Abstract
In the gibberellin (GA) biosynthesis pathway, 20-oxidase catalyzes the oxidation and elimination of carbon-20 to give rise to C19-GAs. All bioactive GAs are C19-GAs. We have overexpressed a cDNA encoding 20-oxidase isolated from Arabidopsis seedlings in transgenic Arabidopsis plants. These transgenic plants display a phenotype that may be attributed to the overproduction of GA. The phenotype includes a longer hypocotyl, lighter-green leaves, increased stem elongation, earlier flowering, and decreased seed dormancy. However, the fertility of the transgenic plants is not affected. Increased levels of endogenous GA1, GA9, and GA20 were detected in seedlings of the transgenic line examined. GA4, which is thought to be the predominantly active GA in Arabidopsis, was not present at increased levels in this line. These results suggest that the overexpression of this 20-oxidase increases the levels of some endogenous GAs in transgenic seedlings, which causes the GA-overproduction phenotype.
During developmental transitions, differentiation, or response to environmental changes, plants regulate their growth by varying the levels of endogenous phytohormones. One class of phytohormones, GAs, are found in young tissues of the shoot and developing seeds, and have regulatory roles in seed germination, stem elongation, flowering, and fruit set (for review, see Crozier, 1983). Whereas tissue responsiveness is an important factor that also contributes to GA-mediated developmental processes, these processes are directly responsive to the concentration of bioactive GAs present.
It is believed that de novo biosynthesis is the main source of bioactive GAs in growing tissues. Thus, enzymes involved in GA biosynthesis are likely to be the regulators of GA-related growth and can serve as targets for the manipulation of plant growth and development through genetic engineering. In Arabidopsis there are at least five loci involved in GA biosynthesis: GA1, GA2, GA3, GA4, and GA5. These loci were identified in mutants that have a GA requirement for normal growth (Koornneef and van der Veen, 1980). By quantifying endogenous GAs and applying various GAs and GA precursors to these mutants, each GA locus has been assigned an enzymatic function in the GA biosynthetic pathway (for review, see Finkelstein and Zeevaart, 1984). The first committed reaction of the GA biosynthesis pathway is the cyclization of geranylgeranyl pyrophosphate to ent-kaurene, a two-step conversion. Copalyl diphosphate synthase, formerly ent-kaurene synthetase A, the enzyme responsible for the first part of the reaction, is encoded by the GA1 locus and has been cloned (Sun et al., 1992; Sun and Kamiya, 1994). The GA2 locus encodes ent-kaurene synthase, which completes the conversion of geranylgeranyl pyrophosphate to ent-kaurene (Yamaguchi et al., 1998). Plants bearing mutations in the GA3 locus show a growth response only to intermediates after ent-kaurenal in the GA biosynthesis pathway. Therefore, the GA3 locus probably encodes a Cyt P450 monooxygenase, which catalyzes the oxidation of ent-kaurene to ent-kaurenoic acid. The endogenous concentration of various GAs in ga4 and ga5 mutant plants was carefully measured (Talon et al., 1990), and the results suggested that GA5 and GA4 encode GA 20-oxidase and 3β-hydroxylase, respectively. Both genes have been cloned, and GA5 protein produced in vitro exhibits GA 20-oxidase activity (Chiang et al., 1995; Xu et al., 1995).
Oxidation at carbon-20 of GAs is thought to be an important aspect of regulation in the GA biosynthetic pathway. In spinach the enhanced oxidation activity is associated with the bolting response brought on by exposure to long days (Gilmour et al., 1986). In maize seedlings evidence suggests that the 20-oxidase activities are down-regulated as a result of feedback control by GA (Hedden and Croker, 1992). The cloning of the GA5 locus in Arabidopsis allowed the study of the regulation of 20-oxidase at the molecular level (Phillips et al., 1995; Xu et al., 1995), and these studies showed that expression of GA5 increases when plants are transferred from short-day to long-day conditions and decreases when plants are treated with bioactive GA. Furthermore, there are at least three different 20-oxidase genes in Arabidopsis, and their expression patterns are differentially regulated (Phillips et al., 1995). Therefore, the developmental and environmental regulation of 20-oxidase gene expression appears to affect the level of endogenous GAs, which influences plant growth.
To study the effects of 20-oxidase expression in vivo, we isolated a full-length cDNA fragment from Arabidopsis seedling RNA by reverse-transcriptase PCR and overexpressed it under the control of a constitutive promoter in Arabidopsis. The transgenic plants that overexpressed the 20-oxidase cDNA exhibited a “GA-overproduction” phenotype. This phenotype includes longer hypocotyl length, lighter-colored leaves, longer and accelerated stem elongation, earlier flowering, and decreased seed dormancy. We also examined the endogenous GA levels of the transgenic plants. The results suggest that the phenotype is the result of increased endogenous GA levels caused by overexpression of the 20-oxidase cDNA.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis ecotype Columbia were obtained from Lehle Seeds (Tucson, AZ). Plants were grown in potting soil (Scotts, Marysville, OH) in a growth chamber at 24°C with a 16-h photoperiod (120 μE m−2 s−1). In the seedling growth and germination studies, seeds were surface sterilized and germinated on Murashige and Skoog medium (M0404, Sigma) supplemented with Suc (1%) and Mes (0.5 g L−1) at pH 5.8.
Cloning of 20-Oxidase cDNA and Transformation Plasmid Construction
The PCR primers 5′-ATGGCCGTAAGTTTCGTAAC-3′ and 5′-TTAGATGGGTTTGGTGAGCC-3′, located at the start and end of the 20-oxidase reading frame, were designed based on the reported sequence of GA5 (Xu et al., 1995). These primers were used in a reverse-transcriptase-PCR reaction to amplify a 1.1-kb cDNA fragment from RNA extracted from 5-d-old Arabidopsis seedlings. The PCR product was inserted into the TA cloning vector (Invitrogen, San Diego, CA) for further manipulation. To detect any mutations generated by PCR, several independent reverse-transcriptase-PCR-amplified cDNAs were sequenced and compared. The consensus GA5 cDNA was then inserted into a binary transformation vector for Agrobacterium tumefaciens-mediated transformation into Arabidopsis. The resulting plasmid, pMON29925, contains the 20-oxidase cDNA under transcriptional control of an enhanced 35S promoter. The −90 to −342 region of the original 35S promoter (Fang et al., 1989) was duplicated to produce the enhanced 35S promoter. Arabidopsis plants were transformed according to the method of Bechtold et al. (1993).
Identification of T2 Plants Containing the Transgene by PCR
Genomic DNA was isolated from a single rosette leaf disc using the hexadecyltrimethyl-ammonium bromide protocol (Stewart and Via, 1993). The same PCR primers used to prepare the cDNA were used to analyze the genomic DNA of individual T2 plants for the presence or absence of the transgene. T2 plants positive for the presence of the transgene were determined to be hemizygous or homozygous for the transgene locus by the segregation ratios of kanamycin-resistant T3 progeny; T2 plants lacking the transgene and also showing uniform kanamycin sensitivity in T3 progeny were selected as the isogenic wild-type controls.
Isolation of Genomic DNA and DNA Gel-Blot Analysis
Genomic DNA was isolated from 0.25 g of T2 seedlings from each transgenic line tested as described previously (Coleman and Kao, 1992). Genomic DNA (5 μg) was digested with SpeI, separated on a 0.8% (w/v) agarose gel, and transferred to a nylon membrane (Qiagen, Chatsworth, CA). Prehybridization, hybridization, washing, and detection of the membrane were conducted by using the nonradioactive DIG system from Boehringer Mannheim following the manufacturer's protocols.
Isolation of Total RNA and RNA-Blot Analysis
Total RNA was isolated from 5-d-old T2 seedlings of each transgenic line tested. Only kanamycin-resistant seedlings were used for RNA isolation by TRIzol Reagent (Life Technologies) following the manufacturer's protocols. Total RNA samples (20 μg) were electrophoresed on 1.2% (w/v) agarose/formaldehyde gels and transferred to a nylon membrane (Qiagen). The blot was analyzed by the same DIG system that was used in the DNA gel-blot analysis.
Measurement of Endogenous GA Levels
Approximately 1 g fresh weight of Arabidopsis tissue (7-d-old seedling) was frozen with liquid nitrogen and ground into a fine powder with a mortar and pestle. The frozen powder was transferred to a glass 40-mL centrifuge tube and homogenized (Pro300D, Pro Scientific, Monroe, CT) in 80% (v/v) methanol. Deuterated GA standards (17,17-d2-GA1, 17,17-d2-GA4, 17,17-d2-GA9, and 17,17-d2-GA20; L. Mander, Australian National University) were added to a level of 0.2 ng/mL before homogenization. The homogenate was filtered (Whatman no. 42 filter paper) and the filtrate was added to a 6-mL C18 chromatography column (Bakerbond spe, J.T. Baker). GAs were eluted with 4 mL of 80% (v/v) methanol and the methanol was evaporated under vacuum. The remaining aqueous phase was adjusted to pH 3.0 with HCl and partitioned three times against hydrated ethyl acetate. The combined ethyl acetate fractions were evaporated under a vacuum, resuspended in 35% methanol containing 0.05% (v/v) acetic acid, and filtered (0.25 μm, 25 mm Nylon Acrodisc, Gelman Sciences, Ann Arbor, MI). The filtered extract was injected onto a C18 reverse-phase column (Xpertek Spherisorb ODS-2, 5 μm, 4.6 mm × 250 mm) and eluted at a flow rate of 1 mL/min with a 40-min linear gradient from 35% to 100% (v/v) acidified methanol controlled by a gradient controller (model 680, Waters).
One-milliliter fractions were collected and pooled according to the expected retention of GAs of interest, as determined by previous chromatography of tritiated standards for GA1, GA4, and GA9 (obtained from R. Pharis, University of Calgary, Alberta, Canada). Pooled HPLC fractions were evaporated under a vacuum and resuspended in 300 μL of methanol. Each sample (100 μL) was methylated with diazomethane (10–20 μL) in a 1-mL glass vial at room temperature. Excess diazomethane and its solvent were removed with a stream of nitrogen. Each methylated sample (except GA9) received 1 μL of pyridine and 50 μL of N,O-bis(trimethylsilyl)trifluroracetamide and was heated at 70°C for 45 min. Excess N,O-bis(trimethylsilyl)trifluroracetamide was removed with a stream of nitrogen. An aliquot of each sample was injected into a gas chromatograph for GC-selected ion monitoring-MS. The gas chromatograph was typically programmed from 100°C to 300°C at 10°C/min. The MS signal-peak-height method for endogenous GAs was used, and the di-deuterated GAs were chosen for quantitation.
RESULTS
Transformation of Arabidopsis Plants with a 20-Oxidase cDNA Driven by an Enhanced 35S Promoter
Primers were designed based on the reported Arabidopsis 20-oxidase sequence from Xu et al. (1995). These primers were used to isolate a 20-oxidase full-length cDNA fragment from 4-d-old Arabidopsis seedling RNA by reverse-transcriptase PCR. The sequence of the PCR-amplified 20-oxidase cDNA was identical to that of GA5 (accession no. U20872) except for four base-pair changes. Three were silent substitutions and one resulted in the conversion of Lys-310 to Glu. These base-pair changes were unlikely to be mutations resulting from PCR because they appeared in several independent PCR-amplified 20-oxidase cDNA sequences. One possible explanation for the discrepancies is the difference in ecotypes of Arabidopsis used for the DNA preparation. The GA5 sequence was isolated from the Landsberg erecta ecotype.
The binary Ti plasmid (pMON29925) containing the 20-oxidase cDNA driven by an enhanced 35S promoter was introduced into Arabidopsis via A. tumefaciens-mediated vacuum infiltration (Bechtold et al., 1993). Transgenic T1 lines were selected by growth on kanamycin-containing medium. Among the 36 transgenic T1 plants identified, 33 were considerably taller than the control plants, which had been transformed with another binary Ti plasmid containing GUS driven by the same promoter. All of the 33 T1 plants showed phenotypic segregation in their T2 progeny. We randomly chose four segregating T1 lines (25-1, 25-2, 25-3, and 25-24) for further studies.
Phenotypic and Molecular Characterization of Four Transgenic Lines
Approximately 30 T2 seeds from each of the four T1 lines were planted, and leaf samples collected from each T2 plant were tested for the presence of the transgene by PCR. Each plant in the segregating T2 population was placed into one of two groups, depending on the presence or absence of the transgene. The physical characteristics were then measured and compared between these two groups from each line. Table I summarizes the data obtained from all of the T2 populations. In all four T2 populations examined, the T2 plants containing the transgene were taller than their isogenic counterparts by at least 15% and up to 30%, as seen in lines 25-2 and 25-3. The increased height of all four transgenic lines is attributable to increased internode length. The plants containing the transgene also had lighter-green leaves, which was more pronounced in lines 25-2 and 25-3.
Table I.
Phenotypic evaluation of 20-oxidase-overexpressing lines
| Line | PCR Result | Plants | Terminal Heighta | Average Internode Lengthb | (kanr/kans)c | χ2 (P at 3:1) |
|---|---|---|---|---|---|---|
| no. | cm | |||||
| 25-1 | + | 17 | 39.6 ± 1.3 | 0.98 ± 0.03 | 106 /36 | 0.01 |
| − | 11 | 34.3 ± 0.9 | 0.85 ± 0.04 | (0.80 –0.50) | ||
| 25-2 | + | 31 | 41.1 ± 0.4 | 0.98 ± 0.01 | 119 /48 | 1.25 |
| − | 10 | 30.2 ± 1.1 | 0.84 ± 0.02 | (0.50 –0.20) | ||
| 25-3 | + | 17 | 45.1 ± 0.6 | 0.97 ± 0.02 | 150 (40)d/55 | 0.84 |
| − | 4 | 33.5 ± 1.9 | 0.84 ± 0.03 | (0.50 –0.20) | ||
| 25-24 | + | 25 | 43.6 ± 1.1 | 0.96 ± 0.02 | 216 /79 | 0.50 |
| − | 6 | 36.5 ± 0.6 | 0.85 ± 0.02 | (0.50 –0.20) | ||
| Wild type | 20 | 33.6 ± 0.6 | 0.90 ± 0.00 | |||
T2 seeds from each of the four T1 lines were planted, and leaf samples collected from each T2 plant were tested for the presence of the transgene by PCR. Each plant from the segregating T2 population was placed into one of two groups, depending on the presence or absence of the transgene. The physical characteristics were then measured and compared between these two groups for each line. Data are means ± se.
Height at which the main stem produced at global proliferative arrest.
Terminal height/number of branches (lateral shoots plus siliques).
Media-grown T2 seedlings were assayed for the presence or the absence of the selectable marker kanamycin by assaying for the bleaching effect of this antibiotic on early seedlings. kanr, Kanamycin-resistant; kans, kanamycin-sensitive.
Of the 150 kanr seedlings listed in this line, 40 were albino, possibly as a result of a recessive mutation caused by T-DNA insertion.
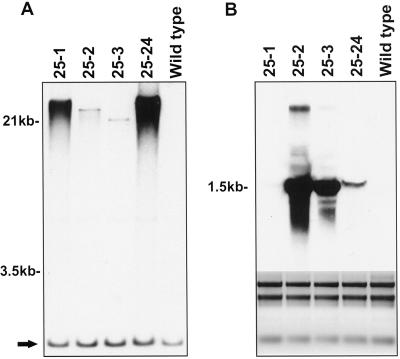
As shown in Table I, the progeny segregation of all four transgenic lines fit a 3:1 ratio, which suggests that they contain an active insertion locus in the genome. This is confirmed by DNA gel-blot analysis (Fig. 1A). When genomic DNA was digested with a restriction enzyme, which cuts outside the T-DNA region, the blot showed only two bands on each lane. The lower bands present in all lines represent endogenous 20-oxidase genes (indicated by the arrow) and the higher-Mr bands are the transgenes. Three more bands appeared on each lane of the blot when a less-stringent hybridization condition was used (data not shown); these were most likely the other 20-oxidase genes described previously in Arabidopsis (Phillips et al., 1995). Although all four lines showed a single insertion locus, lines 25-1 and 25-24 had much stronger signals than the other two lines. This is most likely attributable to tandem repeats of transgenes at the insertion locus in each of two lines; this was verified by repeating the DNA gel-blot analysis with other restriction digestions (data not shown).
Figure 1.
DNA and RNA gel-blot analysis of 20-oxidase-overexpressing plants. A, DNA gel blot containing SpeI digests of genomic DNA (5 μg per lane) isolated from transgenic lines 25-1, 25-2, 25-3, 25-24, and a nontransgenic plant was hybridized with a 20-oxidase cDNA probe. The arrow marks a lower common band that is probably the endogenous 20-oxidase gene. B, RNA gel blot containing total seedling RNA (20 μg per lane) isolated from transgenic lines 25-1, 25-2, 25-3, 25-24, and a nontransgenic plant was hybridized with a 20-oxidase cDNA probe. On the bottom is an ethidium bromide stain of an agarose gel that was loaded with the RNA (1 μg per lane) used in the gel blot.
Total RNA isolated from kanamycin-resistant T2 seedlings of four T1 lines was used for RNA gel-blot analysis. The results show a strong correlation between the amount of transgene mRNA accumulated and the GA-overproduction phenotype (Fig. 1B; lines 25-2 and 25-3). When the blot was exposed for longer periods of time, the more weakly expressing line, 25-1, began to show a similar banding pattern to the other lines; however, endogenous expression of 20-oxidase was still not detectable under these conditions. The lines containing many copies of the transgene per locus (25-1 and 25-24) had lower steady-state RNA levels than lines 25-2 and 25-3, which had fewer copies of the transgene. This may be attributable to gene silencing, a phenomenon that has been observed in other transgenic plant systems (Meyer, 1996).
In each line, seeds produced by each homozygous transgenic plant were collected and pooled to obtain a homozygous transgenic T3 population. All of the studies described below were based on the homozygous transgenic T3 population of each line, except for line 25-3, in which hemizygous plants were used. The integration of the transgene in line 25-3 caused a recessive albino mutation.
Elongated Hypocotyl in Transgenic Seedlings
Many plant species have abnormally elongated stems when treated with exogenous GA. A comparable phenomenon was observed in transgenic lines even at the seedling stage. Transgenic and wild-type seeds were germinated on vertical plates, as shown in Figure 2A, so that their hypocotyl and root lengths could be more easily measured. The seedlings were grown under either 16 h of light or total darkness with or without GA in the medium. All transgenic seedlings were taller than wild-type seedlings when grown in the light. The hypocotyls of 7-d-old seedlings of lines 25-2 and 25-3 were approximately twice the length of the wild-type hypocotyls (4.2 and 3.8 versus 2.0 mm), whereas the root lengths remained the same (Table II). Both transgenic and wild-type seedlings were responsive to exogenous GA3, but the differences in hypocotyl length were not as dramatic under these conditions. Root lengths of transgenic and wild-type seedlings were not influenced by exogenous GA3. When seeds were germinated in the dark, the hypocotyls did not show a significant difference between transgenic and wild-type seedlings regardless of the presence or absence of GA3 in the medium. The T3 generations of lines 25-2 and 25-3 retained the strong GA-overproduction phenotype, whereas lines 25-1 and 25-24 displayed only a slight phenotype.
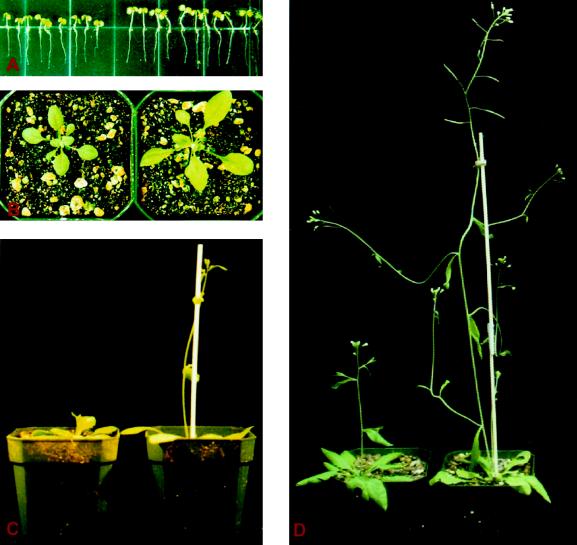
Figure 2.
Promoted growth of 20-oxidase-overexpressing plants. The pictures compare the growth of transgenic (right) and wild-type (left) plants at 1 week (A), 3 weeks (B), 3.5 weeks (C), and 4 weeks of age (D). The plants shown are from the T3 generation of transgenic line 25-2. The transgenic plants are homozygous for the transgene and the wild-type plants are their nontransgenic siblings.
Table II.
The effect of light and GA3 on 7-d-old T3 seedlings
| Line | Light-Grown
Seedlings
|
Dark-Grown Seedlings
|
||
|---|---|---|---|---|
| −GA | +GA | −GA | +GA | |
| Hypocotyl length | mm | |||
| 25-1 | 2.4 ± 0.09 | 4.4 ± 0.10 | 20.1 ± 0.29 | 19.0 ± 0.48 |
| 25-2 | 4.2 ± 0.10 | 5.1 ± 0.37 | 21.0 ± 0.29 | 18.3 ± 0.42 |
| 25-3a | 3.8 ± 0.10 | 4.6 ± 0.21 | 20.2 ± 0.40 | 22.1 ± 0.50 |
| 25-24 | 2.6 ± 0.09 | 3.7 ± 0.10 | 20.5 ± 0.37 | 20.2 ± 0.39 |
| Wild type | 2.0 ± 0.10 | 4.3 ± 0.20 | 21.2 ± 0.34 | 19.9 ± 0.28 |
| Root length | ||||
| 25-1 | 17.2 ± 0.70 | 20.6 ± 0.55 | 13.7 ± 0.34 | 12.9 ± 0.48 |
| 25-2 | 18.0 ± 0.64 | 18.4 ± 0.59 | 14.2 ± 0.40 | 13.4 ± 0.43 |
| 25-3a | 17.5 ± 0.91 | 22.9 ± 0.56 | 17.8 ± 0.39 | 15.8 ± 0.35 |
| 25-24 | 19.0 ± 0.56 | 20.0 ± 0.65 | 15.0 ± 0.43 | 13.8 ± 0.30 |
| Wild type | 18.7 ± 0.77 | 19.1 ± 0.67 | 13.7 ± 0.42 | 13.7 ± 0.30 |
Transgenic and wild-type seeds were germinated on vertical plates, as shown in Figure 2A. These seedlings were grown under either 16 h of light or total darkness with (+) or without (−) GA3 (10 μm) in the medium. Data are means ± se (n < 30).
The integration of the transgene in line 25-3 resulted in a recessive albino mutation; therefore, hemizygous transgenic plants of line 25-3 were used to generate data for this and the following tables.
Growth Cycle of Transgenic Plants
GAs have been shown to induce flowering, especially in rosette plants (for review, see Zeevaart, 1983), and we tested whether plants overexpressing 20-oxidase would flower earlier than their wild-type counterparts. In experiments done in the T3 generation, transgenic plants of lines 25-2 and 25-3 bolted 4 to 5 d (18.5 and 18.3 versus 23.0 d) earlier and had fewer rosette leaves than wild-type plants (Table III). However, consistent with the vegetative data, plants of lines 25-1 and 25-24 were indistinguishable from wild-type plants. The addition of exogenous GA3 accelerated flowering in transgenic lines 25-1, 25-24, and wild-type plants, with these plants approaching the floral-timing pattern of the 25-2 and 25-3 plants. Thus, the addition of exogenous GA3 to wild-type plants can phenocopy the phenotype observed in transgenic lines 25-2 and 25-3 for both floral timing and hypocotyl length (Tables II and III).
Table III.
The flowering time of transgenic plants under long days
| Line | Without GA
|
With
GAa
|
||
|---|---|---|---|---|
| Days | Leaf no. | Days | Leaf no. | |
| 25-1 | 23.6 ± 0.2 | 11.1 ± 0.3 | 20.7 ± 0.3 | 7.5 ± 0.2 |
| 25-2 | 18.5 ± 0.3 | 7.4 ± 0.2 | 18.6 ± 0.3 | 6.9 ± 0.3 |
| 25-3 | 18.3 ± 0.2 | 7.7 ± 0.2 | 18.4 ± 0.2 | 7.3 ± 0.2 |
| 25-24 | 23.4 ± 0.2 | 10.5 ± 0.2 | 19.8 ± 0.2 | 7.8 ± 0.2 |
| Wild type | 23.0 ± 0.2 | 10.4 ± 0.3 | 19.0 ± 0.3 | 7.3 ± 0.2 |
Transgenic and wild-type plants were grown in alternating periods of 16 h of light and 8 h of dark, and the number of days for primary bolt to reach a height of 1 cm for each plant was recorded. Data are means ± se (n = 16).
10 μm GA3 spread daily.
Observations of the overall pattern of growth in the T3 generation of these 20-oxidase-overexpressing plants revealed not only increased stature but also a shortened growth cycle in comparison with wild-type plants. As indicated in Table IV and Figure 2, height differences between transgenic lines 25-2 and 25-3 and wild-type plants are obvious throughout the entire growth cycle. Because the 25-2 and 25-3 lines flower earlier than the wild-type plants, the differences in height measurements by days were inflated. However, height measurements at the terminal stage clearly indicated that lines 25-2 and 25-3 were significantly taller than wild-type plants. Plants of lines 25-2 and 25-3 reach 47.2 and 49.5 cm at global proliferative arrest of the inflorescence meristems (Hensel et al., 1994) in comparison with 40.2 cm for the wild-type plants. It is interesting that the time required to reach these heights was 37.4 and 37.2 d for the plants of lines 25-2 and 25-3, respectively, versus 39.4 d for the wild type. In addition, these transgenic plants had fewer tertiary inflorescence branches than wild-type plants. These observations suggest that GA promotes primary growth and accelerates development throughout the plant's life cycle, probably at the expense of reducing tertiary branches. Transgenic lines 25-1 and 25-24 grew similarly and responded to exogenous GA3 like wild-type plants. In summary, these results suggest that the phenotype displayed by two lines of the transgenic plants (lines 25-2 and 25-3) is inheritable, and similar to that of wild-type plants, with elevated GA levels attributable to exogenous application of GA3. The two other transgenic lines (lines 25-1 and 25-24) lost their phenotype at the T3 generation. However, this so-called “gene-silencing” phenomenon is often observed in other transgenic plant systems (Meyer, 1996), especially in transgenic plants containing multiple copies of transgenes and being carried through multiple generations.
Table IV.
Phenotypes of transgenic plants throughout the T3 growth cycle
| Line | Height
|
Average Speed Set | |||
|---|---|---|---|---|---|
| 24 d | 28 d | 31 d | Terminal (days needed) | ||
| cm | mg/planta | ||||
| Without GA | |||||
| 25-1 | 2.0 ± 0.4 | 10.1 ± 0.8 | 18.5 ± 0.8 | 36.1 ± 0.9 (39.9 ± 0.4) | 38.3 |
| 25-2 | 18.8 ± 0.8 | 31.2 ± 0.8 | 39.9 ± 1.0 | 47.2 ± 1.2 (37.4 ± 0.3) | 72.1 |
| 25-3 | 19.3 ± 0.8 | 32.2 ± 0.8 | 40.3 ± 0.7 | 49.5 ± 0.5 (37.2 ± 0.2) | 68.7 |
| 25-24 | 2.1 ± 0.3 | 10.9 ± 0.8 | 18.6 ± 0.9 | 36.2 ± 0.8 (40.2 ± 0.4) | 49.7 |
| Wild type | 2.8 ± 0.4 | 12.3 ± 0.8 | 21.5 ± 0.6 | 40.2 ± 0.6 (39.4 ± 0.3) | 42.2 |
| With GAb | |||||
| 25-1 | 8.5 ± 0.9 | 19.6 ± 1.2 | 27.7 ± 1.2 | 42.4 ± 1.0 (38.1 ± 0.3) | 46.6 |
| 25-2 | 17.2 ± 1.1 | 28.8 ± 1.1 | 38.5 ± 0.8 | 50.2 ± 0.7 (37.3 ± 0.2) | 39.6 |
| 25-3 | 19.5 ± 0.8 | 32.1 ± 0.8 | 38.7 ± 1.5 | 53.2 ± 0.7 (37.4 ± 0.3) | 72.6 |
| 25-24 | 14.6 ± 0.7 | 26.6 ± 0.8 | 35.8 ± 0.9 | 50.5 ± 0.7 (38.4 ± 0.2) | 77.7 |
| Wild type | 15.7 ± 0.8 | 26.8 ± 0.9 | 35.0 ± 0.8 | 44.5 ± 0.7 (37.4 ± 0.2) | 47.0 |
Data are means ± se (n = 16).
Total amount of seeds harvested 2 weeks after reaching terminal height divided by number of plants.
10 μm GA3 spread daily.
Plants of lines 25-2 and 25-3 produced more seeds than plants of the wild type and lines 25-1 and 25-24, which had lost their phenotype (Table IV). The data seem to suggest that overexpressing 20-oxidase in Arabidopsis plants increases the number of seeds produced. However, when GA3 was applied to these plants, the effect on seed yields was inconclusive (Table IV).
Reduced Seed Dormancy in Transgenic Seeds
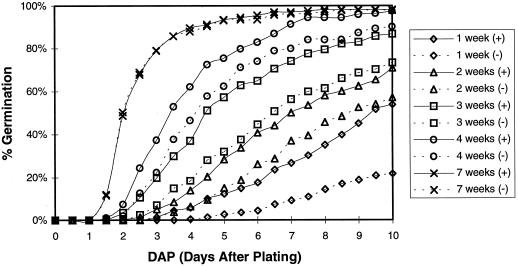
Although seed germination in Arabidopsis can be influenced by a variety of extrinsic factors, GA biosynthesis is a requirement. Several Arabidopsis mutants impaired in the GA biosynthetic pathway are unable to germinate independently and need to be supplied with exogenous GA (Koorneef and van der Veen, 1980). Freshly collected T4 seeds of line 25-2 and wild-type seeds were air dried and stored at room temperature. A small portion of these seeds was sown on germination medium at weekly intervals, and the fraction that had germinated was recorded every 12 h (Fig. 3). After 1 week at room temperature, transgenic seeds began to germinate 3 d after planting and 54% germinated within 10 d. In contrast, wild-type seeds did not germinate until 4 d after planting and only 22% germinated within 10 d. As the seeds aged, dormancy decreased, as expected. Transgenic seeds consistently germinated better than wild-type seeds until the seed had been stored for 7 weeks. At this time, wild-type seeds germinated as well as transgenic seeds and both seeds had nearly 100% germination efficiencies a few days later. However, when plated seeds were cold treated for 4 d before being incubated for germination, both transgenic and wild-type seeds germinated well, even with less than 1 week of storage at room temperature.
Figure 3.
Germination efficiency comparison of freshly collected transgenic line 25-2 T4 and wild-type seeds. Germination efficiencies of seeds collected from the T3 generation of transgenic plants (+) and wild- type (−) plants are shown. Seeds were plated on medium for germination at 1 (⋄), 2 (▵), 3 (□), 4 (○), and 7 (×) weeks after harvesting. Transgenic plants are indicated by solid lines and wild-type plants are indicated by dashed lines.
Analysis of Endogenous GA Levels in Transgenic Seedlings
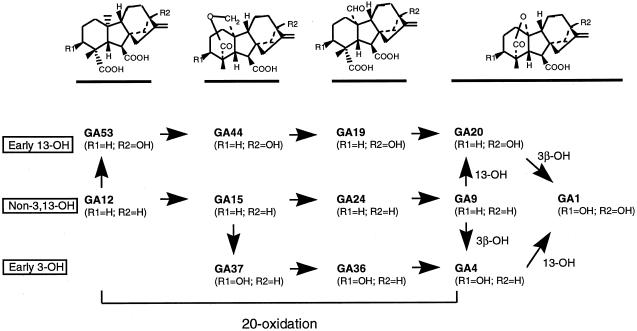
In Arabidopsis three parallel steps of 20-oxidation in the GA biosynthesis pathway have been suggested to produce GA4, GA9, and GA20 (Talon et al., 1990; Zeevaart and Talon, 1992). Subsequently, GA20 can be 3β-hydroxylated or GA4 can be 13-hydroxylated to produce GA1 (Fig. 4). To help differentiate between these biosynthetic routes, we measured endogenous concentrations of GA1, GA4, GA9, and GA20 in T4 seedlings of line 25-2 and wild-type seedlings (Fig. 5). In 7-d-old plants there were higher concentrations of GA1 and GA20 in transgenic than in wild-type seedlings, whereas the concentration of GA4 remained about the same. These results have confirmed that 20-oxidase-overexpressing seedlings have higher 20-oxidase activity, mainly in the early 13-hydroxylation pathway of the GA biosynthetic pathway (Fig. 4). However, we cannot conclude that the 20-oxidase activity also increases in the other pathways. The endogenous level of GA9 increased slightly, but the level of GA4 in the seedlings under these conditions did not change (Fig. 5).
Figure 4.
Proposed three endogenous GA 20-oxidations in Arabidopsis. Different derivatives of C20-GAs can be oxidized by endogenous 20-oxidases to produce C19-GAs. The early 13-hydroxylation pathway leads to GA20; the non-3,13-hydroxylation pathway leads to GA9; and the early 3-hydroxylation pathway leads to GA4. GA9 can be 13-hydroxylated and 3β-hydroxylated to give GA20 and GA4. GA4 can be 13-hydroxylated and GA20 can be 3β-hydroxylated to give to GA1.
Figure 5.
Endogenous GA levels in line 25-2 T4 and wild-type seedlings. Endogenous GA1, GA4, GA9, and GA20 levels in 7-d-old seedling were measured by GC-MS after GA extraction. The amounts are indicated by nanograms of GA in 1 g of seedlings (fresh weight).
DISCUSSION
20-Oxidase activity and expression have previously been shown to be regulated by environmental or physiological changes (Gilmour et al., 1986; Hedden and Croker, 1992; Xu et al., 1995), suggesting that oxidation at carbon-20 of GA is a key regulatory step in the GA biosynthetic pathway. We have confirmed these results by overexpressing the GA5 cDNA in Arabidopsis, which gives rise to a GA-overproduction phenotype in transgenic plants. The phenotype includes elongated stems, early flowering, reduced seed dormancy, and an accelerated growth cycle. This phenotype is similar to spy mutants, whose mutations turn on the GA signal pathway constitutively (Jacobsen and Olszewski, 1993), and the wild-type plants repeatedly treated with exogenous GA. These similarities strongly suggest that the phenotype in 20-oxidase-overexpressing lines is a result of elevated endogenous GA levels.
The transgenic GA-overproduction phenotype can be further enhanced by the addition of exogenous GA, which is also observed in spy mutants (Jacobsen and Olszewski, 1993). As shown in Table II, transgenic plants exposed to exogenous GA3 exhibited further increased seedling hypocotyl length. However, unlike spy mutants, which have greatly reduced seed set, the 20-oxidase-overexpressing transgenic plants are fully fertile and even display an increase in seed set. This could be attributable to differences between the response of floral tissues to constitutive activation of the GA signal transduction pathway and their response to elevated endogenous GA levels. Alternatively, the observed differences may be the result of reduced expression of the enhanced 35S promoter driving 20-oxidase in reproductive tissues.
Although the 20-oxidase-overexpressing plants accumulate more GA than wild-type plants and display a GA-overproduction phenotype under normal growth conditions, there was no obvious phenotypic difference between them when they were grown under conditions known to increase GA sensitivity in the plants. For example, when seedlings were grown in the dark, there was little difference in hypocotyl length between transgenic and wild-type seedlings either in the presence or absence of exogenous GA in the medium (Table II). This may be attributable to the saturation of the GA response in dark-grown seedlings, a hypothesis supported by the observation that Arabidopsis phyB mutant seedlings are more sensitive to GA (Reed et al., 1996). Presumably, the endogenous GA of wild-type plants exerts a maximal effect on hypocotyl elongation when the seedlings are grown in the dark, and the excess GA produced in transgenic plants or supplied exogenously does not exaggerate the extended hypocotyl phenotype. Similarly, freshly collected transgenic seeds germinated better than wild-type seeds of the same age, but no difference was apparent between transgenic and wild-type seeds stored at room temperature for more than 7 weeks or cold treated for 4 d. It has been suggested that GA sensitivity is independent of GA biosynthesis during Arabidopsis seed germination and that aging and chilling increase responsiveness (Koornneef and Karssen, 1984). The 20-oxidase-overexpressing transgenic seeds accumulate more GA and, therefore, are able to germinate at a less GA-sensitive stage, when higher GA levels are required to stimulate the response. Once seeds are ripened or stratified, their increased GA sensitivity allows wild-type and transgenic seeds to germinate equally well (even though there is more GA produced in transgenic seeds).
GA1 is detected in Arabidopsis shoots, but GA4 is thought to be the primary active GA in Arabidopsis (Talon et al., 1990; Zeevaart and Talon, 1992). Also, Phillips et al. (1995) showed that when expressed in Escherichia coli, 20-oxidases preferred GA12 (non-13-hydroxylated GAs) to GA53 (13-hydroxylated GAs). In our studies, we detected increased levels of GA1 but not GA4 in the plants of the 25-2 line. The accumulation of GA20 suggests that the overproduced GA1 in transgenic plants is derived from 3β-hydroxylation of GA20 rather than from 13-hydroxylation of GA4 (Fig. 4). This finding seems to indicate that the overexpression of this 20-oxidase cDNA isolated from seedlings promotes the 13-hydroxylation GA biosynthesis pathway in transgenic seedlings, and that the accumulation of GA1, the end product of that pathway, causes the phenotype. However, with only one transgenic line and four GAs analyzed, these results are preliminary. It is difficult to characterize the kinetics of overall GA biosynthesis without additional transgenic lines and endogenous GAs being analyzed.
The phenotype exhibited in these transgenic plants is of agronomic interest, particularly the accelerated growth cycle that promotes primary stem growth with no reduction in seed set. Such characteristics may be useful, for example, in forest cultivation, in which the growth rate and primary stem mass are of greater concern, or in areas in which a rapid-cycling cultivar would allow multiple plantings and harvests. In fact, application of GA has long been used as a way to increase the economic value of agriculture (Carlson and Crovetti, 1990). Here we provide an alternative approach to GA application by genetic engineering.
In summary, our findings suggest that 20-oxidase is the rate-limiting step in the GA biosynthesis pathway in Arabidopsis. We demonstrate the feasibility of manipulating the endogenous GA levels through genetic engineering. The phenotypes displayed by these transgenic plants not only have interesting agricultural implications but are also useful for basic GA research.
ACKNOWLEDGMENTS
We thank Debbie Stone and Wendi Zumwalt for plant transformation and Greg Heck for valuable comments on the manuscript.
LITERATURE CITED
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Carlson RD, Crovetti AJ. Commercial uses of gibberellins and cytokinins and new areas of applied research. In: Pharis RP, Rood SB, editors. Plant Growth Substances. Heidelberg, Germany: Springer Verlag; 1990. pp. 604–610. [Google Scholar]
- Chiang HH, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Kao TH. The flanking regions of two Petunia inflata S alleles are heterogeneous and contain repetitive sequences. Plant Mol Biol. 1992;18:725–737. doi: 10.1007/BF00020014. [DOI] [PubMed] [Google Scholar]
- Crozier A (1983) The Biochemistry and Physiology of Gibberellins, Vol 2. Praeger, New York
- Fang RX, Nagy F, Sivasubramaniam S, Chua NH. Multiple cis regulatory elements for maximal expression of cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell. 1989;1:141–150. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Zeevaart JAD. Gibberellins and abscisic acid. In: Somerville CR, Meyerowitz EM, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1984. pp. 523–553. [Google Scholar]
- Gilmour SJ, Zeevaart JAD, Schwenen L, Graebe JE. Gibberellin metabolism in cell-free extracts from spinach leaves in relation to photoperiod. Plant Physiol. 1986;82:190–195. doi: 10.1104/pp.82.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Croker SJ. Regulation of gibberellin biosynthesis in maize seedling. In: Karssen CM, Van Loon LC, Weugdenhil D, editors. Progress in Plant Growth Regulation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 534–544. [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB. The fate of inflorescence meristem is controlled by developing fruits in Arabidopsis. Plant Physiol. 1994;106:863–876. doi: 10.1104/pp.106.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM. Seed dormancy and germination. In: Somerville CR, Meyerowitz EM, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1984. pp. 313–334. [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana L. Heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Meyer P. Repeat-induced gene silencing: common mechanisms in plants and fungi. Biol Chem Hoppe-Seyler. 1996;377:87–95. [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1059. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J. Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 1996;112:337–342. doi: 10.1104/pp.112.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JCN, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR application. BioTechniques. 1993;14:748–751. [PubMed] [Google Scholar]
- Sun TP, Goodman HM, Ausubel FM. Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell. 1992;4:119–128. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci USA. 1990;87:7983–7987. doi: 10.1073/pnas.87.20.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase. Molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Sun TP, Kawaide H, Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998;116:1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD (1983) Gibberellins and flowering. In A Crozier, ed, The Biochemistry and Physiology of Gibberellins, Vol 2. Praeger, New York, pp 333–374
- Zeevaart JAD, Talon M (1992) Gibberellin mutants in Arabidopsis thaliana. In CM Karssen, LC Van Loon, D Weugdenhil, eds, Progress in Plant Growth Regulation. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 34–41