Abstract
The development and coordination of complex tissues in eukaryotes requires precise spatial control of fate-specifying genes. Although investigations of such control have traditionally focused on mechanisms of transcriptional activation, transcriptional repression has emerged as being equally important in the establishment of gene expression territories. In the angiosperm flower, specification of lateral organ fate relies on the spatial regulation of the ABC floral organ identity genes. Our understanding of how the boundaries of these expression domains are controlled is not complete. Here, we report that the A-class organ identity gene APETALA2 (AP2), which is known to repress the C-class gene AGAMOUS, also regulates the expression borders of the B-class genes APETALA3 and PISTILLATA, and the E-class gene SEPALLATA3. We show that AP2 represses its target genes by physically recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. These results demonstrate that AP2 plays a broad role in flower development by controlling the expression domains of numerous floral organ identity genes.
Keywords: APETALA2, Floral development, Transcriptional repression, Arabidopsis
INTRODUCTION
In multicellular organisms, complex functional structures often require precise spatial arrangements of individual tissues. How the identities of these different tissues are established is a central question in developmental biology, and usually involves domain-specific expression patterns of fate-specifying genes. Although early studies on eukaryotic gene expression focused on mechanisms of transcriptional activation, it has become clear that transcriptional repression is equally important in the regulation of cell identity (Gray and Levine, 1996). Gene expression boundaries underlying many patterning processes rely on DNA-binding transcription factors that negatively regulate target genes by interacting with co-repressors. These co-repressors recruit proteins that confer active transcriptional repression, including chromatin-modifying factors (Courey and Jia, 2001). Co-repressors of the Groucho (Gro)/Transducin-like enhancer of split (TLE) family are well characterized in animal systems and have been implicated in a myriad of developmental events, including Drosophila dorsoventral patterning and vertebrate anteroposterior patterning (Buscarlet and Stifani, 2007).
In plants, the flower is a useful model for investigating transcriptional mechanisms of fate specification. It comprises four types of lateral organs (sepals, petals, stamens and carpels) arranged in concentric whorls in eudicots (numbered 1 to 4 from the peripheral sepals to the central carpels). Floral organ identity genes specify the fate of each whorl, and their misregulation can lead to homeotic conversions of one organ type to another. Such conversions are produced by a number of classic mutants in model angiosperm species such as Arabidopsis and Antirrhinum, the study of which led to the establishment of the ABC model of flower development over two decades ago (Bowman et al., 1991; Coen and Meyerowitz, 1991). This model posits that three functions, termed A, B and C, act alone or in combination to specify the identities of the floral lateral organs. A-function specifies sepal fate, A and B together confer petal identity, B and C jointly specify stamen fate, and C confers carpel identity in the central whorl. Additionally, A and C were hypothesized to operate antagonistically by restricting each other's functional domains. In Arabidopsis, these activities were originally attributed to four genes based on genetic analyses: APETALA2 (AP2) provided A-function; APETALA3 (AP3) and PISTILLATA (PI) conferred B-function; and AGAMOUS (AG) provided C-function. APETALA1 (AP1) was later identified as another A-function gene (Bowman et al., 1993), and more recently, an E-function has been defined by the SEPALLATA (SEP)1-4 family (Pelaz et al., 2000; Ditta et al., 2004). Like the ABC factors (with the exception of AP2), SEPs are MADS-domain proteins, and they facilitate the formation of ABCE transcriptional complexes that specify floral organ fate (Honma and Goto, 2001). Quadruple loss-of-function sep mutants have flowers consisting solely of leaf-like organs (Ditta et al., 2004), and ectopic expression of SEP and ABC genes is sufficient to convert leaves into floral organs (Honma and Goto, 2001; Pelaz et al., 2001). Unlike the ABC genes, expression of which is generally confined to two neighboring whorls, most SEP genes are expressed in all floral organ primordia. SEP3 is an exception, as its expression is mainly restricted to the inner three whorls (Mandel and Yanofsky, 1998).
Although ABCE genes are initially activated by floral meristem identity genes (Weigel et al., 1992; Bowman et al., 1993; Weigel and Meyerowitz, 1993; Lee et al., 1997), their functional domains are refined by cadastral regulators that establish the boundaries of their expression. AP2 restricts AG expression to whorls 3 and 4, and in strong ap2 mutants, AG spreads throughout the flower resulting in carpel-like whorl 1 organs and loss of whorl 2 organs (Bowman et al., 1991; Drews et al., 1991). Other factors have been shown to contribute to the spatial restriction of AG (Byzova et al., 1999; Bao et al., 2004; Takeda et al., 2004; Krizek et al., 2006), including the transcriptional co-repressor LEUNIG (LUG) and its interacting partner SEUSS (SEU) (Liu and Meyerowitz, 1995; Conner and Liu, 2000; Franks et al., 2002). SUPERMAN (SUP), a C2H2-type zinc finger protein, is necessary for maintaining the inner boundary of B-class gene expression between whorls 3 and 4, as AP3 expression expands into the central region of sup flowers, producing ectopic stamens at the expense of carpels (Schultz et al., 1991; Bowman et al., 1992; Sakai et al., 1995). A small RNA (miR172) that regulates AP2 also aids in the establishment of this border, as overexpression of an miRNA-resistant AP2 transgene results in expansion of AP3 and PI towards the center of the floral meristem (Park et al., 2002; Aukerman and Sakai, 2003; Chen, 2004; Zhao et al., 2007). Despite these advances, many questions regarding the spatial control of the ABCE genes remain. For instance, the mechanisms underlying many of these regulatory relationships, including the inhibition of AG by AP2, have yet to be fully characterized. Additionally, it is unknown whether cadastral regulators directly restrict the peripheral expansion of B- and E-class (SEP3) genes in the outermost whorl.
Here, we report one molecular mechanism underlying the repression of AG by AP2, a genetic relationship that contributed to one of the original principles of the ABC model (antagonism of C-by A-function). Specifically, we show that AP2 directly represses AG by recruiting a transcriptional co-repressor and a histone deacetylase. This AP2 repressor complex also targets B- and E-class genes to restrict their expression in whorl 1, elucidating new regulatory roles for AP2 not previously defined in the ABC model. Collectively, these results show that AP2 plays a broad role in flower development, spatially controlling the expression domains of multiple floral organ identity genes.
MATERIALS AND METHODS
Plant material
Plants were grown on soil in greenhouse conditions under a 16-hour light/8-hour dark cycle. The Landsberg erecta (Ler) ecotype of Arabidopsis was used as wild type. Genetic analyses utilized the previously described mutants tpl-1 (Long et al., 2002), tpl-2, tpr1, tpr3, tpr4, hda19-1 (Long et al., 2006), ap2-2, pi-1, ag-1 (Bowman et al., 1989), ap3-3 (Jack et al., 1992) and sep3-2 (Pelaz et al., 2000). tpr2 is a T-DNA insertion allele (SALK_112730) (Alonso et al., 2003). Owing to the unavailability of Ler sep3 alleles, tpl-1 was introgressed into the Col-0 ecotype and then crossed to the sep3-2 mutant for genetic analyses. To genotype plants for tpl-1, hda19-1, ap2-2, ap3-3, pi-1, ag-1 and sep3-2 mutant alleles, DNA extractions were used as template in PCR with primers given in supplementary material Table S1.
RNA in situ hybridization
RNA in situ hybridizations with digoxigenin-labeled riboprobes were performed as described at http://pbio.salk.edu/pbiol/in_situ_protocol.html. Probes used for detecting expression of AP3 (Jack et al., 1992), PI (Goto and Meyerowitz, 1994), AG (Yanofsky et al., 1990) and HDA19 (Long et al., 2006) have been detailed previously. The entire coding region of TPL was used as probe. Three different SEP3 probes were used and all produced similar expression patterns. These included the full-length coding sequence and two probes previously described (Mandel and Yanofsky, 1998; Liu et al., 2009). Stages attributed to flowers, as described in figure legends, are based on standard convention (Smyth et al., 1990).
Protein interaction analyses
Yeast two-hybrid (Y2H) assays were performed with yeast strain MaV203 using Invitrogen ProQuest expression vectors and product protocols. Semi-in vivo pull-down experiments with TPL and TPL N176H GST fusions, including expression and purification of recombinant proteins, were carried out as previously reported (Szemenyei et al., 2008) with minor modifications. Specifically, unopened floral buds from stages 1 to 12 (Smyth et al., 1990), along with the inflorescence meristem, were harvested from stably transformed Arabidopsis lines and processed to produce plant lysates. TPL Y2H and GST fusion constructs have been described (Szemenyei et al., 2008). PCR-generated AP2 cDNAs were cloned in-frame into SalI-NotI sites of pDBLeu and pEXP-AD502 Y2H vectors (Invitrogen) or into XbaI-SalI sites of GST expression vector pGEX-KG. Conserved leucines within the EAR motif of AP2 were converted to alanines (CTCGATTTGAGCTTG to gcCGATgcGAGCgcG) by site-directed mutagenesis. Bimolecular fluorescence complementation (BiFC) experiments were performed in a tobacco transient expression system as previously detailed (Szemenyei et al., 2008). A full-length HDA19 genomic clone (XhoI-KpnI) was used to generate BiFC expression constructs.
Chromatin immunoprecipitation (ChIP) and western blots
ChIP experiments were performed as previously described (Bowler et al., 2004; Smith and Long, 2010) with the following modifications. Starting material consisted of unopened floral buds from stages 1 to 12 (Smyth et al., 1990), along with the inflorescence meristem. Protein G Dynabeads (Invitrogen) were used with anti-HA (Covance HA.11), anti-GFP (Abcam ab290), or anti-H4K16Ac (Active Motif 39167) antibodies in immunoprecipitations. Tissues expressing epitope-tagged HDA19 were pre-treated with 1.5 mM ethylene glycol bis(succinimidylsuccinate) (EGS) (Thermo Scientific) for 1 hour under vacuum. Pre-treatment of non-transgenic wild-type (Ler) tissue with EGS did not significantly affect ChIP enrichment values relative to non-EGS-treated control tissue. Therefore, EGS-treated Ler control values are not presented in Fig. 2I, Fig. 3O, Fig. 5L and supplementary material Fig. S4. Anti-H3K9Ac (Abcam ab4441), anti-H4K5Ac (AbD Serotec AHP962) and anti-H3 (Abcam ab1791) antibodies were used for western blotting. Western signal intensities were quantified using ImageJ software (National Institutes of Health, http://imagej.nih.gov/ij).
Fig. 2.
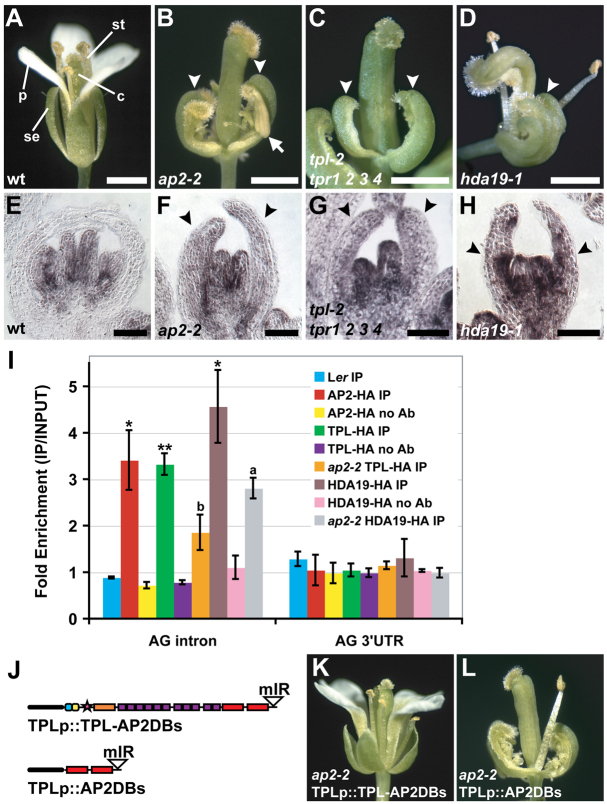
AP2, TPL and HDA19 cooperatively repress the C-class gene AG in outer whorl floral organs. (A) Wild-type Arabidopsis flower composed of four sepals (se), four petals (p), six stamens (st) and two fused carpels (c). (B-D) Mutant flowers exhibiting carpel-like whorl 1 organs (arrowheads) with stigmatic tissue at their margins. Arrow in B designates stamen-like identity. (E-H) AG in situ hybridizations on stage 7-8 flowers. (E) Wild-type flower with expression restricted to stamen and carpel primordia. (F-H) Mutants expressing AG throughout the flower (arrowheads). (I) Anti-HA ChIP showing specific binding of AP2, TPL and HDA19 to the second AG intron. Control ChIPs were performed on non-transgenic wild-type tissue (Ler) or without antibody (no Ab). Data were normalized relative to input and ACT2 abundance. Data are represented as mean ± s.e.m. of at least two biological replicates. Student's t-test was used to determine the significance of target enrichment relative to Ler IP (*P≤0.05, **P≤0.005) and the significance of decreased binding in ap2-2 (a, P≤0.08; b, P≤0.05). (J) Transgenes used in K,L. Domains are described in Fig. 1A. The TPL promoter is depicted as a black oval. (K) TPLp::TPL-AP2DBs rescues ap2-2 floral defects. Compare with A,B. (L) TPLp::AP2DBs is insufficient to rescue ap2-2. Scale bars: 1 mm in A-D; 50 μm in E-H.
Fig. 3.
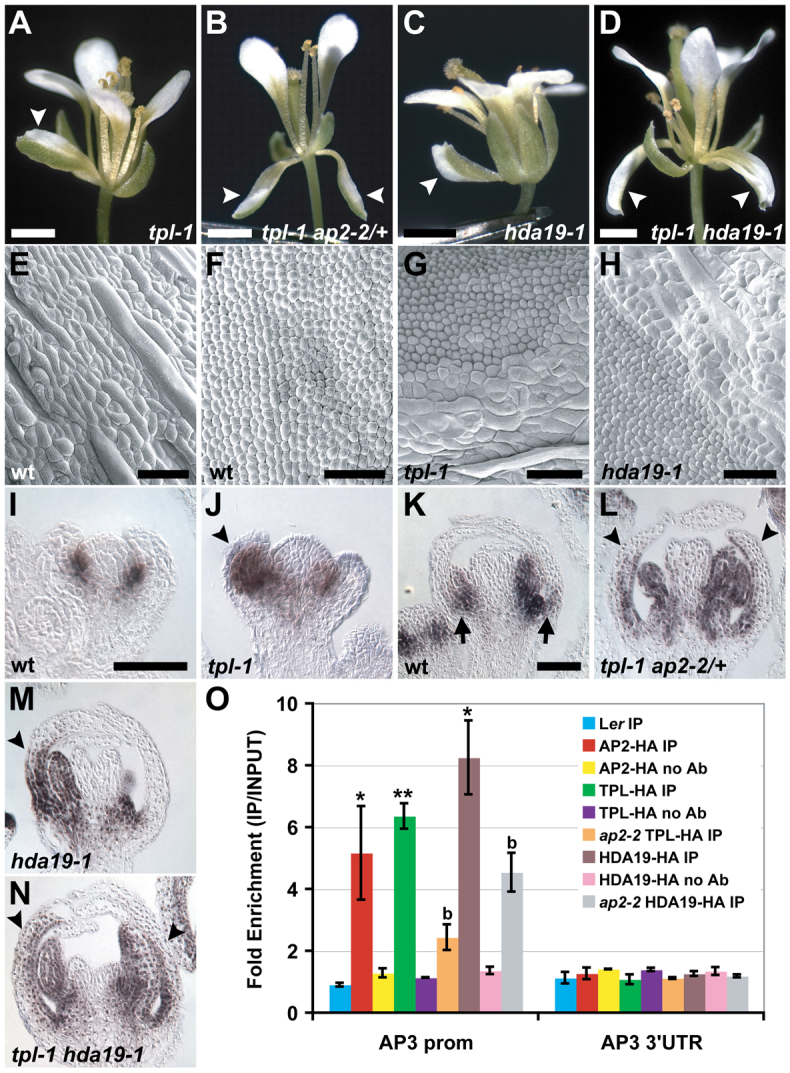
AP2, TPL and HDA19 repress the B-class gene AP3 in whorl 1 organs. (A-D) Mutant Arabidopsis flowers with sepal-to-petal homeotic conversions (arrowheads). Compare with wild type in Fig. 2A. (E-H) Scanning electron micrographs of abaxial surfaces of floral organs. (E) Elongated epidermal cells of wild-type sepals are variable in size. (F) Slightly conical cells of wild-type petals are relatively uniform in size. (G) tpl-1 and (H) hda19-1 mosaic whorl 1 organs with a sharp boundary between sepal and petal identity. (I-N) AP3 in situ hybridizations on stage 4 (I,J) and stage 7-8 (K-N) flowers. (I) AP3 is not expressed in young wild-type sepal primordia. (J) Ectopic AP3 expression in a young tpl-1 whorl 1 organ (arrowhead). (K) AP3 is detectable in later-stage wild-type sepals, but only at the organ base (arrows). (L-N) AP3 expression domains are expanded in mutant whorl 1 organs (arrowheads). (O) Anti-HA ChIP showing AP2, TPL and HDA19 specifically bind the AP3 promoter. Control ChIPs were performed on non-transgenic wild-type tissue (Ler) or without antibody (no Ab). Data were normalized relative to input and ACT2 abundance. Data are represented as mean ± s.e.m. of at least two biological replicates. Student's t-test was used to determine the significance of target enrichment relative to Ler IP (*P≤0.05, **P≤0.005) and the significance of decreased binding in ap2-2 (b, P≤0.05). Scale bars: 1 mm in A-D; 100 μm in E-H; 50 μm in I-N.
Fig. 5.
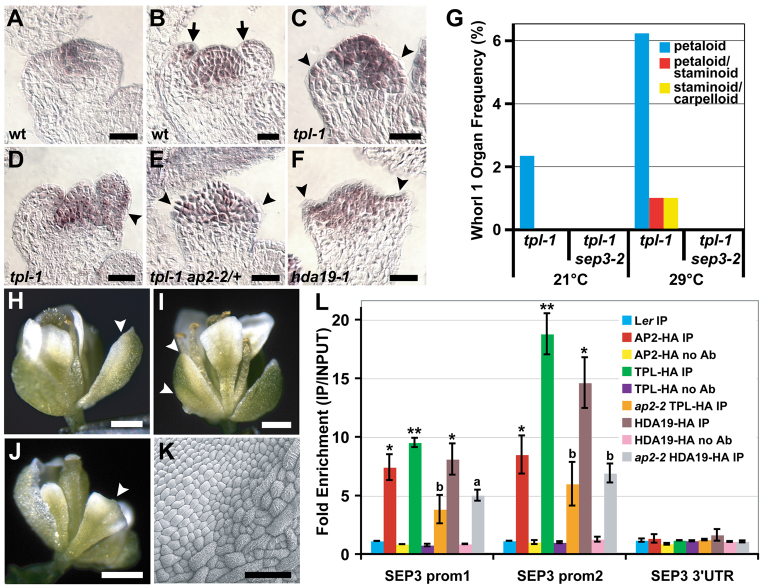
The E-class gene SEP3 is repressed in whorl 1 organ primordia by AP2, TPL and HDA19. (A-F) SEP3 in situ hybridizations on stage 3 (A,C,E,F) and stage 4 (B,D) Arabidopsis flowers. (A) SEP3 is not expressed in newly initiated outer whorl organ primordia in wild type. (B) By stage 4, SEP3 expression is detectable in wild-type sepals, but only at the distal tips of the organs (arrows). (C,E,F) SEP3 is ectopically expressed in mutant outer whorl organ primordia (arrowheads). (D) Sepal of a stage 4 tpl-1 flower showing an expanded SEP3 expression domain (arrowhead). (G) sep3-2 suppresses whorl 1 homeotic conversions of tpl-1 at 21°C and 29°C. (H-J) 2x35Sp::SEP3 flowers with sepals exhibiting petal identity (arrowheads) on one margin (H), both margins (I) or throughout the entire organ (J). (K) Scanning electron micrograph of a mosaic 2x35Sp::SEP3 sepal with ectopic petal epidermal identity (upper left; compare with Fig. 3E,F). (L) Anti-HA ChIP showing binding of AP2, TPL and HDA19 to two regions of the SEP3 promoter. Control ChIPs were performed on non-transgenic wild-type tissue (Ler) or without antibody (no Ab). Data were normalized relative to input and ACT2 abundance. Data are represented as mean ± s.e.m. of at least two biological replicates. Student's t-test was used to determine the significance of target enrichment relative to Ler IP (*P≤0.05, **P≤0.005) and the significance of decreased binding in ap2-2 (a, P≤0.08; b, P≤0.05). Scale bars: 20 μm in A-F; 500 μm in H-J; 50 μm in K.
Real-time PCR
Real-time PCR on ChIP samples was performed with a Bio-Rad CFX96 Real-time PCR system using Bio-Rad SsoFast EvaGreen Supermix reagents according to manufacturer protocols. Data analysis, including calculation of primer pair reaction efficiencies and Ct values, was carried out by Bio-Rad CFX Manager software. Enrichment was calculated as a ratio of the signal from ChIP samples to that from input samples. Fold enrichment was presented based on normalization with ACT2 data.
Transgenic plant lines
Arabidopsis transgenic lines were generated by the Agrobacterium-mediated floral dip method into the Ler ecotype (Clough and Bent, 1998). HDA19p::HDA19-HA was made using a PCR-generated HDA19 genomic clone (including 716 bp of promoter sequence) that was introduced into the XhoI site of plasmid pBJ36 (Eshed et al., 2001) upstream of a 6xHA epitope tag. This fusion gene was subcloned into the NotI site of binary vector pART27 (Gleave, 1992), transformed into hda19-1, and fully complemented the mutant. AP2 and AP2mEAR cDNAs were cloned as PCR-generated SalI-KpnI fragments downstream of the 2x35S promoter and upstream of GFP in vector pBJ36. Each AP2 fusion was subcloned into a sulfadiazine resistance binary vector (Heisler et al., 2010) and transformed into hda19-1 HDA19p::HDA19-HA for use in pull-down assays. The TPLp::TPL-HA transgenic line employed in pull-down and ChIP experiments has been described (Szemenyei et al., 2008). TPLp::TPL-GFP, HDA19p::HDA19-GFP and AP2p::AP2-2xYFP-3xHA transgenes used in ChIP assays have been published previously (Long et al., 2006; Wollmann et al., 2010).
TPLp::TPL-AP2DBs is a translational fusion between a full-length TPL cDNA and the coding sequence for the DNA-binding domains of AP2, followed by the recognition sequence for the small RNA miR172. This transgene was cloned into vector pBJ36 behind 4.13 kb of TPL promoter sequence and upstream of GFP. The TPL coding sequence was removed from this construct and a start codon was introduced into AP2DBs to produce the control transgene TPLp::AP2DBs. PCR was used to generate AP2DBs fragments with SmaI restriction sites at their termini to facilitate cloning (primer sequences in supplementary material Table S1). Both transgenes were introduced into the NotI site of the binary vector pMLBART (Eshed et al., 2001) for plant transformation.
To generate 2x35Sp::SEP3, a full-length SEP3 cDNA was amplified by PCR (as a ClaI-BamHI fragment) and cloned downstream of a 2x35S promoter and upstream of YFP-3xHA in plasmid pBJ36. This transgene was transferred into the NotI site of binary vector pMLBART and transformed into a Ler wild-type background.
Transcriptional repression assay
The transcriptional repression assay employing the β-glucuronidase (GUS) reporter was performed as previously described (Szemenyei et al., 2008). 2x35Sp::AP2-GAL4DB-3xHA was generated by fusing a full-length AP2 cDNA clone (SalI-KpnI) to GAL4DB-3xHA in pBJ36. This transgene was then transferred into the NotI site of pMLBART for plant transformation.
Microscopy
RNA in situ hybridizations were imaged using a Leica DM5000B compound microscope. Whole flowers and floral organs were imaged with a Leica MZ FLIII stereomicroscope. Scanning electron micrographs were acquired using a Zeiss EVO LS 15 analytical environmental scanning electron microscope.
RESULTS
APETALA2 physically interacts with a co-repressor and histone deacetylase
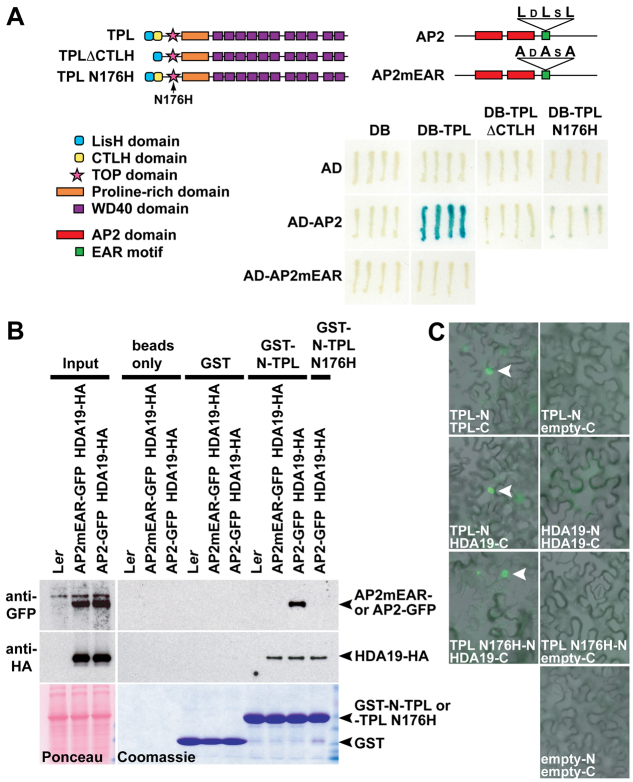
Although it is well established that AP2 represses AG, the molecular mechanism behind this control is not known (Drews et al., 1991; Yant et al., 2010). Using a yeast two-hybrid (Y2H) screen, we determined that AP2 physically interacts with the Gro/Tup1-type co-repressor TOPLESS (TPL), a result confirmed in a recent report (Causier et al., 2012). This interaction depended on the C-Terminal to Lissencephaly Homology (CTLH) domain of TPL (Fig. 1A), which is required for other reported TPL protein interactions (Szemenyei et al., 2008; Gallavotti et al., 2010). AP2 contains a consensus ERF-associated amphiphilic repression (EAR) motif (LxLxL) (Fig. 1A) (Ohta et al., 2001; Kagale et al., 2010), which has been implicated in the physical recruitment of TPL by various proteins (Szemenyei et al., 2008; Gallavotti et al., 2010; Pauwels et al., 2010). Mutation of the AP2 EAR motif (mEAR) abolished interaction with TPL in yeast, showing that this motif is necessary for AP2-TPL association (Fig. 1A). The tpl-1 missense mutation (N176H), which acts as a dominant negative for the TOPLESS RELATED (TPR) family (Long et al., 2006), also disrupted binding with AP2 in yeast (Fig. 1A). We verified these results using semi-in vivo pull-down assays (Szemenyei et al., 2008), testing the ability of recombinant GST-tagged TPL or TPL N176H amino-terminal fragments to bind GFP fusions of wild-type AP2 or AP2mEAR expressed in stable transgenic Arabidopsis lines. GST-N-TPL, but not GST-N-TPL N176H, interacted with wild-type AP2-GFP (Fig. 1B), whereas both GST-TPL variants were able to bind to full-length TPL (supplementary material Fig. S1A) (Szemenyei et al., 2008). Consistent with our Y2H data, GST-N-TPL was unable to pull-down AP2mEAR (Fig. 1B).
Fig. 1.
Physical association of AP2, TPL and HDA19. (A) Yeast two-hybrid (Y2H) assays. Protein domains of TPL and AP2 are depicted. TPL and AP2 interact by Y2H (bottom right) as indicated by blue coloration. Deletion of the CTLH domain or introduction of the N176H missense mutation into the TOP domain of TPL disrupts interaction with AP2. Mutation of the conserved leucines within the EAR motif of AP2 prevents binding to TPL. (B) In a semi-in vivo pull-down assay, GST N-TPL binds AP2-GFP from floral bud lysates but not AP2 mEAR-GFP. GST N-TPL N176H does not bind AP2-GFP. Both GST N-TPL and GST N-TPL N176H interact with HDA19-HA. Ponceau Red and Coomassie Blue staining shows equal protein loading and efficient GST protein expression, respectively. (C) Bimolecular fluorescence complementation (BiFC) assays. Nuclear fluorescence (arrowheads) shows physical association between HDA19 and both TPL and TPL N176H in transiently transfected tobacco cells. Homodimerization of TPL serves as a positive control; the right column depicts negative control combinations.
Co-repressors commonly recruit chromatin-remodeling factors to actively repress gene expression, and prior genetic analyses implicated the RPD3-like HISTONE DEACETYLASE 19 (HDA19) in TPL-dependent transcriptional regulation (Long et al., 2006). Therefore, we assessed the ability of TPL to pull-down HDA19-HA, which was expressed in the same transgenic Arabidopsis lines described above. Unlike AP2, HDA19 associated with both GST-N-TPL and GST-N-TPL N176H (Fig. 1B), and this was confirmed in planta using bimolecular fluorescence complementation (BiFC) (Fig. 1C). From these assays, we cannot determine whether the TPL-HDA19 interaction is direct, as adapter proteins may facilitate association between the two factors. Regardless, these findings suggest a potential mechanism for the dominant-negative nature of tpl-1: the TPL N176H mutant protein cannot bind AP2 but can interact with both TPL and HDA19, thereby ‘poisoning’ TPL/TPR-HDA19 complexes and preventing their recruitment to genomic loci by transcription factors such as AP2 (supplementary material Fig. S1B). In summary, our interaction data indicates that AP2 regulates target gene expression as part of a transcriptional repression complex that includes the co-repressor TPL and histone deacetylase HDA19.
The AP2-TPL-HDA19 repressor complex negatively regulates the C-class gene AG
We next sought to determine whether AP2 recruits TPL and HDA19 to regulate the C-class gene AG, which is expressed in whorls 3 and 4 and specifies the identities of the reproductive organs (Yanofsky et al., 1990). We performed RNA in situ hybridizations with TPL and HDA19 probes and found that both factors are broadly expressed in floral development and overlap with AP2 expression in outer whorl floral organ primordia, where AG is not normally expressed (supplementary material Fig. S2) (Wollmann et al., 2010). Additionally, genetic analyses showed that tpl-2 tpr1 tpr2 tpr3 tpr4 quintuple loss-of-function mutants and late-arising flowers of hda19-1 exhibit carpelloid outer whorl organs resembling those of strong ap2-2 mutants (Fig. 2B-D). All of these genetic backgrounds display AG misexpression throughout the flower (Fig. 2F-H) (Drews et al., 1991), indicating that AG is repressed not only by AP2, but also by redundantly acting members of the TPL/TPR family and HDA19. In the case of hda19-1, flowers begin to display carpelloid whorl 1 organs after ~20 flowers have been initiated. This is well before the termination of the inflorescence meristem, thereby distinguishing hda19-1 flowers from the ‘terminal carpelloid flower’ defects displayed by other mutants (Bao et al., 2004).
AP2 was recently shown to directly bind the second intron of AG (Yant et al., 2010; Dinh et al., 2012). Using chromatin immunoprecipitation (ChIP), we showed that this region of the second AG intron (supplementary material Fig. S3A) was specifically enriched in TPL and HDA19 immunoprecipitated samples, as well as in AP2 control ChIP experiments (Fig. 2I; supplementary material Fig. S4A). Enrichment was diminished in ap2-2, a protein-null allele (Chen, 2004) but not absent (Fig. 2I), potentially owing to the ability of TPL to interact physically with redundantly functioning AP2 homologs (Causier et al., 2012). This decrease in enrichment was not due to a reduction of TPL or HDA19 levels (supplementary material Fig. S3D).
To test further the functional importance of AP2-mediated recruitment of TPL, we fused TPL to the DNA-binding domains of AP2 along with the recognition sequence of miR172 (which controls AP2 distribution) (Aukerman and Sakai, 2003; Chen, 2004). This transgene rescued ap2-2 floral defects (Fig. 2J,K), indicating that recruitment of TPL to AP2 target genes can functionally substitute for AP2. This also suggests that to correctly pattern perianth (sepal and petal) whorls, AP2 predominantly acts as a transcriptional repressor. In agreement with this, AP2 negatively regulated the expression of a reporter gene in perianth organs in a transcriptional repression assay (supplementary material Fig. S5). Collectively, our results indicate that AP2 functions as a transcriptional repressor in floral patterning, and does so by recruiting TPL and, in turn, HDA19. Moreover, this reveals a molecular mechanism for AP2-mediated repression of AG, as all complex members associate with the second intron of AG during floral development and repress its transcription in outer whorls.
AP2, TPL and HDA19 restrict petal fate by regulating the outer boundary of B-class gene expression
Whereas tpl-2 tpr1 tpr2 tpr3 tpr4 quintuple mutant flowers resemble those of strong ap2 alleles, the tpl-1 mutant displays partial sepal-to-petal homeotic conversions (Fig. 3A,G). Such conversions are also found in tpl-2 tpr1 tpr3 tpr4 quadruple loss-of-function mutant flowers (supplementary material Fig. S6A) indicating that tpl-1 is not a complete dominant negative for the TPL/TPR family during floral patterning, as it is during embryogenesis (Long et al., 2006). Flowers of tpl-1 ap2-2/+ mutants show a much higher frequency of sepal-to-petal conversions compared with tpl-1 alone (Fig. 3B; supplementary material Table S2). Therefore, TPL and AP2 appear to cooperatively repress petal identity in whorl 1, revealing a new role for AP2 in floral patterning that is not predicted by the ABC model. HDA19 is also involved in this repression, as similar homeotic transformations occur in early-arising flowers of hda19-1 (Fig. 3C,H), and the frequency of these is greatly increased in tpl-1 hda19-1 mutants (Fig. 3D; supplementary material Table S2). Finally, like ap2-2/+, hda19-1/+ heterozygous mutations enhance the frequency of tpl-1 sepal-to-petal conversions (supplementary material Table S2).
The sepal-to-petal conversions displayed by the mutant combinations described above suggest misregulation of the B-class genes AP3 and PI in the first floral whorl. In wild type, both genes are predominantly expressed in whorls 2 and 3 where they cooperatively specify petal and stamen fate (Fig. 3I,K; Fig. 4A,C) (Jack et al., 1992; Goto and Meyerowitz, 1994). AP3 is also expressed at the base of wild-type sepals in later stages of their development (Fig. 3K) (Weigel and Meyerowitz, 1993). PI expression is not detectable in whorl 1 organs at any stage (Goto and Meyerowitz, 1994). In flowers of tpl-1, ap2-2 and hda19-1 mutant combinations, however, PI is ectopically expressed in outer whorl organs (Fig. 4B,D-F), and AP3 is expressed precociously in newly initiated sepal primordia and in a much larger domain in older whorl 1 organs (Fig. 3J,L-N). The whorl 1 homeotic conversions displayed by these mutants not only correlate with this B-class gene misregulation but depend upon it, as either ap3-3 or pi-1 completely suppresses sepal-to-petal transformations (supplementary material Table S2).
Fig. 4.
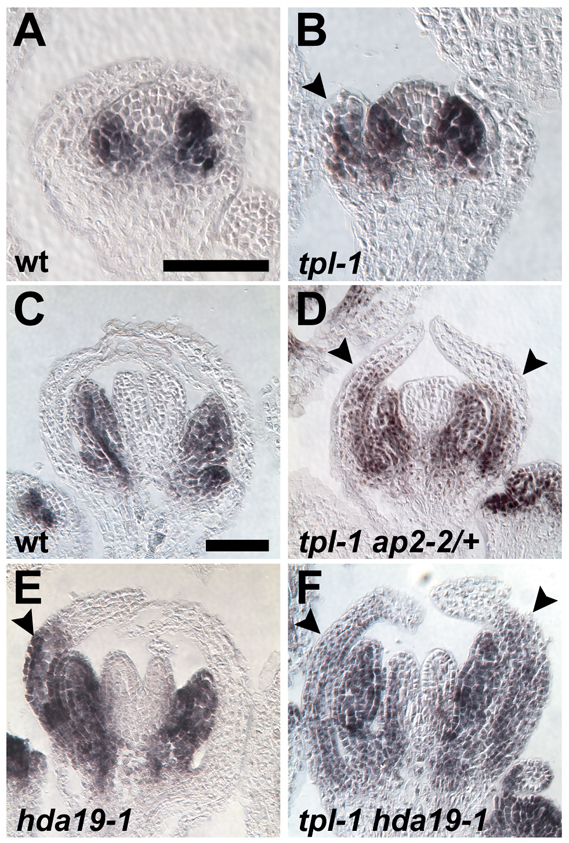
The B-class gene PI is misexpressed in the outer whorl of tpl-1 and hda19-1 mutant backgrounds. (A-F) PI in situ hybridizations. (A,B) Stage 4 and (C-F) stage 7-8 Arabidopsis flowers. (A,C) PI is not expressed in whorl 1 organs in either stage of floral development. (B,D-F) PI is ectopically expressed in mutant whorl 1 organs (arrowheads). Scale bars: 50 μm.
To determine whether AP2 recruits TPL and HDA19 to directly repress B-class genes, we performed ChIP experiments on all three factors. Consistent with published AP2 ChIP with high-throughput sequencing (ChIP-seq) data (Yant et al., 2010), we did not detect binding of AP2 to the PI promoter. However, AP2, TPL and HDA19 did bind a common promoter region of AP3 (Fig. 3O; supplementary material Fig. S3B, Fig. S4B), and binding by TPL and HDA19 was reduced in ap2-2 (Fig. 3O). Although AP3 was not reported as being significantly bound by AP2 in ChIP-seq analyses (Yant et al., 2010), evidence of AP2 binding to this region of the AP3 promoter could be detected in the raw data available from this study. Therefore, the role of AP2 in restriction of petal fate appears to involve the direct repression of AP3 and, probably, the indirect repression of PI.
Based on these regulatory relationships, ectopic B-class gene expression would be predicted in an ap2 mutant background. Indeed, AP3 and PI are variably misexpressed in whorl 1 organs of ap2-2 flowers (supplementary material Fig. S6B,C) (Goto and Meyerowitz, 1994). Consistent with this misexpression, ap2-2 carpelloid whorl 1 organs often produce tissue with stamen identity at their margins (Fig. 2B) (Bowman et al., 1991). By contrast, AP3 and PI levels are vastly reduced in their normal domains of expression in strong ap2 mutants (supplementary material Fig. S6B,C) (Jack et al., 1992; Goto and Meyerowitz, 1994). This apparent contradiction can be explained by the loss of whorl 2 and 3 organs in ap2 due to the spread of C-function, which plays a role in meristem determinacy (Jack et al., 1992; Goto and Meyerowitz, 1994). As a result, decreased expression of B-class genes might simply be an indirect consequence of reduced cellular proliferation within the floral whorls in which they act. In agreement with this, whorl 2 and 3 organ production and the expression domains of AP3 and PI are restored in an ap2 ag double mutant background (supplementary material Fig. S6E,F) (Zhao et al., 2007).
AP2, TPL and HDA19 directly repress expression of the E-class gene SEP3 in the outer floral whorl
The absence of evidence for direct regulation of PI by AP2 prompted us to consider that AP2 might negatively regulate PI indirectly in first whorl organs by repressing an upstream activator. One such candidate is the AP2 target SEP3 (Yant et al., 2010), a direct positive regulator of floral organ identity genes, including PI (Kaufmann et al., 2009). Unlike other SEP genes, SEP3 is not normally expressed in newly initiated outer whorl organ primordia (Fig. 5A) (Mandel and Yanofsky, 1998). However, it expands to this domain in ap2-2 flowers (supplementary material Fig. S6D), as well as in tpl-1, tpl-1 ap2-2/+ and hda19-1 flowers (Fig. 5C,E,F). Stage 4 tpl-1 flowers variably display SEP3 expression throughout sepal primordia, whereas at this stage in wild type, SEP3 is restricted to the distal tips (Fig. 5B,D) (Kaufmann et al., 2009). To test whether SEP3 misexpression is required for sepal-to-petal conversions, we introduced the sep3-2 mutation (Pelaz et al., 2000) into the tpl-1 background, which shows homeotic changes in first whorl organs in a temperature-dependent manner. At both temperatures analyzed, sep3-2 completely suppressed tpl-1 homeotic conversions in the first whorl (Fig. 5G; supplementary material Table S2). To determine whether SEP3 expression in the first whorl is sufficient to alter sepal fate, we overexpressed SEP3 and saw varying degrees of sepal-to-petal conversions in the flowers of multiple independent transgenic lines (Fig. 5H-K). These observations suggest that when AP2, TPL and HDA19 function is compromised, SEP3 is misexpressed in whorl 1 and promotes ectopic petal identity. Consistent with this, both TPL and HDA19 associated with the same two promoter regions of SEP3 that AP2 has been shown to bind (Fig. 5L; supplementary material Fig. S3C, Fig. S4C) (Yant et al., 2010), and binding by TPL and HDA19 was reduced in ap2-2 (Fig. 5L). Therefore, AP2 restricts the outer boundary of petal fate not only by controlling the expression domains of B-class genes, but also by directly repressing the E-class gene SEP3 in whorl 1.
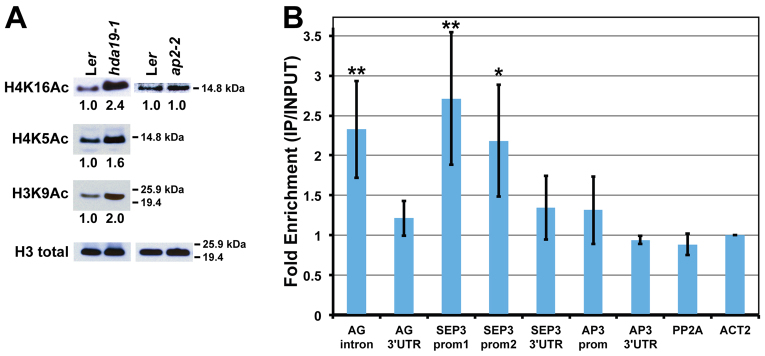
Histone acetylation levels at AP2 target genes increase in an ap2-2 background
Because AP2 appears to recruit HDA19 to repress the transcription of its target genes, an increase in histone acetylation levels at these loci would be predicted in an ap2-2 mutant background. To test this hypothesis, we first sought to determine which histone acetylation mark(s) are likely to be regulated by HDA19 by assessing global acetylation levels in hda19-1 flowers. We tested acetylation marks reportedly affected by hda19 at other developmental stages and/or by RPD3-like histone deacetylases (HDACs) in other systems, including H3K9Ac, H4K5Ac and H4K16Ac (Kadosh and Struhl, 1998; Tian et al., 2005; Benhamed et al., 2006; Miller et al., 2010). All modifications were increased to some extent in hda19-1 flowers compared with wild type, suggesting that HDA19 deacetylates these histone residues in planta (Fig. 6A). Because H4K16Ac showed the greatest increase, we performed ChIP using an antibody against this mark to assess acetylation levels at AP2 target loci in ap2-2 and wild-type flowers (Fig. 6B). Regulatory regions of both AG and SEP3 that were bound by AP2, TPL and HDA19 showed significantly increased H4K16Ac levels in ap2-2 (Fig. 6B), even though global levels of H4K16Ac in this background appeared comparable to wild type (Fig. 6A). These results are consistent with our model in which AP2 recruits a TPL-HDA19 repressor complex, as in the absence of AP2, histone marks controlled by HDA19 increase at AP2 target loci. These observations also suggest that unlike the global effect of hda19-1 on histone acetylation marks, the ap2-2 mutation affects H4K16Ac levels at specific loci. H4K16Ac abundance was only moderately increased at the AP3 promoter in ap2-2 (Fig. 6B), possibly due to the small fraction of cells that misexpress AP3 in the mutant flower.
Fig. 6.
The effects of hda19-1 and ap2-2 mutation on histone acetylation levels. (A) Histone acetylation modifications H4K16Ac, H4K5Ac and H3K9Ac are increased in hda19-1 Arabidopsis flowers compared with wild type (left). H4K16Ac levels in ap2-2 flowers are comparable to that of wild type (right). Band intensities were normalized relative to total histone H3 loading controls. (B) Anti-H4K16Ac ChIP showing enrichment at the second intron of AG and at two promoter regions of SEP3 in ap2-2 flowers. A slight enrichment is seen at the promoter of AP3. Data are represented as mean ± s.e.m. of three biological replicates and are presented as the ratio of enrichment in ap2-2 over that of wild type. Data were normalized relative to INPUT and ACT2 abundance. The constitutively expressed PP2A gene serves as a negative control. Student's t-test was used to determine the significance of target enrichment relative to PP2A (*P≤0.08; **P≤0.05).
DISCUSSION
In this study, we show that, in combination with TPL and HDA19, AP2 acts as a cadastral regulator of multiple floral homeotic genes to pattern the flower correctly (Fig. 7), broadening our knowledge of how gene expression boundaries are maintained in eukaryotic development. This includes the C-class gene AG, the outer expression boundary of which is controlled by an AP2-TPL-HDA19 repression complex. We therefore determine a molecular mechanism underlying a genetic relationship that contributed to the original formulation of the ABC model (the antagonism of AG by AP2).
Fig. 7.
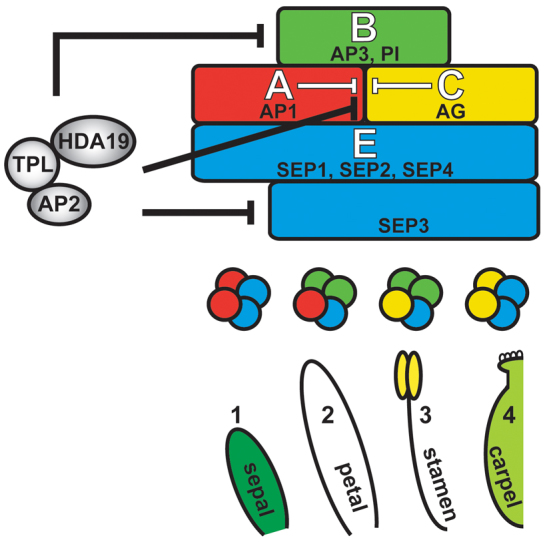
Model of AP2 function during flower development. AP2 controls the outer expression boundaries of B-, C- and E-class genes by recruiting TPL and HDA19. This AP2 repression complex directly represses the C-class gene AG in whorls 1 (sepal) and 2 (petal) and the B-class gene AP3 and E-class gene SEP3 in whorl 1. AP2-mediated repression of the B-class gene PI in whorl 1 may occur through negative regulation of upstream activators, such as SEP3. MADS-domain products of the floral homeotic genes (colored circles) are predicted to interact with one another and with SEP proteins to form quartet complexes that regulate gene expression required for organ identity specification (Honma and Goto, 2001). Intermediary proteins might facilitate TPL and HDA19 association.
We demonstrate further that AP2 recruits TPL and HDA19 to restrict whorl 1 expression of AP3, SEP3 and subsequently PI. This elucidates the presence of a repressor complex that establishes the peripheral expression boundaries of not only B-class genes but also the E-class gene SEP3, for which the restricted domain of activity in young flowers is unique among the SEPs (Mandel and Yanofsky, 1998). Our genetic analyses indicate that this regulation is an important aspect of floral patterning, as it is required to prevent the outward spread of petal fate. This represents a novel role for AP2 not predicted in the original ABC model, and in fact appears to disagree with the designation of AP2 as an A-class gene responsible for the specification of perianth fate (see below). Taking into account these new roles, AP2 has now been shown to directly restrict the expression domains of B-, C- and E-class genes in the flower (Fig. 7). Therefore, it is increasingly apparent that AP2 patterns the flower as a transcriptional repressor, a notion strongly supported by the observation that expression of a fusion protein between the AP2 DNA-binding domains and a transcriptional co-repressor is sufficient to rescue ap2 floral patterning defects (Fig. 2K).
Although AP2, TPL and HDA19 are all expressed in whorls 1 and 2, petal fate is repressed only in the outermost whorl. The underlying cause of this positional difference is unknown, but it is plausible that AP2 constitutively represses SEP3 and B-class genes, but in whorl 2, antagonizing activators of petal fate, such as the F-box protein UNUSUAL FLORAL ORGANS (UFO), overcome this repression (Lee et al., 1997). UFO acts as a co-factor with the meristem identity gene LEAFY (LFY) to activate B-function (Lee et al., 1997). Whereas LFY is ubiquitously expressed in young flowers, UFO is transiently expressed in whorls 2 and 3, conferring domain specificity to LFY activity, which includes upregulation of target genes such as AP3, PI and SEP3 (Lee et al., 1997; Winter et al., 2011). Ectopically expressing UFO throughout the flower results in sepal-to-petal conversions and AP3 misexpression in whorl 1 organs (Lee et al., 1997), showing that UFO activity is epistatic to AP2-mediated repression of B-function. It is therefore not surprising that petal fate arises in whorl 2, where UFO is normally expressed, despite the presence of the AP2 repressor complex. Somewhat paradoxically, data indicate that UFO promotes transcription at the AP3 promoter by targeting LFY (or an associated factor) for degradation (Chae et al., 2008). In light of our results, UFO might function at the AP3 promoter to destabilize a repressor (Samach et al., 1999), such as AP2 and/or one of its complex members.
A balance between transcriptional activation and repression seems to underlie numerous aspects of floral organ fate specification. For example, activators of AG in the central domains of the flower compete with broadly distributed repressors of C-function to elicit region-specific transcription (Sridhar et al., 2006). This antagonism may ‘fine-tune’ AG expression levels to ensure appropriate floral organ identity. Such tuning mechanisms appear to exist in Antirrhinum and Petunia, in which mutations resulting in increased expression levels of C-class genes in their natural domains of expression result in the outward expansion of C-function (Cartolano et al., 2007).
The identification of an AP2-TPL-HDA19 repressor complex involved in the direct repression of AG reveals increased functional redundancy between transcriptional regulators known to inhibit C-function (reviewed by Kaufmann et al., 2010a). For example, LUG, a Gro/Tup1-type co-repressor structurally similar to TPL, and its interacting partner SEU appear to be recruited by the A-class regulator AP1 (and SEP3) to repress AG (Sridhar et al., 2006; Liu and Karmarkar, 2008). LUG has been shown to interact physically with HDA19 (Gonzalez et al., 2007), indicating that AP1-SEU-LUG and AP2-TPL repressor complexes might share chromatin-modifying factors to repress AG in outer whorl organs. Additionally, LUG and SEU might be recruited by AP2 to negatively regulate target genes not examined in the present work (Grigorova et al., 2011). These observations indicate that physical interactions between factors operating redundantly in the repression of AG may be elaborate.
The transcriptional regulation of the B-class gene AP3 also appears to be complex in nature. AP3 promoter analyses have identified cis-acting elements required for expression in early floral development (stage 3-5), along with elements necessary for later expression in petals and stamens (Hill et al., 1998; Tilly et al., 1998). Three MADS-domain binding sequences (CArG) reside in the AP3 promoter, the most proximal of which is apparently involved in the recruitment of a negative regulator affecting overall expression levels (Tilly et al., 1998). Promoter deletions, however, failed to identify elements potentially bound by AP2 or other repressors that restrict the outward spread of AP3 expression. The close proximity of our elucidated AP2-TPL-HDA19 binding site to activating CArG sites in the AP3 promoter (supplementary material Fig. S3B) might have complicated such analyses.
Although AP2 is clearly an important cadastral regulator of multiple floral organ identity genes, it is not obvious how AP2 acts as a floral organ identity gene itself (as described in the ABC model). Based on its classification as A-function, one of the proposed roles of AP2 is the specification of perianth fate. It is difficult to reconcile how AP2 can play an instructive role in perianth fate while negatively regulating genes (B-class and SEP3) responsible for promoting petal identity. It is possible that AP2 represses petal fate in whorl 1 and promotes it in whorl 2. Indeed, the normal domain of B-class gene expression is reduced in ap2; however, previous reports attribute this reduction to organ loss resulting from C-function spread (Jack et al., 1992; Goto and Meyerowitz, 1994). The restoration of AP3 and PI expression in whorls 2 and 3 of ap2 ag shows that AP2 is not necessary for B-class gene activation (supplementary material Fig. S6E,F). These restored organs show a mixture of petal and stamen fate (Bowman et al., 1991), further indicating that AP2 is not strictly required for petal identity. If AP2 were required, the ABC model predicts that its expression would expand into the central region of indeterminate ag flowers (which display reiterations of perianth organs), but this does not occur (Wollmann et al., 2010). Together, these results indicate that AP2 does not directly specify perianth fate and appears not to fit its original A-function designation. This is in agreement with other reports suggesting that the disruption of perianth identity in ap2 mutants is an indirect effect reflecting an earlier role for AP2 in floral meristem specification (Okamuro et al., 1997; Litt, 2007; Causier et al., 2010). In this context, AP2 first contributes to floral meristem identity (required for the production of the perianth) and later regulates the expression domains of B-, C- and E-class genes that specify floral organ fate. This interpretation does not exclude the possibility that other factors confer true A-function in Arabidopsis and other plant species.
A future challenge will be to determine how well conserved the novel protein interactions and transcriptional processes revealed in this study are in floral patterning events outside of Arabidopsis. SEP3 and B-class gene expression is commonly excluded from first whorl floral organs of angiosperms (Malcomber and Kellogg, 2005; Zahn et al., 2005a; Zahn et al., 2005b), and it will be of great interest to determine the role of AP2, TPL and HDA19 orthologs in this regulation. The perianths of some monocots and basal eudicots consist of two outer whorls of petaloid organs that express SEP3 or B-class genes (Kramer et al., 2003; Malcomber and Kellogg, 2005; Theissen and Melzer, 2007) and variations in AP2, TPL and HDA19 function in these species might contribute to this outward spread of petal fate. Characterizing such variation will provide valuable insight into the transcriptional mechanisms underlying the seemingly endless floral diversity of angiosperms.
Supplementary Material
Acknowledgements
We thank members of the Long laboratory, J. Chory, B. Chow, B. Crawford, F. Wellmer and M. Yanofsky for comments on the manuscript; M. Yanofsky for sep3-2 seeds and vectors for in situ hybridization probes; M. Joens and J. Fitzpatrick for technical assistance with scanning electron microscopy; D. Nusinow for technical assistance with ChIP; and the Arabidopsis Biological Resource Center for the insertion line SALK_112730.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant GM072764 to J.A.L.]; the National Science Foundation [grant IOS0822411 to J.A.L.]; and a Natural Sciences and Engineering Research Council Postdoctoral Fellowship and San Diego Foundation Blasker Science and Technology Grant [to N.T.K.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.085407/-/DC1
References
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653-657 [DOI] [PubMed] [Google Scholar]
- Aukerman M. J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15, 2730-2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X., Franks R. G., Levin J. Z., Liu Z. (2004). Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16, 1478-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.-X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18, 2893-2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A. V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39, 776-789 [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1-20 [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Sakai H., Jack T., Weigel D., Mayer U., Meyerowitz E. M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599-615 [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Alvarez J., Weigel D., Meyerowitz E. M., Smyth D. R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721-743 [Google Scholar]
- Buscarlet M., Stifani S. (2007). The ‘Marx’ of Groucho on development and disease. Trends Cell Biol. 17, 353-361 [DOI] [PubMed] [Google Scholar]
- Busch M. A., Bomblies K., Weigel D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285, 585-587 [DOI] [PubMed] [Google Scholar]
- Byzova M. V., Franken J., Aarts M. G. M., de Almeida-Engler J., Engler G., Mariani C., Van Lookeren Campagne M. M., Angenent G. C. (1999). Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev. 13, 1002-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartolano M., Castillo R., Efremova N., Kuckenberg M., Zethof J., Gerats T., Schwarz-Sommer Z., Vandenbussche M. (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 39, 901-905 [DOI] [PubMed] [Google Scholar]
- Causier B., Schwarz-Sommer Z., Davies B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73-79 [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158, 423-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E., Tan Q. K., Hill T. A., Irish V. F. (2008). An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135, 1235-1245 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022-2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743 [DOI] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31-37 [DOI] [PubMed] [Google Scholar]
- Conner J., Liu Z. (2000). LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97, 12902-12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Jia S. (2001). Transcriptional repression: the long and the short of it. Genes Dev. 15, 2786-2796 [DOI] [PubMed] [Google Scholar]
- Dinh T. T., Girke T., Liu X., Yant L., Schmid M., Chen X. (2012). The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139, 1978-1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935-1940 [DOI] [PubMed] [Google Scholar]
- Drews G. N., Bowman J. L., Meyerowitz E. M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991-1002 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S. F., Perea J. V., Bowman J. L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251-1260 [DOI] [PubMed] [Google Scholar]
- Franks R. G., Wang C., Levin J. Z., Liu Z. (2002). SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129, 253-263 [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Long J. A., Stanfield S., Yang X., Jackson D., Vollbrecht E., Schmidt R. J. (2010). The control of axillary meristem fate in the maize ramosa pathway. Development 137, 2849-2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A. P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203-1207 [DOI] [PubMed] [Google Scholar]
- Gonzalez D., Bowen A. J., Carroll T. S., Conlan R. S. (2007). The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 27, 5306-5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548-1560 [DOI] [PubMed] [Google Scholar]
- Gray S., Levine M. (1996). Transcriptional repression in development. Curr. Opin. Cell Biol. 8, 358-364 [DOI] [PubMed] [Google Scholar]
- Grigorova B., Mara C., Hollender C., Sijacic P., Chen X., Liu Z. (2011). LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138, 2451-2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M. G., Hamant O., Krupinski P., Uyttewaal M., Ohno C., Jönsson H., Traas J., Meyerowitz E. M. (2010). Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 8, e1000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. A., Day C. D., Zondlo S. C., Thackeray A. G., Irish V. F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125, 1711-1721 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525-529 [DOI] [PubMed] [Google Scholar]
- Jack T., Brockman L. L., Meyerowitz E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683-697 [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K. (1998). Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18, 5121-5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Links M. G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152, 1109-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J. M., Jauregui R., Airoldi C. A., Smaczniak C., Krajewski P., Angenent G. C. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7, e1000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Pajoro A., Angenent G. C. (2010a). Regulation of transcription in plants: mechanisms controlling developmental switches. Nat. Rev. Genet. 11, 830-842 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J. M., Ferrier T., Wuest S. E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E. M., et al. (2010b). Orchestration of floral initiation by APETALA1. Science 328, 85-89 [DOI] [PubMed] [Google Scholar]
- Kramer E. M., Di Stilio V. S., Schluter P. M. (2003). Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int. J. Plant Sci. 164, 1-11 [Google Scholar]
- Krizek B. A., Lewis M. W., Fletcher J. C. (2006). RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J. 45, 369-383 [DOI] [PubMed] [Google Scholar]
- Lamb R. S., Hill T. A., Tan Q. K., Irish V. F. (2002). Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129, 2079-2086 [DOI] [PubMed] [Google Scholar]
- Lee I., Wolfe D. S., Nilsson O., Weigel D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7, 95-104 [DOI] [PubMed] [Google Scholar]
- Litt A. (2007). An evaluation of A-function: evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 168, 73-91 [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711-722 [DOI] [PubMed] [Google Scholar]
- Liu Z., Meyerowitz E. M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975-991 [DOI] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13, 137-144 [DOI] [PubMed] [Google Scholar]
- Lohmann J. U., Hong R. L., Hobe M., Busch M. A., Parcy F., Simon R., Weigel D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793-803 [DOI] [PubMed] [Google Scholar]
- Long J. A., Woody S., Poethig S., Meyerowitz E. M., Barton M. K. (2002). Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129, 2797-2806 [DOI] [PubMed] [Google Scholar]
- Long J. A., Ohno C., Smith Z. R., Meyerowitz E. M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520-1523 [DOI] [PubMed] [Google Scholar]
- Malcomber S. T., Kellogg E. A. (2005). SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 10, 427-435 [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Yanofsky M. F. (1998). The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22-28 [Google Scholar]
- Miller K. M., Tjeertes J. V., Coates J., Legube G., Polo S. E., Britton S., Jackson S. P. (2010). Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17, 1144-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M., Yanofsky M. F. (2001). Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13, 739-753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959-1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro J. K., Szeto W., Lotys-Prass C., Jofuku K. D. (1997). Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell 9, 37-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484-1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Barbero G. F., Geerinck J., Tilleman S., Grunewald W., Pérez A. C., Chico J. M., Bossche R. V., Sewell J., Gil E., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G. S., Baumann E., Wisman E., Yanofsky M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200-203 [DOI] [PubMed] [Google Scholar]
- Pelaz S., Tapia-López R., Alvarez-Buylla E. R., Yanofsky M. F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11, 182-184 [DOI] [PubMed] [Google Scholar]
- Sakai H., Medrano L. J., Meyerowitz E. M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199-203 [DOI] [PubMed] [Google Scholar]
- Samach A., Klenz J. E., Kohalmi S. E., Risseeuw E., Haughn G. W., Crosby W. L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433-445 [DOI] [PubMed] [Google Scholar]
- Schultz E. A., Pickett F. B., Haughn G. W. (1991). The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell 3, 1221-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z. R., Long J. A. (2010). Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464, 423-426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V. V., Surendrarao A., Liu Z. (2006). APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133, 3159-3166 [DOI] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384-1386 [DOI] [PubMed] [Google Scholar]
- Takeda S., Matsumoto N., Okada K. (2004). RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131, 425-434 [DOI] [PubMed] [Google Scholar]
- Theissen G., Melzer R. (2007). Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. 100, 603-619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Fong M. P., Wang J. J., Wei N. E., Jiang H., Doerge R. W., Chen Z. J. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169, 337-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly J. J., Allen D. W., Jack T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125, 1647-1657 [DOI] [PubMed] [Google Scholar]
- Weigel D., Meyerowitz E. M. (1993). Activation of floral homeotic genes in Arabidopsis. Science 261, 1723-1726 [DOI] [PubMed] [Google Scholar]
- Weigel D., Alvarez J., Smyth D. R., Yanofsky M. F., Meyerowitz E. M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843-859 [DOI] [PubMed] [Google Scholar]
- Winter C. M., Austin R. S., Blanvillain-Baufumé S., Reback M. A., Monniaux M., Wu M.-F., Sang Y., Yamaguchi A., Yamaguchi N., Parker J. E., et al. (2011). LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20, 430-443 [DOI] [PubMed] [Google Scholar]
- Wollmann H., Mica E., Todesco M., Long J. A., Weigel D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633-3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35-39 [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T. T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L. M., Kong H., Leebens-Mack J. H., Kim S., Soltis P. S., Landherr L. L., Soltis D. E., Depamphilis C. W., Ma H. (2005a). The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209-2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L. M., Leebens-Mack J., DePamphilis C. W., Ma H., Theissen G. (2005b). To B or Not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J. Hered. 96, 225-240 [DOI] [PubMed] [Google Scholar]
- Zhao L., Kim Y., Dinh T. T., Chen X. (2007). miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 51, 840-849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Xu F., Zhang Y., Cheng Y. T., Wiermer M., Li X., Zhang Y. (2010). Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. USA 107, 13960-13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.