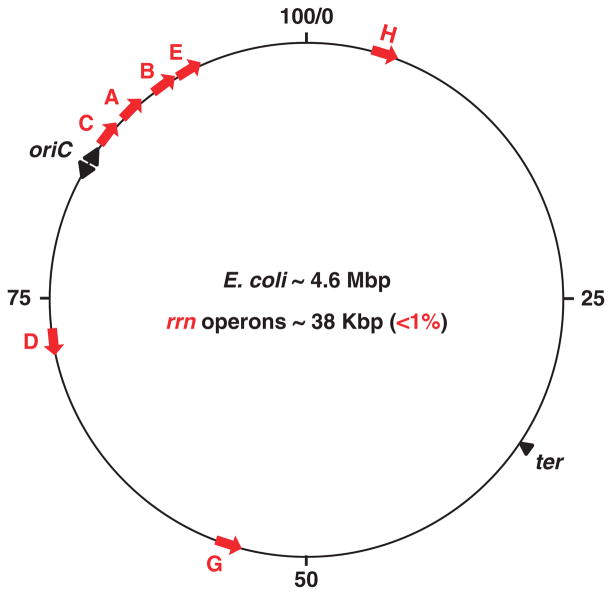
There are seven almost identical rRNA operons in the E. coli chromosome. The genome contains about 4.6 million base pairs (bp) of DNA. Each of the rRNA operons is named rrn with a capital letter (Fig. 1). Four of them, rrnCABE, are clustered near the origin of DNA replication (ori) and the other three are located in other regions within the ori half of the chromosome. Together all seven rrn operons encompass about 38 kbp, <1% of the genome size. The synthesis of rRNA has to compete with the remaining 99% of the genome for RNAP in response to environmental cues. Several features associated with the special location and organization of rRNA operons are important for cell growth and global gene regulation.
Fig. 1.
Location of seven rrn operons as indicated by capital letters (red) in Escherichia coli chromosome map. The numbers indicate the minute in the map, oriC for the origin of chromosome replication and ter for the terminus of replication. The direction of rrn transcription is indicated by arrows (red) and the bidirectional replication by arrowheads. Only a tiny fraction of E. coli genome encodes for the seven rrn operons.
Clustering with the genes for r-proteins
Most of the genes/operons encoding the 55 r-proteins are clustered near the rrn operons, which facilitate the coordination of the production of the components of the ribosome (Nomura et al., 1984). Certain r-proteins act as translational repressors by binding to their own mRNAs if they are in excess of the capacity for ribosome assembly, for example, under conditions when rRNA synthesis is reduced. Thus, the synthesis of rRNA coordinates the production of r-proteins by a translational feedback regulation mechanism (Nomura et al., 1980). Conceivably, the clustering of the rrn operons and the genes for r-proteins would also facilitate ribosome assembly, as those gene products are expressed in close proximity in the cell.
Codirection of rrn transcription and DNA replication
In E. coli DNA replication starts bidirectionally at the ori, from which the resulting replication forks propagate. The transcription direction of all seven rrn operons is the same as the direction of DNA replication (Ellwood & Nomura, 1982). This feature is critical as it minimizes the collision between the machineries of transcription and DNA replication (Brewer, 1988; Liu & Alberts, 1995; Pomerantz & O’Donnell, 2010a, b), particularly during rapid growth, because each of actively transcribing RNAP molecules along the rrn operons is a powerful biological motor that can exert considerable force (Yin et al., 1995). Transcription factor DksA, which is involved in regulation of rRNA synthesis, has an important role in preventing the collisions (Tehranchi et al., 2010). Collisions between DNA and RNAP can lead to DNA breakage and chromosomal instability (Vilette et al., 1996; Srivatsan et al., 2010); thus, the growth advantage of codirectionality of rrn transcription and DNA replication is evolutionarily selected. Note also that expression of essential genes tends to be codirectional with rRNA synthesis and DNA replication in bacteria (Rocha & Danchin, 2003).
Gene dosage effect
In E. coli, the time needed for one round of DNA replication is fixed at 40 min while the time needed for cell division varies depending on growth rate; consequently, multiple replication forks are located at different locations in chromosome before cell division in a fast-growing cell (Bremer & Dennis, 1996). Because the four rrnCABE operons are close to the ori, the number of rrn operons is significantly larger in a fast-growing cell than in a slow-growing cell. For example, it is estimated that a wild-type E. coli cell, with a doubling time of about 23 min, will have an equivalent of 38 rrn operons (Bremer & Dennis, 1996), whereas, a cell with a doubling time of 36 min will have an equivalent of 22 rrn operons. The rate of rRNA synthesis thus will be higher in a fast-growing cell than in a slow-growing cell due to their special locations and increased gene dosage. The gene dosage effect of the rrn operons, coupled with the long rrn transcripts, explains why synthesis of rRNA prominently influences RNAP distribution in the cell.
Nucleolus-like structures
Results from cell biology of RNAP (Cabrera & Jin, 2003; Jin & Cabrera, 2006) demonstrate that in rapidly growing cells cultured in nutrient rich Luria–Bertani (LB), concentrated RNAP form a few (two to three on average per nucleoid) dominant transcription foci. These transcription foci are proposed to be special transcription factories (Cook, 1999) and structures analogous to the eukaryotic nucleolus, where most of the cellular RNAP molecules are engaged in the synthesis of rRNA (Fig. 2a). It is further inferred that under optimal growth conditions, multiple rrn operons are folded together to be in proximity three dimensionally forming the putative nucleolus-like structure, and the nucleoid structure is relatively compact (Fig. 2b). In fast-growing cells, the nucleolus-like structures would considerably facilitate RNAP recycling and recruitment for active rRNA synthesis locally, which in turn would facilitate rRNA processing and ribosome assembly in the proximity. These hyperstructures (Norris et al., 2007), however, are dynamic and sensitive to the environment. For example, the transcription foci and, possibly, nucleolus-like structures disappear leading to an expanded nucleoid when growth is arrested, such as by amino acid starvation, which induces the stringent response, or with rifampicin treatment, which inhibits all transcription initiation (Cabrera & Jin, 2003). Studying the formation and degeneration of the transcription foci and understanding the nature of the nucleolus-like structures for rRNA synthesis will provide another dimension for growth rate regulation in E. coli.
Fig. 2.
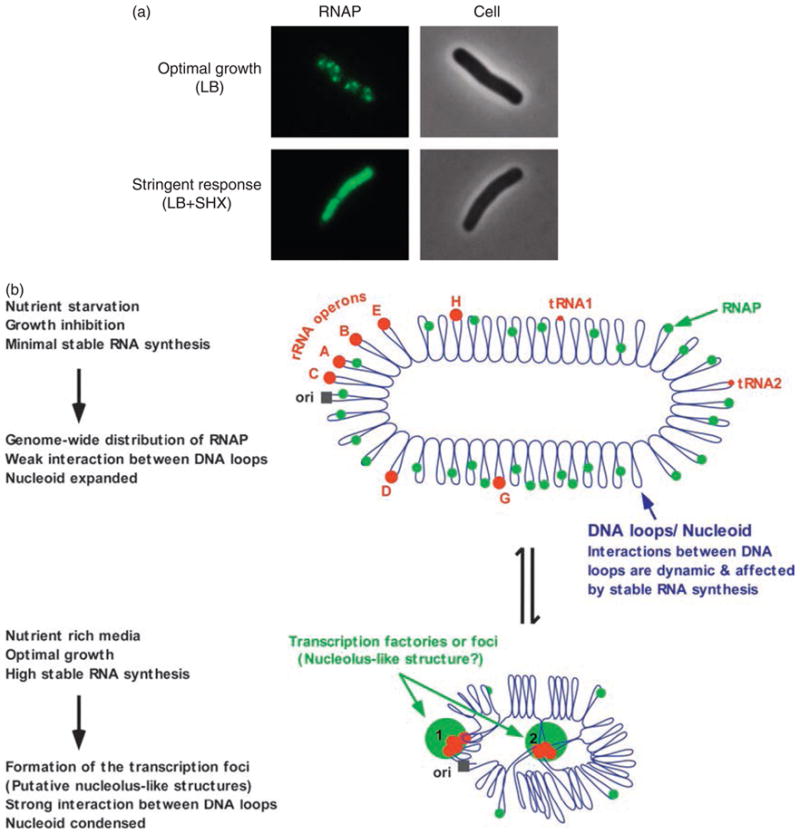
(a) The distribution of RNAP is sensitive to environmental cues. During optimal growth, concentrated RNAP (pseudo-colored green) forms dominant transcription foci, which are proposed to be special transcription factories and the nucleolus-like structures, where rRNA is being actively synthesized, in a mid-log cell grown in nutrient rich LB. With the addition of serine hydroxamate (SHX), which caused cell starvation for amino acid and induced the stringent response, RNAP is redistributed relatively homogeneously in the nucleoids. The RNAP is fused with a fluorescent protein and imaged with a fluorescent microscope with the corresponding cells in phase contrast as described (Cabrera & Jin, 2003). (b) Model illustrating the dynamics of the transcription factories or foci and the putative nucleolus-like structures under the two extreme growth conditions. The Escherichia coli chromosome is represented as blue lines folded in loops, the ori of replication as a black square, the seven rRNA operons as large red circles with letters, and two representative tRNA operons as small red circles. The RNAP molecules are represented as small green circles. For simplicity, during optimal growth only two transcription factories/foci and putative nucleolus-like structures, which make the nucleoid more compact by pulling different stable RNA operons into proximity, are indicated (bottom part of the diagram, large green circles encompassing multiple large red cycles labeled 1 and 2) (Adapted from the study by Cabrera & Jin, 2003).
Regulatory determinants of rRNA operons
Two elements of rrn operons are important for the growth regulation of rRNA synthesis (Fig. 3). The first is the long transcripts of rrn operons, which are among the longest transcripts in the cell. The other is the unique properties of the rrn promoters.
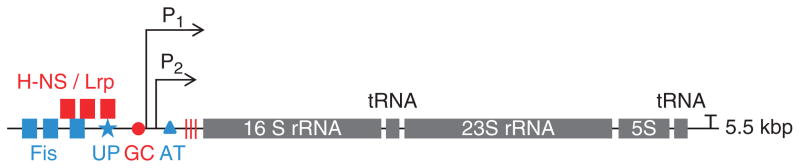
Fig. 3.
A typical rrn operon is schematically illustrated. Two promoters (P1 and P2) and terminators (T) for the long rrn transcript are indicated. Blue symbols indicate positive elements for the regulation of rrn, including Fis-binding sites (box) and UP element (UP, star) at the extended P1 promoter region, and antitermination system (AT, triangle) after the P2 promoter. Red symbols indicate negative elements, including H-NS and/or Lrp binding site (box), G/C-rich ‘discrimination sequence’ (GC, circle) at the extended promoter region, and multiple pausing sites (vertical line) before the 16S RNA gene. The illustration is not drawn to scale.
Structure of rrn operons
All seven rrn operons have a similar structure as illustrated in Fig. 3. In addition to two tandem promoters, they all have the boxB-boxA-boxC elements of the rrn antitermination system (Theissen et al., 1990; Condon et al., 1995b; Torres et al., 2001; Greive et al., 2005; Quan et al., 2005), which are located downstream from the second promoter, and two strong Rho-independent terminators at the end of the operons. The rrn antitermination system is required for maintaining a higher elongation rate compared with rates of elongation of mRNAs, and for preventing premature termination, particularly because untranslated rrn transcripts tend to form secondary and/or tertiary structures, which are termination-prone. The almost identical structure of different rrn operons explains why all rrn operons are subject to the same or similar regulatory mechanisms, although there are some fine-tuning differences (Condon et al., 1992). Only five of the seven rrn operons are necessary to support near-optimal growth; however, all seven rrn operons are required for rapid adaptation to nutrient changes (Condon et al., 1995a), suggesting an evolutionary advantage of having extra rrn operons in changing environments during growth for bacteria (Klappenbach et al., 2000). For rRNA synthesis, regulation at processing/maturation and elongation/antitermination are important; however, the primary regulatory mechanism is focused at the transcription initiation step (Gourse et al., 1986).
Each rrn operon encodes a long transcript with an average length of about 5500 nt, which is subsequently processed into three mature rRNA species, 16S, 23S and 5S RNA genes (Dunn & Studier, 1973; Srivastava & Schlessinger, 1990; Schaferkordt & Wagner, 2001), as well as some tRNA species, which are located in the spacer region between 16S and 23S RNA genes and/or at the distal ends of some rrn operons (Condon et al., 1995b). The long length of the rrn transcripts allows visible identification of rRNA synthesis in the chromosome and the estimation of the number of RNAP engaged in rRNA synthesis. From electron micrographs of chromatin spreads of cells grown in rich medium LB, active synthesis of the nascent rrn transcript can be readily identified as a double Christmas tree morphology (Miller et al., 1970), and the gap between the two Christmas trees reflects the RNase III processed 16S and 23S RNA genes (Hofmann & Miller, 1977). In contrast, under suboptimal conditions leading to slower growth rates, the nascent rrn transcript is difficult to identify in electron micrographs, as rRNA synthesis is reduced (Hamkalo & Miller, 1973a, b). Under the optimal growth conditions used where active rRNA synthesis occurs, as manifested by the double Christmas tree patterns, RNAP is found to be densely packed. It is estimated that on average there are about 65 RNAP molecules per rrn operon, which corresponds to one RNAP molecule for every 85 bp (French & Miller, 1989). Similar density of RNAP is also found in genes encoding some tRNA and r-proteins. In contrast, there is only one RNAP for each 1 kbp in the rpoBC region and only one RNAP molecule for every 10–20 kbp in many regions of the chromosome in fast-growing cells (French & Miller, 1989). The high density of RNAP in transcribing rrn in a fast-growing cell demonstrates that the rrn promoters have extremely high strength and efficiency compared with other E. coli promoters under the same condition.
Promoter regions
All seven rrn operons have extended promoter regions with similar sequences. Each rrn operon has tandem promoters, P1 and P2, which are separated by about 120 bp and recognized by RNAP holoenzyme containing σ70 (de Boer et al., 1979; Gilbert et al., 1979; Glaser & Cashel, 1979). P1 and P2 have similar activities in vitro (Gilbert et al., 1979; Glaser & Cashel, 1979). However, the two promoters behave differently in vivo. It is the activity of the upstream promoter P1, rather than that of P2, that predominates at fast growth rates and is subject to growth rate regulation and the stringent response (Lund & Dahlberg, 1979; Sarmientos & Cashel, 1983; Sarmientos et al., 1983; Zhang & Bremer, 1995). The differential expression of P1 and P2 in fast-growing cells is likely due to the interference at P2 from the transcriptions of RNAP originating from P1 across P2 (Gafny et al., 1994), a phenomenon known as ‘promoter occlusion’ (Adhya & Gottesman, 1982). At low growth rate and during the stringent response, however, P2 is mainly used (Sarubbi et al., 1988). One of the advantages of having the two promoters is that RNAP is always poised in the extended promoter regions, ready for transcription in response to environmental cues. During rapid growth, multiple rrn promoters are likely to be in proximity in the putative nucleolus-like structures to facilitate the capture and reuse of concentrated RNAP for active synthesis of rRNA.
A typical rrn P1 promoter, as illustrated in Fig. 3, has the following features: (1) There is an UP element (Ross et al., 1993; Aiyar et al., 1998) located at about ‘ − 60 to − 40’ upstream from the transcription initiation site (+1). (2) Nucleoid-associated proteins including Fis (Johnson et al., 1988) and H-NS (Falconi et al., 1988), as well as transcription factor Lrp (Platko et al., 1990), bind upstream of the UP element and/or overlap the element (Nilsson et al., 1990; Ross et al., 1990; Tippner et al., 1994; Bokal et al., 1995; Aiyar et al., 2002; Pul et al., 2005). The binding sites for these proteins are nonidentical, but overlap in the extended promoter region. (3) The DNA sequences of the ‘− 35’ regions as well as the spacing between the ‘− 35’ and ‘− 10’ regions of rrn P1 do not match the consensus of other σ70 promoters in E. coli. In addition, the rrn P1 promoter does not have an extended ‘− 10’ region (Burr et al., 2000). (4) There is a G/C-rich ‘discriminator’ region (Travers, 1980a) located immediately downstream the ‘− 10’ region, but before the initiation site ‘+1.’ It has been reported that the actual sequence of the discriminator’ region, in addition to its being G/C rich, is critical for the regulation (Haugen et al., 2006). The first two initially transcribed nucleotides of rrn transcripts are either AC or AU with the exception of rrnD where it is GU. It is important to emphasize that these several features, together as an integral system, determine the unique properties of the rrn P1 promoter and its regulation.
There are two key properties of the rrn P1 promoter that distinguish it from other typical E. coli promoters. The first is its exceptional strength potential, which means that the promoter has the potential to be the strongest promoter only in the cell grown in nutrient-rich media; the promoter strength weakens in the cell grown in nutrient-poor media. The first property is mainly attributed to the presence of the UP element, which enhances RNAP recruitment by providing additional interaction sites (Gourse et al., 1986; Ross et al., 1993, 1998), similar to the upstream region of the stringent promoter of tyrT, encoding a major tRNATyr species (Travers et al., 1983). The UP element enhances the activity of the P1 core promoter (defined as ‘− 41 to +1’) >30-fold, giving rrn P1 the potential to be the strongest promoter in fast-growing cells. The superb strength may explain why rRNA synthesis is the driving force for the distribution of RNAP in the cell (Cabrera & Jin, 2006).
The second property of rrn P1 is its rapid responsiveness to growth conditions in the cell. This property is its opposition to the robust promoter strength potential and is caused by the initiation complexes (or open complex) of rrn P1 being intrinsically unstable before the formation of the first few initial phosphodiester bonds (Gourse, 1988; Borukhov et al., 1993). This second property is the primary reason that the rrn P1 promoter can be regulated rapidly in response to growth conditions. For example, the activity of rrn P1 changes during the stringent response (Cashel et al., 1996), i.e. from being the strongest and most efficient in fast-growing cells grown in nutrient-rich medium to being almost nonexpressive during growth arrest in nutrient-starved cells. Thus, the promoter is termed growth rate regulated and/or stringently controlled.
In addition to rrn P1, there are a few other stringently controlled promoters (hereafter called stringent promoters) with similar features. In general, rrn P1 and the stringent promoters are regulated similarly (Zhou & Jin, 1998). Other stringent promoters include those of abundant tRNAs (Lamond & Travers, 1985; Emilsson & Kurland, 1990; Emilsson et al., 1993) and the rpoD operon including rpsU and dnaG (Taylor et al., 1984; Grossman et al., 1985), as well as rnpB (Jung & Lee, 1997), a component of RNase P, important for tRNA processing, pyrBI operon encoding pyrimidine biosynthetic enzymes (Donahue & Turnbough, 1990) and flagella genes (Durfee et al., 2008; Lemke et al., 2009). Note that these stringent genes are involved in the translational machinery, synthesis of nucleotides, which are the substrates for RNAP, and the cell motility machinery, the latter having a high demand for energy resources of the cell.
Several features of rrn P1 contribute to its intrinsic instability of the open complexes and the stringent response. The G/C-rich ‘discriminator’ region (Travers, 1980a, b; Zacharias et al., 1991) is critical for the instability of the open complexes. It has been suggested that the DNA duplex of the G/C-rich ‘discriminator’ sequences is extremely stable, thus requiring extra energy to form and/or maintain the open complex (Opel et al., 2004). Mutations at the ‘discriminator’ region of rrn P1 and other stringent promoters are no longer sensitive to the stringent response and growth-rate regulation, and enhance the stability of the open complexes, leading to increased efficiency of initiation (Travers, 1980b; Travers et al., 1986; Josaitis et al., 1995; Pemberton et al., 2000; Barker et al., 2001b; Zhi et al., 2003). Moreover, the nonoptimal nature of the ‘ − 35’ region, as well as the spacing between the ‘− 35’ and ‘− 10’ regions, also contribute to the intrinsic instability of the rrn P1 and other stringently controlled promoters (Lamond & Travers, 1985; Josaitis et al., 1995; Park et al., 2002).
Regulation of rrn transcription in vitro
Biochemical analyses of the rrn P1 promoter and other stringent promoters in vitro have provided many insights into the regulation of rRNA synthesis and the stringent response in the cell (Fig. 4). For nonstringent promoters, formation of open complexes from closed complexes is an irreversible step in general (McClure, 1980; von Hippel et al., 1984). By contrast, the open complexes (σRPo) of stringent promoters are conspicuously unstable kinetically, as if RNAP is in a rapid ON/OFF equilibrium before formation of the first few phosphodiester bonds (Gourse, 1988; Ohlsen & Gralla, 1992a; Borukhov et al., 1993; Zhou & Jin, 1998; Barker et al., 2001b; Paul et al., 2004a) (Fig. 4). Thus, the formation and/or maintenance of the open complexes of rrn P1 and other stringent promoters is a rate-limiting step, and is proposed to be the target for regulation (Zhou & Jin, 1998). This shared regulatory mechanism enables the coordinated expression of these genes in response to the nutrient quality of the environment. There are multiple factors that act at this regulatory step by either increasing or decreasing the efficiency of open complex formation and, thus, controlling transcription activity of rrn P1.
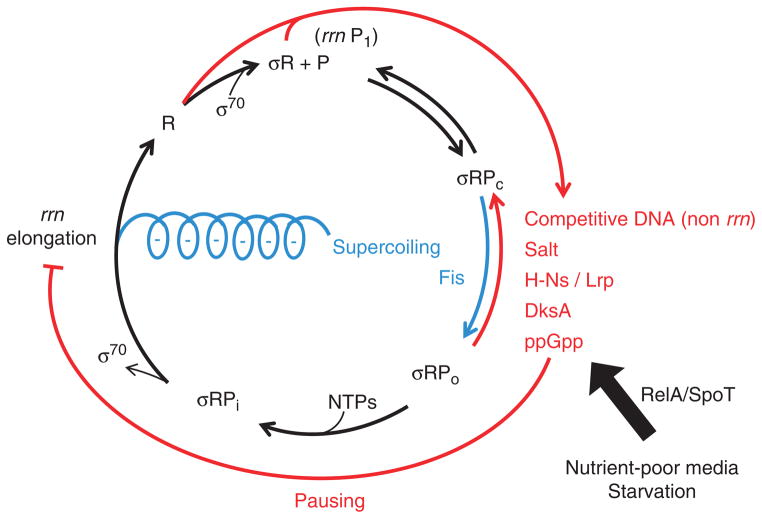
Fig. 4.
Model illustrating regulation of rRNA synthesis at system-level in Escherichia coli. RNAP holoenzyme (σR) binds to the rrn P1 promoter (P), forming multiple initiation complexes, including close complex (σRPc) and open complex (σRPo). Uniquely, the initiation complexes, particularly open complex, are intrinsically unstable and in rapid equilibrium with each other, before the formation of the stable initially transcribing complex σRPi in the presence of NTPs. Many positive (blue) and negative (red) elements as indicated control the expression of rrn P1. The activities of RelA and/or SpoT are responsible for the basal level of ppGpp, which are inversely proportional to the nutrient quality of the media and rapid accumulation of high level of ppGpp during starvation. The effects of ppGpp on rRNA synthesis are twofold, including inhibition of initiation, and enhancing pausing of elongation RNAPs, which in turn decreases elongation-induced supercoiling and causes jamming of RNAP at the rrn P1 promoter. Depending on cell growth conditions, released RNAP (R) from rrn operons either rebinds to rrn P1 for reinitiation or redistributes to other non-rrn genome-wide DNA. See text for details.
Positive regulators in transcription initiation
Two factors, negative supercoiling and the Fis protein, act as positive regulators by stabilizing the open complexes of rrn P1 and other stringent promoters. Both factors are likely to be relevant in vivo. A third factor, the initiating NTPs, is also found to be important in vitro, likely by promoting the rate of formation of the stable initially transcribing complex (σRPi) of rrn P1.
Supercoiling
A positive effect of supercoiling on rRNA synthesis is implied from the reports that the antibiotic novobiocin, which is a gyrase inhibitor, reduces the synthesis of 16S and 23S RNA genes both in vitro and in vivo (Yang et al., 1979; Oostra & Gruber, 1980). Using typical in vitro transcription assays, high efficiency of transcription of the rrn promoters occurs when supercoiled DNA is used; however, only minimal transcription activity of the promoters is detected when linear DNA is used (Glaser et al., 1983). The open complex of rrn P1 on a linear DNA template is more unstable and more sensitive to salt (Gourse, 1988; Ohlsen & Gralla, 1992b, c). The inhibitory effect of salt on transcription of rrn P1 is discussed below. Even with supercoiled DNA, the half-life (seconds) of the open complexes of rrn P1 is limiting in the presence of 40 mM KCl (Zhi et al., 2003), allowing for regulation. Supercoiling can be influenced locally by the transcription process itself because an elongating RNAP generates negative supercoiling behind it and positive supercoiling ahead of it when constrained DNA is used (Liu & Wang, 1987). It is reported that the highest transcription rate associated with rRNA synthesis from bacterial rrn operons produces a very high level of constrained supercoiling density locally (Booker et al., 2010). As proposed (Zhou & Jin, 1998), such a supercoiling feedback mechanism by transcription activity per se at rrn is likely to be an important integral element for the growth regulation of rrn P1 by stabilizing the open complexes.
Fis
Fis is a nucleoid-associated protein with multiple functions (Finkel & Johnson, 1992). As described above, there are multiple Fis-binding sites upstream of the rrn P1 promoter region (Fig. 3). Binding of Fis to these sites activates transcription from the promoter (Ross et al., 1990), by interaction with RNAP (Gosink et al., 1993, 1996; Bokal et al., 1995). It has been suggested that Fis also acts at a step subsequent to closed complex formation (Bartlett et al., 2000), and experiments have demonstrated that Fis stabilizes the open complex of rrn P1, leading to the activation of the promoter (Zhi et al., 2003). How Fis stabilizes the open complex of rrn P1 is still unknown; it is possible that Fis binding results in a translocation of superhelical energy required for the formation and/or maintenance of the open complexes, similar to the action of Fis binding on the leuV promoter, which is a stringent promoter (Opel et al., 2004). The Fis protein thus is a positive modulator important for the growth regulation of rRNA synthesis.
Initiating NTPs
Formation of the first few phosphodiester bonds bypasses the intrinsically unstable open complex step (Gourse, 1988; Ohlsen & Gralla, 1992a, b; Borukhov et al., 1993), indicating the importance of the rate of the initially transcribing complex formation in the equilibrium as proposed (Zhou & Jin, 1998). Similarly, the initiation of rrn P1 is activated in vitro with a high concentration of the first nucleotide when other NTPs are constant (Gaal et al., 1997), likely due to promoting the formation of the initially transcribing complex. Potentially, any element affecting the rate of formation of the initially transcribing complex containing the first few phosphodiester bonds should influence the transcription of rrn P1. The role of NTPs on the rRNA synthesis in vivo has been reported and proposed (Gaal et al., 1997; Schneider et al., 2002). Subsequent work showed that the concentration of ATP, which is the first nucleotide for most of the rrn transcripts, changes little, if at all, with growth rate; thus, NTP sensing is most likely not responsible for growth rate regulation of rRNA synthesis in E. coli (Petersen & Moller, 2000; Schneider & Gourse, 2004). However, ATP concentration increases significantly during cells’ outgrowth from stationary phases, which correlates with the expression of rrn P1 (Murray et al., 2003), indicating that NTP sensing works under some growth conditions.
Negative regulators in transcription initiation
In contrast to the actions of supercoiling and Fis, there are also several negative modulators, which act in the opposite direction of the equilibrium; collectively, they reduce the efficiency of the open complex formation at rrn P1 and other stringent promoters.
Salt and competitive DNA
The interaction between RNAP and promoter in general is sensitive to salt concentration (Record et al., 1978); however, the interaction between RNAP and rrn P1 is particularly weakened by salt due to the intrinsic instability of the open complex of the promoter (Gourse, 1988; Zhi et al., 2003; Gralla & Vargas, 2006).
A high concentration of salt decreases transcription of rrn P1 (Kajitani & Ishihama, 1984; Zhi et al., 2003) by reducing the efficiency of open complex formation at rrn P1 in vitro (Ohlsen & Gralla, 1992c). Regardless of the template topology, the open complexes of rrn P1 are very sensitive to salt; however, as discussed above, the salt effect is much more severe on the open complex of rrn P1 on a linear DNA template than that on a supercoiled DNA. Although it is not known whether there are changes in salt concentration in cells with different growth rates, it is established that cellular salts change when cells are exposed to high salt in the environment (Dinnbier et al., 1988; Cayley et al., 1991; Cayley & Record, 2003). The effect of salt on initiation of rrn P1 is biologically significant because rRNA synthesis is significantly inhibited during hyperosmotic response (Gralla & Vargas, 2006).
Heparin competes for RNAP and decreases the efficiency of open complex formation at the rrn P1 and other stringent promoters (Ohlsen & Gralla, 1992b). This is the expected result as the intrinsic instability of the open complex of rrn P1 is only manifested in the presence of a DNA competitor. Analogous to the action of heparin, nonspecific DNA also competes effectively for RNAP on preformed initiation complexes of rrn P1, which leads to inhibition of transcription from the promoter (Zhou & Jin, 1998). Because there is an overwhelming presence of non-rrn DNA (>99%) in the genome, this negative effect of nonspecific competitive DNA is likely relevant to the regulation of RNA synthesis in cells during the stringent response or under nutrient-poor conditions. Under these stress conditions, RNAP would be easily titrated out from the rrn P1 promoter by the competitive genomic DNA.
ppGpp
Immediately following its discovery >40 years ago, ppGpp was thought to be a negative regulator of rRNA synthesis (Cashel & Gallant, 1969). Since then, many studies have analyzed the effect of ppGpp on transcription from the rrn promoters, but with inconsistent results (for comprehensive references, see Cashel et al., 1996). The literature, however, reveals that some of those contradictory results could be reconciled if the different experimental conditions used are considered. Notably, inhibitory effects of ppGpp on the initiation of rrn P1 are consistently found under conditions in which the efficiency of open complex formation at rrn P1 is reduced, such as with a linear DNA template (Gilbert et al., 1979; Kingston et al., 1981a), in the presence of nonspecific competitor DNA (Kajitani & Ishihama, 1984), or in the presence of a relatively moderate concentration of salt (50 mM KCl) (Glaser et al., 1983). These results are also consistent with reports that ppGpp is ineffective on open complexes of rrn P1 with supercoiled DNA and low salt (Ohlsen & Gralla, 1992c), conditions known to stabilize the open complexes of rrn P1. When ppGpp is effective, it significantly decreases further the half-life of the open complex of rrn P1 (Barker et al., 2001b). Inhibition of initiation of rrn P1 in vitro requires a relatively high concentration of ppGpp. It appears that a key role of ppGpp is to effectively affect the lifetime of initiation complexes and shift the equilibrium toward the closed complex conformation during initiation of rrn P1 under conditions in which formation and/or maintenance of the open complex or rrn P1 are constrained. Importantly, the effect of ppGpp is significantly enhanced by DksA.
Several derivatives of ppGpp bind to RNAP (Owens et al., 1987; Reddy et al., 1995; Toulokhonov et al., 2001), indicating that the target for ppGpp is RNAP. Although structures of RNAP•ppGpp complexes have been determined (Artsimovitch et al., 2004), mutational analysis of RNAP based on these structures does not support the structural basis, and the true interaction site(s) between RNAP and ppGpp remains unresolved (Vrentas et al., 2008).
DksA
DksA (Kang & Craig, 1990), an RNAP-associated protein, is also discovered to play a key role in the regulation of rRNA synthesis by Gourse and colleagues in 2004 (Paul et al., 2004a). DksA decreases the half-life of the open complex of rrn P1; moreover, DksA acts synergistically with ppGpp to further reduce the half-life of the open complex of rrn P1. The DksA structure is similar to the transcript cleavage factor GreA (Perederina et al., 2004); thus, it is likely to regulate transcription through the secondary channel of RNAP where GreA binds.
H-NS and Lrp
H-NS, a nucleoid-associated protein, binds upstream of the rrn P1 promoter and inhibits transcription of rrn P1 (Tippner et al., 1994; Afflerbach et al., 1998). H-NS antagonizes the activation function of Fis in a concentration-dependent manner. In the absence of Fis, H-NS traps the bound RNAP at the rrn P1 promoter by bridging or looping the upstream and downstream sequences of the promoter (Schroder & Wagner, 2000; Dame et al., 2002). The binding of H-NS to rrn P1 is enhanced by the leucine-responsive regulatory protein Lrp (Pul et al., 2005). Lrp alone inhibits transcription initiation from rrn P1 by binding to the extended promoter region, similar to H-NS. However, in the presence of leucine, Lrp loses its ability to bind to the promoter, leading to the derepression of the transcription of rrn P1.
Inhibition of transcription elongation by ppGpp
In addition to its inhibitory effect in initiation, ppGpp also enhances pausing of RNAP at several sites during transcription of rrn operons on either linear or supercoiled DNA template (Kingston & Chamberlin, 1981; Krohn et al., 1992). NusA, which binds to core RNAP after the release of σ70 (Greenblatt & Li, 1981), further enhances pausing and/or termination at some sites in the leader regions of the 16S RNA gene (Kingston & Chamberlin, 1981). Although ppGpp also enhances pausing of RNAP during transcription of other non-rrn DNA templates (Kingston et al., 1981b), there is evidence that ppGpp shows the strongest pausing enhancement during RNA chain elongation from rrn P1 and other stringent promoters with the G/C-rich ‘discriminator’ sequences (Krohn & Wagner, 1996). It has been shown that ppGpp inhibits mRNA chain elongation in vivo; however, elongation of a truncated rrn operon, in which about 1 kb DNA encompassing the rrn P1 promoter, pausing sites and about half of 16S RNA gene are deleted, appears to be unaffected by ppGpp in the cell (Vogel & Jensen, 1994). The effect of ppGpp on rRNA chain elongation in the native context in vivo needs to be further investigated.
Regulation of rRNA synthesis in the cell
From what is described in the above sections (Fig. 4), it is understood that regulation of rRNA synthesis involves a concerted action of multiple factors in response to growth conditions. The key regulatory step is at the efficiency of the open complex formation, which is positively controlled by a proposed transcription-coupled supercoiling mechanism and Fis, as well as being negatively controlled by salt, noncompetitive DNA and most importantly, by a concert action of ppGpp/DksA. The role of ppGpp is special because it affects both initiation and elongation; the collective effects of ppGpp are interconnected and amplified. The nucleoid-associated protein H-NS and transcriptional regulator Lrp also negatively control initiation of rrn P1. In this section, we discuss mechanisms that have been proposed for the regulation of rRNA synthesis. Although controversies still remain, some of which could be explained by variables used in the studies, overwhelming evidence underscores the interconnections of different mechanisms.
Establishing active rRNA synthesis by Fis during nutrient upshift
The role of Fis in rRNA synthesis was first identified in 1990 (Nilsson et al., 1990; Ross et al., 1990). Many nucleoid-associated proteins are growth phase-dependent; however, Fis is unique in that it is the first one to accumulate dramatically upon nutrient upshift (Nilsson et al., 1992b; Ali Azam et al., 1999). For example, immediately after stationary cells are subcultured into fresh nutrient-rich LB, Fis increases rapidly and approaches its peak level (about 50 000–100 000 molecules per cell) within the first cell division; shortly afterward, however, the level of Fis decreases rapidly and is undetectable in stationary phase cells (Ball et al., 1992). The burst of the Fis level during the initial outgrowth corresponds to an immediate rapid increase in rRNA synthesis from a level undetectable in stationary phase cells, and leads immediately to exponential growth (Zhi et al., 2003). In contrast to wild type (MG1655), in the isogenic fis mutant there is a significant lag time (>30 min) before exponential growth accompanied by a continued twofold reduction of rRNA synthesis. After the prolonged lag time, however, the fis mutant has a growth rate and rate of rRNA synthesis comparable to those of the wild type during exponential growth. These results demonstrate that Fis is a key effector for the establishment of active rRNA synthesis during initial outgrowth from stationary phase and/or nutrient upshift.
How does Fis promote the establishment of active rRNA synthesis from a basal level expression of rrn during nutrient upshift? It is likely attributed to the role of Fis in both the recruitment of RNAP and stabilization of open complexes of rrn P1, as described in the above section. This positive effect is particularly critical, as DNA supercoiling is reduced in nutrient-starved stationary cells, leading to decreased efficiency of the open complex formation at rrn P1 (Ohlsen & Gralla, 1992c). During the outgrowth, the ATP level also increases, which activates rRNA synthesis (Murray et al., 2003). By working together, transcription from the rrn P1 promoter increases from the basal level. Afterward, elongation of rRNA chain by RNAP would generate ‘waves’ of negative supercoiling at the upstream promoter region, which in turn would further propagate the formation and maintenance of the open complexes of rrn P1 by the proposed supercoiling feedback mechanism described above. Thus, an initial activation of rRNA synthesis promoted by Fis and ATP would reinforce and amplify subsequent initiations of rrn P1, further activating rRNA synthesis. This hypothesized supercoiling feedback mechanism by the rrn transcription per se could explain why active rRNA synthesis would sustain its momentum after the establishment of the rrn transcription. Consequently, Fis would no longer be needed shortly after the establishment of rRNA synthesis. The effects of Fis on rRNA synthesis and growth rate underscore the importance of kinetic assays in assessing the function of Fis in the cell. Different types of assays used could therefore account for variable results in the magnitude of the effects of Fis reported in the literature (Ross et al., 1990; Condon et al., 1992; Nilsson et al., 1992a; Zhang & Bremer, 1996; Afflerbach et al., 1998; Bartlett et al., 2000; Zhi et al., 2003).
Fis antagonizes the H-NS negative effect on rrn P1 (Afflerbach et al., 1998) as the binding sites for Fis and H-NS overlap with each other and the UP element of the rrn P1 promoter. Changes in the binding pattern of these nucleoid-associated proteins at the regulatory region of rrn would affect the local architectural structure of the nucleoid, thus affecting transcription of rrn P1. The level of H-NS reaches a peak value of about 20 000 molecules in the mid-exponential phase when Fis declines rapidly, and gradually decreases to only about half of the peak value in stationary phase when Fis level becomes minimal (Ali Azam et al., 1999; Hansen et al., 2005). The differential levels of Fis and H-NS at different growth phase provide a basis for competition of the two regulators binding to the rrn P1 promoter. Because H-NS is a global repressor for gene expression in the cell (Atlung & Ingmer, 1997; Dorman, 2007; Dillon & Dorman, 2010), a genome-wide silencing of gene expression by H-NS would also facilitate allocation of RNAP to the rRNA synthesis in exponential cells grown in a nutrient-rich medium, assuming that RNAP is limiting in the cell.
The stringent control and growth rate regulation by ppGpp/DksA
In E. coli, ppGpp is produced by two ppGpp synthetases, RelA and SpoT; SpoT also has an additional ppGpp hydrolase activity (for a review, see Cashel et al., 1996). RelA and SpoT, however, sense different signals for nutrient richness or quality of growth media. RelA, a ribosome-associated protein, is the major ppGpp synthetase; the ppGpp synthetase activity of RelA is activated when an increased ratio of uncharged to charged tRNA is present due to starvation for amino acid(s). Accumulation of ppGpp by SpoT can be due to either increasing ppGpp synthetases activity or decreasing ppGpp hydrolase activity. Both activities of SpoT are stimulated by other starvations, such as carbon source, fatty acid, phosphate, iron and energy depletion (Xiao et al., 1991; Spira et al., 1995; Murray & Bremer, 1996; Vinella et al., 2005; Battesti & Bouveret, 2006); however, exactly how the two SpoT activities are regulated in the cell remains unclear. Collectively, RelA and SpoT are responsible for any accumulation of ppGpp in the cell in response to growth conditions, as no ppGpp is produced (ppGppo) in a double null relA spoT mutant (Hernandez & Bremer, 1991; Xiao et al., 1991).
The accumulation of ppGpp inversely corresponds to the nutrient richness or quality of a growth medium, indicating an intimate link between the steady-state growth rate regulation in bacteria afforded by growth medium and the basal levels of ppGpp in the cell (Zacharias et al., 1989; Bremer & Dennis, 1996). Even with the same growth medium, there is an inverse linear relationship between cells’ steady-state growth rates and the amounts of ppGpp accumulated in different spoT mutants; in parallel, there is an inverse linear relationship between the transcription of rrn P1 and the ppGpp level in those cells (Sarubbi et al., 1988). If ppGpp is the major source of growth rate control in E. coli, it would be expected that the regulation is abolished in a ppGppo mutant. Indeed, in contrast to wild type, in the ppGppo mutant RNA/DNA and RNA/protein ratios remain constantly high, independent of the steady-state growth rates (Potrykus et al., 2010); however, contradictory results have been reported when β-galactosidase activities are measured from different rrn P1-lacZ fusions (Gaal & Gourse, 1990; Hernandez & Bremer, 1993). The cause for the differences in the results from different groups is not clear; however, it is known that the double null relA spoT mutation easily accumulates suppressors that conferred growth advantage over the ppGppo mutant (Xiao et al., 1991; Potrykus et al., 2010).
The effect of the stringent response or ppGpp on the rRNA synthesis is universally accepted. When a fast-growing cell is subject to amino acid starvation, which induces the stringent response (Cashel et al., 1996), a rapid accumulation of ppGpp (over 100-fold increase from the basal level) is accompanied by an immediate shutoff of rRNA synthesis and growth arrest (Cashel, 1969). Such an inhibitory effect of the stringent response/ppGpp on rRNA synthesis during amino acid starvation can be visualized by a rapid disintegration of the special transcription factories, where concentrated RNAP engages in active rRNA synthesis during optimal growth (Fig. 2). It should be noted that complex effects of ppGpp have been reported (for reviews, see Chatterji & Ojha, 2001; Nystrom, 2004; Potrykus & Cashel, 2008; Srivatsan & Wang, 2008), particularly because the stringent response invokes reprogramming of global gene expression patterns in the cell (Durfee et al., 2008; Traxler et al., 2008, 2011). However, the consensus is that ppGpp directly inhibits rRNA synthesis and transcription of the stringent genes in the cell.
The inhibitory effect of ppGpp on rRNA synthesis is potentiated by the DksA protein, as the inhibition is significantly reduced upon nutrient starvation in the ΔdksA mutant (Paul et al., 2004a). The structure of DksA suggests that it interacts with RNAP at the secondary channel of RNAP (Perederina et al., 2004). Mutational analyses of DksA and RNAP have identified the regions important for the interaction (Blankschien et al., 2009; Rutherford et al., 2009). Similar to the ppGppo mutant, the growth rate regulation is abolished in the ΔdksA mutant as RNA/protein ratios remain constantly high, independent of the steady-state growth rates (Paul et al., 2004a). Unlike ppGpp, however, the level of DksA remains unchanged with different growth rates and growth phases (Brown et al., 2002; Paul et al., 2004a). Further, only high nonphysiological levels of DksA can substitute for ppGpp in the growth rate control in the ppGppo mutant (Potrykus et al., 2010). Together, these results are consistent with the conclusion that ppGpp is the primary effector in the regulation of rRNA synthesis.
How does ppGpp control rRNA synthesis in the cell? From the in vitro effects of ppGpp on the transcription of rrn described above, it is proposed that the regulation of rRNA synthesis by ppGpp involves a cascade of events during the stringent response (Fig. 4). The target for ppGpp is at both initiation and elongation of rrn, probably simultaneously; such synergistic inhibitory effect of ppGpp on transcription of rrn would effectively turn down rRNA synthesis immediately after nutrient starvation. On the one hand, it is likely that ppGpp increases pausing of elongating RNAP on rrn operons, which would promptly decrease the propagation of supercoiling at the rrn promoters region due to the proposed supercoiling feedback mechanism described above, reducing the efficiency of the open complex formation at the rrn P1 promoter. The pausing effect would also cause an immediate blockage of escape from the rrn P1 promoter by the jamming of RNAP downstream, particularly when RNAP molecules are densely packed. On the other hand, ppGpp and DksA act in concert synergistically to destabilize the weakened initiation complexes of rrn P1. This promotes RNAP dissociation from rrn P1, resulting in inhibition of initiation of the promoter. The synergistic effect of ppGpp and DksA could be augmented further by the challenge of vast amounts of non-rrn genomic DNA, which would effectively titrate RNAP out from the rrn P1 promoter into other parts of the nucleoid in the cell. Decreased rRNA synthesis would facilitate the binding of the H-NS proteins at the rrn P1 regulatory region, preventing further RNAP initiation from the rrn P1 promoter. Lrp (in the absence of leucine) binding in the region can further stimulate the interaction of H-HS in the rrn P1 promoter region. Lrp is a global feast/famine regulator of metabolism in the cell (Newman et al., 1992; Calvo & Matthews, 1994; Yokoyama et al., 2006). Expression of Lrp is stimulated by ppGpp (Landgraf et al., 1996). Note also that H-NS, Lrp and DksA are modulators of chromosomal supercoiling (Hardy & Cozzarelli, 2005). Together, this mode of action of ppGpp in vivo is consistent with the known effects of various factors on the rrn transcription in vitro described in the above section. However, the in vivo effect of ppGpp in the scheme needs to be further studied.
The inhibitory effect of ppGpp on rRNA synthesis and bacterial growth is likely a continuum depending on the level of the inhibitor. As described above, bacterial growth rate is negatively correlated with the basal levels of ppGpp (Sarubbi et al., 1988; Zacharias et al., 1989; Bremer & Dennis, 1996). Similarly, stable RNA synthesis is inversely related to the concentration of ppGpp, ranging >100-fold from basal level to high level during the stringent response (Baracchini & Bremer, 1988). Moreover, mutations in the G/C-rich discriminator regions of the rrn and tyrT promoters affect both the stringent response and growth-rate regulation simultaneously (Travers et al., 1986; Zacharias et al., 1989). Altogether, these results strongly support the argument that both the stringent and growth-rate control of rRNA synthesis are mediated by ppGpp and DksA. Conceivably, basal levels of ppGpp work similarly for the growth rate regulation, but to lower extents compared with the effect caused by high levels of ppGpp during the stringent response.
Promoting growth by polyphosphate/ppGpp during nutrient downshift
After E. coli cells grown in a nutrient-rich medium are subcultured into a fresh nutrient-poor minimal medium, bacteria assume exponential growth after a lag time. In contrast to wild type, however, the lag time is extended significantly to several hours in a strain carrying a null mutation in the ppk gene, which encodes the inorganic polyphosphate kinase (PPK), demonstrating that inorganic polyphosphate is important for bacterial to adapt to growth during nutrient downshift (Kuroda et al., 1999). The rapid accumulation of polyphosphate in starved cells depends on the inhibition of exopolyphosphatase (PPX) by ppGpp (Kuroda et al., 1997), indicating another role of ppGpp in growth regulation. Polyphosphate binds the Lon protease and stimulates the protease activity to degrade r-proteins (Kuroda et al., 2001), providing amino acids to support growth of starved cells during the nutrient limitation. Connection of ppGpp and polyphosphate demonstrates an important role of the two molecules in the nutrient starvation response. Recently, it has been proposed that polypho-sphate could act as a second messenger by binding to the major sigma factor of RNAP in Helicobacter pylori and possibly, in other human pathogens during nutrient starvation (Yang et al., 2010).
The distribution of RNAP and the regulation of rRNA synthesis
Regulation of rRNA synthesis, in essence, reflects the competition of RNAP between transcription of rrn and that of other non-rrn (or nonstringent) genes under different growth conditions. This competition indicates the fact that RNAP is limiting in the cell. In E. coli, it is estimated that there are about 2000 RNAP molecules per genome equivalent (Ishihama, 2000), or about 2600 RNAP molecules per cell in cells grown in LB (Piper et al., 2009), although a higher number of about 5000 RNAP molecules per genome equivalent has also been reported (Grigorova et al., 2006). The E. coli genome encodes about 4500 genes, which are organized into about 2390 operons, many of which have more than one promoter (Blattner et al., 1997; Riley et al., 2006). Moreover, it is likely that during cell growth more RNAP molecules are engaged in elongation than initiation in the cell. These estimates also argue for the notion that RNAP is limiting for genome-wide transcription.
The limitation of RNAP for genome-wide transcription is exacerbated in a fast-growing cell. It is estimated that the rate of rRNA synthesis approaches one initiation per second per rrn operon in a cell under optimal growth conditions (Bremer & Dennis, 1996; Voulgaris et al., 1999). The long length of the rrn transcripts coupled with the gene dosage effect described above has clear implications for the distribution of RNAP in the genome in response to cell growth. For example, rRNA synthesis of an equivalent of 38 rrn operons, which is calculated to be the copy number for these operons in a mid-log growing cell with a doubling time of 23 min, would capture about 2500 RNAP molecules. This amounts to almost all of the RNAP in the cell if using the lowest estimate. Under a suboptimal growth condition with minimal medium, however, the rate of rRNA synthesis decreases to about one initiation every 15 s per rrn operon, and during nutrient starvation such as amino acid starvation leading to the stringent response, the rate of synthesis of rRNA is even more reduced. Growth rate regulation in bacteria is largely about how RNAP is allocated for rRNA synthesis in response to environmental cues.
The rate of rRNA synthesis will determine the differential allocation of RNAP between rrn and non-rrn (and non-stringent) genes in the genome, which in turn will have profound consequences on global gene regulation and the structure of the nucleoid in response to growth conditions (Jin & Cabrera, 2006). During optimal growth, active rRNA synthesis consumes the majority of RNAP molecules; thus, RNAP available for transcription of genome-wide non-rrn genes is limited. Conversely, during the stringent response, more RNAP molecules become free and available for genome-wide transcription due to the amounts of RNAP released from rRNA synthesis. Such an RNAP redistribution (or allocation) concept during the stringent response is originally suggested by the analysis of stringent RNAP mutants that are defective in transcription of rrn P1 and other stringent promoters both in vitro and in vivo (Zhou & Jin, 1998). These RNAP mutants exhibit the stringent response phenotype (thus they are named the stringent RNAP) even when they are grown in nutrient-rich LB. They have slower growth rates compared with wild type; and as expected, the rates of RNA synthesis are reduced and the dominant transcription foci are not evident in these stringent RNAP mutants when grown in LB (Zhou & Jin, 1997; Cabrera & Jin, 2003). Results from biochemical studies, transcriptional profiling and cell biology analysis are consistent with the model underlying the redistribution of RNAP by ppGpp during the stringent response and carbon source limitation (Barker et al., 2001a; Cabrera & Jin, 2003; Liu et al., 2005; Durfee et al., 2008). The direction for the newly available RNAP engaging in genome-wide transcription is provided by ppGpp/DksA, transcription factors or the concentration of free RNAP per se, which is a topic beyond the scope of this review.
Two other models addressing the effect of ppGpp on RNAP have been proposed. One model (Baracchini & Bremer, 1988) proposes that RNAP is partitioned into two forms: one ppGpp-bound, which is unable to participate in initiation from rrn P1, but is able to for other non-rrn promoters, and the other ppGpp free, which is competent for initiation at all promoters. The other model (Jensen & Pedersen, 1990) proposes that RNAP is sequestered by ppGpp-mediated pausing during elongation, leading to reduced RNAP available for global gene expression during the stringent response. Both models were attractive at the time they were proposed and helped us understand the regulation of rRNA synthesis by ppGpp; however, each model mainly emphasizes either the effect of ppGpp on initiation or elongation. As discussed above, there are multiple effects of ppGpp on transcription that can be explained by the RNAP redistribution (allocation) model, which takes into account the effects of ppGpp on both initiation and elongation.
The RNAP redistribution (allocation) model could also explain and/or reconcile some results related to the regulation of rRNA synthesis. For example, a ribosome feedback model has been proposed to account for reduced rRNA synthesis by the presence of a plasmid-borne rrn operon in the cell (Jinks-Robertson et al., 1983); the effect of the extrachromosomal copies of rrnB on rRNA synthesis has been attributed to components involved in translation initiation machinery (Cole et al., 1987), an increased level of ppGpp (Baracchini, 1991) or about a 20% reduction of ATP and no change in ppGpp level (Schneider, 2003). Studies from cell biology of RNAP have shown that RNAP is exclusively colocalized with the nucleoid under normal conditions, whereas, in the presence of a plasmid-borne rrn operon RNAP is located both in the nucleoid and in the cytoplasmic space. Under this condition when RNAP is driven into the cytoplasmic space by the plasmid-borne rrn operon, the dominant transcription foci are diminished in the nucleoid (Cabrera & Jin, 2006). Thus, reduced RNAP in the nucleoid by the plasmid-borne rrn operon could account for the decreased rRNA synthesis in the chromosome (Voulgaris et al., 1999). Also, it has been reported that E. coli mutants, which deleted one to two rrn operons in the chromosome, have a growth rate comparable to the wild type (Condon et al., 1993, 1995a; Asai et al., 1999). The total RNA synthesis in these rrn deletion mutants is unchanged; however, the rate of rRNA synthesis for the remaining intact rrn operons is increased compared with the wild type, as demonstrated by electron micrographs (Condon et al., 1993). These results can be explained by the RNAP redistribution model as the same amount of RNAP in these mutants will be redistributed to the remaining intact rrn operons, leading to an increased frequency of RNAP occupying each of the remaining rrn operons without the need for a reduced ppGpp level in the cells.
Summary and future prospects
Regulation of rRNA synthesis in response to growth conditions involves multiple factors and affects the allocation of RNAP for global gene expression and the structure of the nucleoid in E. coli. The magic spot ppGpp, which acts in concert with DksA, is the major source of growth rate regulation and the stringent response in the bacterium. In the future, identifying the sites in RNAP that are involved in the regulation of rRNA synthesis, particularly defining the ppGpp-binding sites in RNAP, will shed light on the mechanisms of ppGpp in transcription. In addition, studying the dynamics of the dominant transcription foci and the putative nucleolus-like structures will be important to help our understanding of the link between rRNA synthesis, global gene expression, and chromosome structure in this model system.
Acknowledgments
We thank our colleagues for comments on the manuscript. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- Afflerbach H, Schroder O, Wagner R. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol Microbiol. 1998;28:641–653. doi: 10.1046/j.1365-2958.1998.00829.x. [DOI] [PubMed] [Google Scholar]
- Aiyar SE, Gourse RL, Ross W. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. P Natl Acad Sci USA. 1998;95:14652–14657. doi: 10.1073/pnas.95.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar SE, McLeod SM, Ross W, Hirvonen CA, Thomas MS, Johnson RC, Gourse RL. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J Mol Biol. 2002;316:501–516. doi: 10.1006/jmbi.2001.5390. [DOI] [PubMed] [Google Scholar]
- Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Patlan V, Sekine S, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- Asai T, Condon C, Voulgaris J, et al. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracchini E, Bremer H. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J Biol Chem. 1988;263:2597–2602. [PubMed] [Google Scholar]
- Baracchini E, Bremer H. Control of rRNA synthesis in Escherichia coli at increased rrn gene dosage. Role of guanosine tetraphosphate and ribosome feedback. J Biol Chem. 1991;266:11753–11760. [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001a;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001b;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Bartlett MS, Gaal T, Ross W, Gourse RL. Regulation of rRNA transcription is remarkably robust: FIS compensates for altered nucleoside triphosphate sensing by mutant RNA polymerases at Escherichia coli rrn P1 promoters. J Bacteriol. 2000;182:1969–1977. doi: 10.1128/jb.182.7.1969-1977.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- Blankschien MD, Lee JH, Grace ED, et al. Super DksAs: substitutions in DksA enhancing its effects on transcription initiation. EMBO J. 2009;28:1720–1731. doi: 10.1038/emboj.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bokal AJt, Ross W, Gourse RL. The transcriptional activator protein FIS: DNA interactions and cooperative interactions with RNA polymerase at the Escherichia coli rrnB P1 promoter. J Mol Biol. 1995;245:197–207. doi: 10.1006/jmbi.1994.0016. [DOI] [PubMed] [Google Scholar]
- Booker BM, Deng S, Higgins NP. DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2010;78:1348–1364. doi: 10.1111/j.1365-2958.2010.07394.x. [DOI] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Josaitis CA, Gourse RL, Goldfarb A. Two modes of transcription initiation in vitro at the rrnB P1 promoter of Escherichia coli. J Biol Chem. 1993;268:23477–23482. [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Vol. 2. ASM Press; Washington, DC: 1996. pp. 1553–1569. [Google Scholar]
- Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Brown L, Gentry D, Elliott T, Cashel M. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol. 2002;184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr T, Mitchell J, Kolb A, Minchin S, Busby S. DNA sequence elements located immediately upstream of the − 10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 2000;28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JE, Jin DJ. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- Cabrera JE, Jin DJ. Active transcription of rRNA operons is a driving force for the distribution of RNA polymerase in bacteria: effect of extrachromosomal copies of rrnB on the in vivo localization of RNA polymerase. J Bacteriol. 2006;188:4007–4014. doi: 10.1128/JB.01893-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Vol. 1. ASM Press; Washington, DC: 1996. pp. 1458–1496. [Google Scholar]
- Cayley S, Record MT., Jr Roles of cytoplasmic osmolytes, water, and crowding in the response of Escherichia coli to osmotic stress: biophysical basis of osmoprotection by glycine betaine. Biochemistry. 2003;42:12596–12609. doi: 10.1021/bi0347297. [DOI] [PubMed] [Google Scholar]
- Cayley S, Lewis BA, Guttman HJ, Record MT., Jr Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein–DNA interactions in vivo. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- Cole JR, Olsson CL, Hershey JW, Grunberg-Manago M, Nomura M. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J Mol Biol. 1987;198:383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- Condon C, Philips J, Fu ZY, Squires C, Squires CL. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 1992;11:4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, French S, Squires C, Squires CL. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 1993;12:4305–4315. doi: 10.1002/j.1460-2075.1993.tb06115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Liveris D, Squires C, Schwartz I, Squires CL. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J Bacteriol. 1995a;177:4152–4156. doi: 10.1128/jb.177.14.4152-4156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995b;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Wurm R, Wagner R, Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J Biol Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- de Boer HA, Gilbert SF, Nomura M. DNA sequences of promoter regions for rRNA operons rrnE and rrnA in E. coli. Cell. 1979;17:201–209. doi: 10.1016/0092-8674(79)90308-8. [DOI] [PubMed] [Google Scholar]
- Dennis PP, Ehrenberg M, Bremer H. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol Mol Biol R. 2004;68:639–668. doi: 10.1128/MMBR.68.4.639-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Dinnbier U, Limpinsel E, Schmid R, Bakker EP. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch Microbiol. 1988;150:348–357. doi: 10.1007/BF00408306. [DOI] [PubMed] [Google Scholar]
- Donahue JP, Turnbough CL., Jr Characterization of transcriptional initiation from promoters P1 and P2 of the pyrBI operon of Escherichia coli K12. J Biol Chem. 1990;265:19091–19099. [PubMed] [Google Scholar]
- Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. P Natl Acad Sci USA. 1973;70:3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood M, Nomura M. Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J Bacteriol. 1982;149:458–468. doi: 10.1128/jb.149.2.458-468.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Kurland CG. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J. 1990;9:4359–4366. doi: 10.1002/j.1460-2075.1990.tb07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Naslund AK, Kurland CG. Growth-rate-dependent accumulation of twelve tRNA species in Escherichia coli. J Mol Biol. 1993;230:483–491. doi: 10.1006/jmbi.1993.1165. [DOI] [PubMed] [Google Scholar]
- Falconi M, Gualtieri MT, La Teana A, Losso MA, Pon CL. Proteins from the prokaryotic nucleoid: primary and quaternary structure of the 15-kD Escherichia coli DNA binding protein H-NS. Mol Microbiol. 1988;2:323–329. doi: 10.1111/j.1365-2958.1988.tb00035.x. [DOI] [PubMed] [Google Scholar]
- Finkel SE, Johnson RC. The Fis protein: it’s not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- French SL, Miller OL., Jr Transcription mapping of the Escherichia coli chromosome by electron microscopy. J Bacteriol. 1989;171:4207–4216. doi: 10.1128/jb.171.8.4207-4216.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Gourse RL. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. P Natl Acad Sci USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- Gafny R, Cohen S, Nachaliel N, Glaser G. Isolated P2 rRNA promoters of Escherichia coli are strong promoters that are subject to stringent control. J Mol Biol. 1994;243:152–156. doi: 10.1006/jmbi.1994.1641. [DOI] [PubMed] [Google Scholar]
- Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J Mol Biol. 1977;115:335–354. doi: 10.1016/0022-2836(77)90158-9. [DOI] [PubMed] [Google Scholar]
- Gilbert SF, de Boer HA, Nomura M. Identification of initiation sites for the in vitro transcription of rRNA operons rrnE and rrnA in E. coli. Cell. 1979;17:211–224. doi: 10.1016/0092-8674(79)90309-x. [DOI] [PubMed] [Google Scholar]
- Glaser G, Cashel M. In vitro transcripts from the rrn B ribosomal RNA cistron originate from two tandem promoters. Cell. 1979;16:111–121. doi: 10.1016/0092-8674(79)90192-2. [DOI] [PubMed] [Google Scholar]
- Glaser G, Sarmientos P, Cashel M. Functional interrelationship between two tandem E. coli ribosomal RNA promoters. Nature. 1983;302:74–76. doi: 10.1038/302074a0. [DOI] [PubMed] [Google Scholar]
- Gosink KK, Ross W, Leirmo S, Osuna R, Finkel SE, Johnson RC, Gourse RL. DNA binding and bending are necessary but not sufficient for Fis-dependent activation of rrnB P1. J Bacteriol. 1993;175:1580–1589. doi: 10.1128/jb.175.6.1580-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink KK, Gaal T, Bokal AJt, Gourse RL. A positive control mutant of the transcription activator protein FIS. J Bacteriol. 1996;178:5182–5187. doi: 10.1128/jb.178.17.5182-5187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, de Boer HA, Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- Gralla JD. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol Microbiol. 2005;55:973–977. doi: 10.1111/j.1365-2958.2004.04455.x. [DOI] [PubMed] [Google Scholar]
- Gralla JD, Vargas DR. Potassium glutamate as a transcriptional inhibitor during bacterial osmoregulation. EMBO J. 2006;25:1515–1521. doi: 10.1038/sj.emboj.7601041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt J, Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation–termination cycle of transcription. Cell. 1981;24:421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- Greive SJ, Lins AF, von Hippel PH. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J Biol Chem. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. P Natl Acad Sci USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AD, Taylor WE, Burton ZF, Burgess RR, Gross CA. Stringent response in Escherichia coli induces expression of heat shock proteins. J Mol Biol. 1985;186:357–365. doi: 10.1016/0022-2836(85)90110-x. [DOI] [PubMed] [Google Scholar]
- Hamkalo BA, Miller OL., Jr Electronmicroscopy of genetic activity. Annu Rev Biochem. 1973a;42:379–396. doi: 10.1146/annurev.bi.42.070173.002115. [DOI] [PubMed] [Google Scholar]
- Hamkalo BA, Miller OL., Jr Visualization of genetic transcription. Basic Life Sci. 1973b;1:63–74. doi: 10.1007/978-1-4684-0877-5_6. [DOI] [PubMed] [Google Scholar]
- Hansen AM, Qiu Y, Yeh N, Blattner FR, Durfee T, Jin DJ. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol Microbiol. 2005;56:719–734. doi: 10.1111/j.1365-2958.2005.04567.x. [DOI] [PubMed] [Google Scholar]
- Hardy CD, Cozzarelli NR. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol. 2005;57:1636–1652. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Hernandez VJ, Bremer H. Escherichia coli ppGpp synthetase II activity requires spoT. J Biol Chem. 1991;266:5991–5999. [PubMed] [Google Scholar]
- Hernandez VJ, Bremer H. Characterization of RNA and DNA synthesis in Escherichia coli strains devoid of ppGpp. J Biol Chem. 1993;268:10851–10862. [PubMed] [Google Scholar]
- Hofmann S, Miller OL., Jr Visualization of ribosomal ribonucleic acid synthesis in a ribonuclease III-Deficient strain of Escherichia coli. J Bacteriol. 1977;132:718–722. doi: 10.1128/jb.132.2.718-722.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- Jensen KF, Pedersen S. Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol Rev. 1990;54:89–100. doi: 10.1128/mr.54.2.89-100.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Cabrera JE. Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J Struct Biol. 2006;156:284–291. doi: 10.1016/j.jsb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S, Gourse RL, Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983;33:865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Ball CA, Pfeffer D, Simon MI. Isolation of the gene encoding the Hin recombinational enhancer binding protein. P Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josaitis CA, Gaal T, Gourse RL. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. P Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YH, Lee Y. Escherichia coli rnpB promoter mutants altered in stringent response. Biochem Bioph Res Co. 1997;230:582–586. doi: 10.1006/bbrc.1996.6005. [DOI] [PubMed] [Google Scholar]
- Kajitani M, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase. Differential stringent control of the multiple promoters from ribosomal RNA and protein operons. J Biol Chem. 1984;259:1951–1957. [PubMed] [Google Scholar]
- Kang PJ, Craig EA. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Vol. 1. ASM Press; Washington, DC: 1996. pp. 1417–1431. [Google Scholar]
- Kingston RE, Chamberlin MJ. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell. 1981;27:523–531. doi: 10.1016/0092-8674(81)90394-9. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Gutell RR, Taylor AR, Chamberlin MJ. Transcriptional mapping of plasmid pKK3535. Quantitation of the effect of guanosine tetraphosphate on binding to the rrnB promoters and a lambda promoter with sequence homologies in the CII binding region. J Mol Biol. 1981a;146:433–449. doi: 10.1016/0022-2836(81)90041-3. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Nierman WC, Chamberlin MJ. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem. 1981b;256:2787–2797. [PubMed] [Google Scholar]
- Kjeldgaard NO, Maaloe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:607–616. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microb. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn M, Wagner R. Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. J Biol Chem. 1996;271:23884–23894. doi: 10.1074/jbc.271.39.23884. [DOI] [PubMed] [Google Scholar]
- Krohn M, Pardon B, Wagner R. Effects of template topology on RNA polymerase pausing during in vitro transcription of the Escherichia coli rrnB leader region. Mol Microbiol. 1992;6:581–589. doi: 10.1111/j.1365-2958.1992.tb01504.x. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Tanaka S, Ikeda T, Kato J, Takiguchi N, Ohtake H. Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. P Natl Acad Sci USA. 1999;96:14264–14269. doi: 10.1073/pnas.96.25.14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Nomura K, Ohtomo R, et al. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Travers AA. Stringent control of bacterial transcription. Cell. 1985;41:6–8. doi: 10.1016/0092-8674(85)90050-9. [DOI] [PubMed] [Google Scholar]
- Landgraf JR, Wu J, Calvo JM. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Alberts BM. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science. 1995;267:1131–1137. doi: 10.1126/science.7855590. [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. P Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ, Blattner FR. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J Biol Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Initiation of Escherichia coli ribosomal RNA synthesis in vivo. P Natl Acad Sci USA. 1979;76:5480–5484. doi: 10.1073/pnas.76.11.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaloe O, Kjeldgaard NO. Control of Macromolecular Synthesis: A Study of DNA, RNA, and Protein Synthesis in Bacteria. W. A. Benjamin, Inc; New York, NY: 1966. [Google Scholar]
- McClure WR. Rate-limiting steps in RNA chain initiation. P Natl Acad Sci USA. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL, Jr, Hamkalo BA, Thomas CA., Jr Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Murray KD, Bremer H. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J Mol Biol. 1996;259:41–57. doi: 10.1006/jmbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Fraenkel DG. Metabolic regulation of RNA synthesis in bacteria. Cold Spring Harb Sym. 1961;26:63–74. doi: 10.1101/sqb.1961.026.01.012. [DOI] [PubMed] [Google Scholar]
- Newman EB, D’Ari R, Lin RT. The leucine-Lrp regulon in E. coli: a global response in search of a raison d’etre. Cell. 1992;68:617–619. doi: 10.1016/0092-8674(92)90135-y. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Vanet A, Vijgenboom E, Bosch L. The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J. 1990;9:727–734. doi: 10.1002/j.1460-2075.1990.tb08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Verbeek H, Hoffmann U, Haupt M, Bosch L. Inactivation of the fis gene leads to reduced growth rate. FEMS Microbiol Lett. 1992a;99:85–88. doi: 10.1016/0378-1097(92)90292-v. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Verbeek H, Vijgenboom E, van Drunen C, Vanet A, Bosch L. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J Bacteriol. 1992b;174:921–929. doi: 10.1128/jb.174.3.921-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Yates JL, Dean D, Post LE. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein MRNA. P Natl Acad Sci USA. 1980;77:7084–7088. doi: 10.1073/pnas.77.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Norris V, den Blaauwen T, Cabin-Flaman A, et al. Functional taxonomy of bacterial hyperstructures. Microbiol Mol Biol R. 2007;71:230–253. doi: 10.1128/MMBR.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- Ohlsen KL, Gralla JD. Melting during steady-state transcription of the rrnB P1 promoter in vivo and in vitro. J Bacteriol. 1992a;174:6071–6075. doi: 10.1128/jb.174.19.6071-6075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsen KL, Gralla JD. DNA melting within stable closed complexes at the Escherichia coli rrnB P1 promoter. J Biol Chem. 1992b;267:19813–19818. [PubMed] [Google Scholar]
- Ohlsen KL, Gralla JD. Interrelated effects of DNA supercoiling, ppGpp, and low salt on melting within the Escherichia coli ribosomal RNA rrnB P1 promoter. Mol Microbiol. 1992c;6:2243–2251. doi: 10.1111/j.1365-2958.1992.tb01400.x. [DOI] [PubMed] [Google Scholar]
- Oostra BA, Gruber M. Involvement of DNA gyrase in the transcription of ribosomal RNA. Nucleic Acids Res. 1980;8:4235–4246. doi: 10.1093/nar/8.18.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel ML, Aeling KA, Holmes WM, Johnson RC, Benham CJ, Hatfield GW. Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism. Mol Microbiol. 2004;53:665–674. doi: 10.1111/j.1365-2958.2004.04147.x. [DOI] [PubMed] [Google Scholar]
- Owens JR, Woody AY, Haley BE. Characterization of the guanosine-′diphosphate-5′-diphosphate binding site on E. coli RNA polymerase using a photoprobe, 8-azidoguanosine-3′-5′-bisphosphate. Biochem Bioph Res Co. 1987;142:964–971. doi: 10.1016/0006-291x(87)91508-7. [DOI] [PubMed] [Google Scholar]
- Park JW, Jung Y, Lee SJ, Jin DJ, Lee Y. Alteration of stringent response of the Escherichia coli rnpB promoter by mutations in the -35 region. Biochem Bioph Res Co. 2002;290:1183–1187. doi: 10.1006/bbrc.2001.6331. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004a;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004b;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Pemberton IK, Muskhelishvili G, Travers AA, Buckle M. The G+C-rich discriminator region of the tyrT promoter antagonises the formation of stable preinitiation complexes. J Mol Biol. 2000;299:859–864. doi: 10.1006/jmbi.2000.3780. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel–structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Petersen C, Moller LB. Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J Biol Chem. 2000;275:3931–3935. doi: 10.1074/jbc.275.6.3931. [DOI] [PubMed] [Google Scholar]
- Piper SE, Mitchell JE, Lee DJ, Busby SJ. A global view of Escherichia coli Rsd protein and its interactions. Mol Biosyst. 2009;5:1943–1947. doi: 10.1039/B904955j. [DOI] [PubMed] [Google Scholar]
- Platko JV, Willins DA, Calvo JM. The ilvIH operon of Escherichia coli is positively regulated. J Bacteriol. 1990;172:4563–4570. doi: 10.1128/jb.172.8.4563-4570.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010a;9:2537–2543. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010b;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Murphy H, Philippe N, Cashel M. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol. 2010;13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pul U, Wurm R, Lux B, Meltzer M, Menzel A, Wagner R. LRP and H-NS – cooperative partners for transcription regulation at Escherichia coli rRNA promoters. Mol Microbiol. 2005;58:864–876. doi: 10.1111/j.1365-2958.2005.04873.x. [DOI] [PubMed] [Google Scholar]
- Quan S, Zhang N, French S, Squires CL. Transcriptional polarity in rRNA operons of Escherichia coli nusA and nusB mutant strains. J Bacteriol. 2005;187:1632–1638. doi: 10.1128/JB.187.5.1632-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record MT, Jr, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Reddy PS, Raghavan A, Chatterji D. Evidence for a ppGpp-binding site on Escherichia coli RNA polymerase: proximity relationship with the rifampicin-binding domain. Mol Microbiol. 1995;15:255–265. doi: 10.1111/j.1365-2958.1995.tb02240.x. [DOI] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- Ross W, Thompson JF, Newlands JT, Gourse RL. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, et al. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Ross W, Aiyar SE, Salomon J, Gourse RL. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmientos P, Cashel M. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: differential control of dual promoters. P Natl Acad Sci USA. 1983;80:7010–7013. doi: 10.1073/pnas.80.22.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmientos P, Sylvester JE, Contente S, Cashel M. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell. 1983;32:1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- Sarubbi E, Rudd KE, Cashel M. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Schaferkordt J, Wagner R. Effects of base change mutations within an Escherichia coli ribosomal RNA leader region on rRNA maturation and ribosome formation. Nucleic Acids Res. 2001;29:3394–3403. doi: 10.1093/nar/29.16.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Gourse RL. Changes in Escherichia coli rRNA promoter activity correlate with changes in initiating nucleoside triphosphate and guanosine 5′ diphosphate 3′-diphosphate concentrations after induction of feedback control of ribosome synthesis. J Bacteriol. 2003;185:6185–6191. doi: 10.1128/JB.185.20.6185-6191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Gourse RL. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J Biol Chem. 2004;279:8262–8268. doi: 10.1074/jbc.M311996200. [DOI] [PubMed] [Google Scholar]
- Schneider DA, Gaal T, Gourse RL. NTP-sensing by rRNA promoters in Escherichia coli is direct. P Natl Acad Sci USA. 2002;99:8602–8607. doi: 10.1073/pnas.132285199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder O, Wagner R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J Mol Biol. 2000;298:737–748. doi: 10.1006/jmbi.2000.3708. [DOI] [PubMed] [Google Scholar]
- Spira B, Silberstein N, Yagil E. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WE, Straus DB, Grossman AD, Burton ZF, Gross CA, Burgess RR. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell. 1984;38:371–381. doi: 10.1016/0092-8674(84)90492-6. [DOI] [PubMed] [Google Scholar]
- Tehranchi AK, Blankschien MD, Zhang Y, et al. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Behrens SE, Wagner R. Functional importance of the Escherichia coli ribosomal RNA leader box A sequence for post-transcriptional events. Mol Microbiol. 1990;4:1667–1678. doi: 10.1111/j.1365-2958.1990.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Tippner D, Afflerbach H, Bradaczek C, Wagner R. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol Microbiol. 1994;11:589–604. doi: 10.1111/j.1365-2958.1994.tb00339.x. [DOI] [PubMed] [Google Scholar]
- Torres M, Condon C, Balada JM, Squires C, Squires CL. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 2001;20:3811–3820. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov II, Shulgina I, Hernandez VJ. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta′-subunit. J Biol Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- Travers AA. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980a;141:973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers AA. A tRNATyr promoter with an altered in vitro response to ppgpp. J Mol Biol. 1980b;141:91–97. doi: 10.1016/s0022-2836(80)80030-1. [DOI] [PubMed] [Google Scholar]
- Travers AA, Lamond AI, Mace HA, Berman ML. RNA polymerase interactions with the upstream region of the E. coli tyrT promoter. Cell. 1983;35:265–273. doi: 10.1016/0092-8674(83)90229-5. [DOI] [PubMed] [Google Scholar]
- Travers AA, Lamond AI, Weeks JR. Alteration of the growth-rate-dependent regulation of Escherichia coli tyrT expression by promoter mutations. J Mol Biol. 1986;189:251–255. doi: 10.1016/0022-2836(86)90397-9. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen HT, Stark SE, Conway T. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 2011;79:830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilette D, Ehrlich SD, Michel B. Transcription-induced deletions in plasmid vectors: M13 DNA replication as a source of instability. Mol Gen Genet. 1996;252:398–403. doi: 10.1007/BF02173004. [DOI] [PubMed] [Google Scholar]
- Vinella D, Albrecht C, Cashel M, D’Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Vogel U, Jensen KF. Effects of guanosine 3′,5′-bisdiphosphate (ppGpp) on rate of transcription elongation in isoleucine-starved Escherichia coli. J Biol Chem. 1994;269:16236–16241. [PubMed] [Google Scholar]
- von Hippel PH, Bear DG, Morgan WD, McSwiggen JA. Protein–nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]
- Voulgaris J, French S, Gourse RL, Squires C, Squires CL. Increased rrn gene dosage causes intermittent transcription of rRNA in Escherichia coli. J Bacteriol. 1999;181:4170–4175. doi: 10.1128/jb.181.14.4170-4175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Ross W, Gourse RL. Still looking for the Magic Spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. Regulation of ribosomal RNA synthesis in E. coli: effects of the global regulator guanosine tetraphosphate (ppGpp) J Mol Microb Biotech. 2002;4:331–340. [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Yang HL, Heller K, Gellert M, Zubay G. Differential sensitivity of gene expression in vitro to inhibitors of DNA gyrase. P Natl Acad Sci USA. 1979;76:3304–3308. doi: 10.1073/pnas.76.7.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZX, Zhou YN, Yang Y, Jin DJ. Polyphosphate binds to the principal sigma factor of RNA polymerase during starvation response in Helicobacter pylori. Mol Microbiol. 2010;77:618–627. doi: 10.1111/j.1365-2958.2010.07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Wang MD, Svoboda K, Landick R, Block SM, Gelles J. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Ishijima SA, Clowney L, et al. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol Rev. 2006;30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- Zacharias M, Goringer HU, Wagner R. Influence of the GCGC discriminator motif introduced into the ribosomal RNA P2- and tac promoter on growth-rate control and stringent sensitivity. EMBO J. 1989;8:3357–3363. doi: 10.1002/j.1460-2075.1989.tb08498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M, Theissen G, Bradaczek C, Wagner R. Analysis of sequence elements important for the synthesis and control of ribosomal RNA in E coli. Biochimie. 1991;73:699–712. doi: 10.1016/0300-9084(91)90050-b. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem. 1995;270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bremer H. Effects of Fis on ribosome synthesis and activity and on rRNA promoter activities in Escherichia coli. J Mol Biol. 1996;259:27–40. doi: 10.1006/jmbi.1996.0299. [DOI] [PubMed] [Google Scholar]
- Zhi H, Wang X, Cabrera JE, Johnson RC, Jin DJ. Fis stabilizes the interaction between RNA polymerase and the ribosomal promoter rrnB P1, leading to transcriptional activation. J Biol Chem. 2003;278:47340–47349. doi: 10.1074/jbc.M305430200. [DOI] [PubMed] [Google Scholar]
- Zhou YN, Jin DJ. RNA polymerase beta mutations have reduced sigma70 synthesis leading to a hyper-temperature-sensitive phenotype of a sigma70 mutant. J Bacteriol. 1997;179:4292–4298. doi: 10.1128/jb.179.13.4292-4298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like ‘stringent’ RNA polymerases in Escherichia coli. P Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]