Abstract
Immunization with whole cells has been used extensively to generate monoclonal antibodies, produce protective immune responses, and discover new disease antigens. While glycans are abundant on cell surfaces, anti-glycan immune responses have not been well-characterized. We used glycan microarrays to profile 49 tumor-binding monoclonal antibodies generated by immunizing mice with whole cancer cells. A substantial proportion (41%) of the tumor binding antibodies bound carbohydrate antigens. The antibodies primarily recognize a group of 5 glycan antigens: Sialyl Lewis A (SLeA), Lewis A (LeA), Lewis X (LeX), blood group A (BG-A), and blood group H on a type 2 chain (BG-H2). The results have important implications for monoclonal antibody production and cancer vaccine development.
Keywords: Glycan Array, Antibody, Carbohydrate, Tumor antigen, Cancer vaccine
Immunization with whole cells is a strategy that has been used frequently for generating monoclonal antibodies, producing protective immune responses, and discovering novel disease antigens.1–6 For example, vaccines composed of irradiated tumor cells have been evaluated in a variety of clinical trials for the treatment of cancer.7–9 Immunization with whole cells has a number of advantages for these applications, such as 1) prior identification of a specific target antigen is not necessary, 2) the antigens are presented to the immune system in a natural context, 3) one can obtain antibodies to a variety of antigens in parallel, and 4) one can discover previously unknown antigens of interest. To develop innovative tumor-targeted diagnostic and therapeutic agents and maximize the information obtained from this approach, identification of the target antigens is critical. Unfortunately, cells display a complex variety of antigens to the immune system, making it difficult to determine which antigens are recognized by the induced antibodies. This problem has been especially true for antibodies that bind carbohydrate antigens, due to the challenges associated with identifying and characterizing carbohydrate-protein interactions. Therefore, new strategies to rapidly identify relevant antigens, especially glycan antigens, are needed.
Our hypothesis was that a large percentage of antibodies produced upon immunization with whole tumor cells would target glycan antigens based on the following facts: 1) glycans are an abundant class of antigens on cells, with numerous carbohydrates displayed in the form of glycoproteins and glycolipids; 2) glycans are often the most exposed determinants on cell surfaces; and 3) the repertoire of glycans produced in cells changes significantly with the onset and progression of cancer.10 Presently, a number of carbohydrates with altered expression on malignant cells have been identified, such as the STn antigen, Globo H, and the Tn antigen. These tumor-associated carbohydrate antigens are being targeted as diagnostic markers and cancer vaccine antigens.11–13
While glycans are potentially an important class of antigens for whole cell vaccination, several factors suggested that anti-glycan antibodies may only be a minor subset of the overall immune response. Glycans on tumor cells are typically over-expressed self antigens or structures that are very similar to self antigens. Therefore, the immune system has evolved tolerance to these antigens. In contrast, glycans found on pathogens, such as bacterial polysaccharides, can be highly immunogenic. As evidence of poor immunogenicity, induction of immune responses to tumor-associated carbohydrates is known to be frustratingly difficult.14 Nevertheless, previous studies have shown that immunization with whole tumor cells can, at least in some cases, produce anti-glycan antibodies (for some examples15–23); however, the spectrum of anti-glycan antibodies that are produced has not been well characterized. In addition, the proportion of antibodies targeting carbohydrates relative to other antigen families is not known.
To address these questions, we combined a functional screening assay with glycan array technology. The strategy involved immunizing mice with a colon cancer cell line, generating a large panel of monoclonal antibodies, screening for antibodies with the ability to bind colon cancer cells, and profiling antibody binding to a large, diverse collection of carbohydrates in parallel using glycan arrays.24–27 It is important to note that this approach focuses on the subset of antibodies with tumor binding ability, rather than providing a general evaluation of the overall response. This strategy was selected to specifically assess the importance of glycan antigens as targets for monoclonal antibody development and cancer vaccine efficacy.
To obtain a library of hybridomas, BALB/c mice were immunized with whole colon cancer NSY cells. These cells have previously been used to successfully obtain monoclonal antibodies that stain human tumors and inhibit tumor cell migration and invasion.20 Briefly, Titermax™ Gold adjuvant (25 μL; Sigma Chemical Co.) and human colon carcinoma NSY cells (1 × 106) were injected intraperitonealy into 6-week-old female BALB/c mice once a week for 4 weeks.28 Three days before euthanasia, mice were boosted with the same doses of adjuvant and tumor cells as used before. Spleen cells from a mouse with a serum titer >4,000X were used for fusion. The hybridoma library was established by fusion of both spleen cells from the immunized BALB/c mice and myeloma cells (P3/x63.Ag8) at 5:1 ratio with polyethylene glycol (PEG)-1500 (Sigma Chemical Co.) following standard procedures.29 The PEG fused cells were seeded into 96 well flat-bottom plates and allowed to grow for 10 to 15 days. The supernatants from the hybridomas were collected for screening.
Next, hybridomas were screened for secretion of antibodies that bind to tumor cells. To select antibodies that target cell surface antigens, an immunofluorescence staining assay was applied to cultured cells without fixation and permeation. Note that living cells with intact cell membranes prevent IgG (150KD) from entering into the cells passively. Briefly, we seeded 5,000 cells/well in 96 well plates and allowed them to continuously culture in an incubator at 37 °C for 48 h. After decanting the culture medium, 50 μL of supernatant from each well of the hybridoma culture plate was added and incubated for 40 min at 37 °C. The plates were washed three times with PBS and 50 μL of 1000X diluted Alexa flour 488-conjugated goat anti-mouse IgG antibody (Molecular Probes) was added to the plates, which were incubated for another 40 min at room temperature. The plates were washed with PBS and observed under a fluorescence microscope.
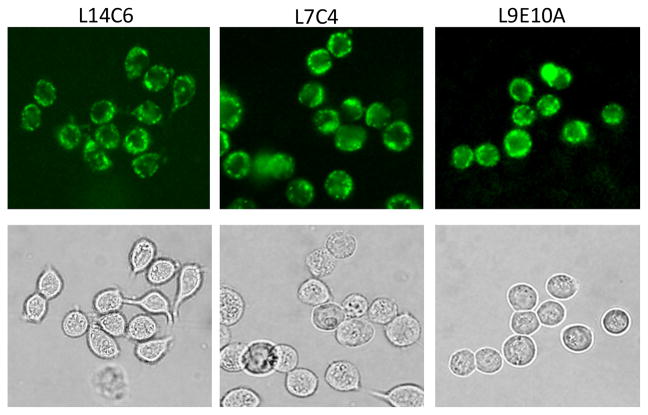
Indeed, antibodies in hybridoma supernatants intensively stained the surface membranes of cultured NSY cancer cells as demonstrated in Figure 1. By using this immunofluorescence staining approach, more than 300 hybridoma clones that secrete antibodies reacting to human colon carcinoma NSY cells were obtained. We randomly chose 49 antibody-containing hybridoma supernatants for glycan array analysis.
Figure 1.
Representative immunofluorescence staining images demonstrating tumor cell membrane staining with L14C6, L7C4, and L9E10A monoclonal antibodies. Cultured human colon cancer NSY cells were stained with supernatants of the hybridomas.
Potential glycan binding was evaluated using a glycan microarray, a microscope slide containing many different carbohydrates immobilized on the surface in a spatially defined arrangement.24–27 Prior to printing, glycans were covalently attached to a protein carrier (typically albumin) to form a neoglycoprotein. The array format and assay have been described previously,30 along with analysis of reproducibility31 and validation with numerous antibodies and lectins.32–37 The particular version of the array used for this study contained 191 array components encompassing a diverse collection of N-linked glycans, O-linked glycans, glycopeptides, and glycolipids. The majority of the components were human glycans, but some non-human glycans (8%) and glycoproteins (15%) were also present. A full list of array components and their abbreviations can be found in the Supporting Information. Positive signals were defined as components producing a signal 3 times higher than the background.38
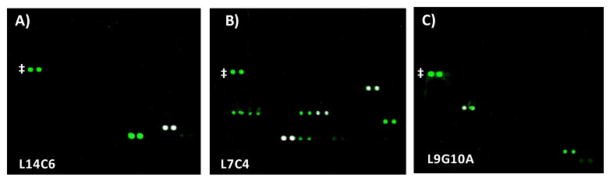
Of the 49 antibodies tested, 20 bound well to at least 1 glycan on the array. As seen in Figure 2, the array images show clear, distinct, and specific fluorescent signals for one to several glycans for each of these antibodies. In addition, the signals were at least 100 fold higher than the background. Of the remaining 29 antibodies, 2 showed relatively weak binding to a broad range of array components.39 Since the signals were low and binding was also observed to recombinant human serum albumin, this binding was considered non-specific and these antibodies were not classified as “glycan binders”. The other 27 antibodies showed no binding at all on our array.
Figure 2.
Representative array images illustrating specific glycan recognition. Antibodies were incubated on the array and binding was detected with Cy3-labeled goat anti-mouse Ig. Each array component was printed in duplicate, resulting in pairs of spots. Control spots, Cy3-labeled BSA, are marked with a ‡. A) Antibody L14C6, which preferentially binds LeA and LeX; B) Antibody L7C4, which preferentially binds BG-A, BG-A1, and glycoproteins displaying blood group A; C) Antibody L9G10A, which binds BG-H2 and maltose.
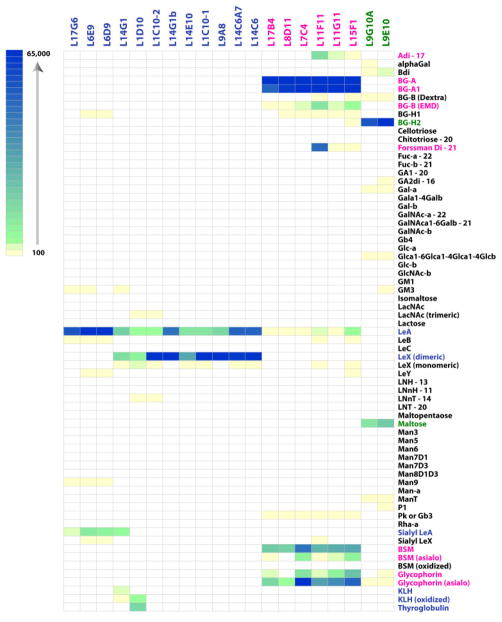
Interestingly, the group of glycan binding antibodies reacted with a relatively small set of glycans (see Figure 3). In particular, all 20 of these antibodies specifically recognized one or more Lewis antigens or blood group antigens (see Figure 4). Interestingly, no binding to other well-known tumor-associated carbohydrate antigens, such as Tn, TF, type 1 precursor, LNT, Gb3 a GM3, was observed for any of the antibodies. For the discussion below, the 20 glycan-binding antibodies were organized into 3 families: Lewis antigen binders, blood group A/A1 binders, and blood group H2 binders.
Figure 3.
Heat Map of Binding Profiles for the 20 Glycan-Binding Antibodies. Data for each antibody is shown in columns. Data for each glycan is shown in rows. Colored boxes correspond to the raw fluorescence units for a given glycan; white boxes indicate no significant signal above the background (signal ≤ 3x the background). Only a subset of the array components is shown (for a full list and complete data, see the Supporting Information). Antibodies have been grouped by family. Glycans are listed alphabetically with the glycoproteins listed at the bottom LeX/LeA/SLeA binders and the corresponding glycans are shown in blue font; BG-A/A1 binders and the corresponding glycans are shown in pink font; and BG-H2 binders and the corresponding glycans are shown in green font.
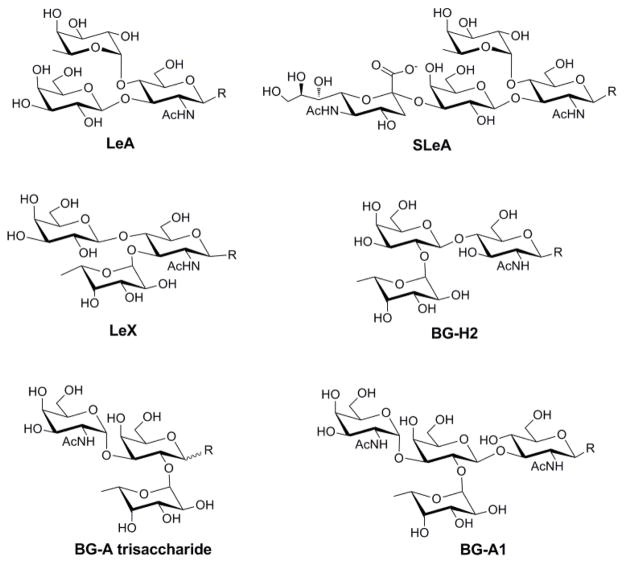
Figure 4.
Structures of selected Lewis and blood group antigens
The majority of the glycan-binding antibodies (12/20) bound 3 closely related Lewis antigens: LeA, LeX, and SLeA. Of these, 8/12 recognized LeA and dimeric Lewis X (LeX dimeric). Interestingly, the antibodies that bound dimeric LeX displayed only weak (>20 fold lower) binding to monomeric LeX. Several antibodies (3/12) bound strongly to LeA and had modest cross-reactivity to SLeA (~7–10 fold lower intensity than LeA). One antibody (L14G1) bound all three antigens.
Expression of Lewis antigens is known to be altered on malignant cells and antibodies to Lewis antigens have been isolated previously after immunization with whole cells.15, 16, 23 SLeA, also known as the CA19-9 antigen, is a well known tumor-associated carbohydrate antigen.11, 23, 40, 41 Monomeric and dimeric forms of LeX are expressed during embryonic development and have increased expression in a number of cancers.11, 15, 42 Dimeric LeX, in particular, has markedly increased expression in colon cancer.42 Antibodies to these glycans likely represent an anti-tumor response. LeA is expressed in a variety of normal tissues, but its expression is often altered in malignant cells.43, 44
The second largest family of antibodies included 6 antibodies that primarily recognized the blood group A antigen (see Figure 3). These antibodies each bound to both the blood group A trisaccharide (BG-A) and the type 1 blood group A tetrasaccharide (BG-A1). The 6 BG-A/A1 antibodies also recognized several glycoproteins, including bovine submaxillary mucin and glycophorin, which are known to display blood group A. Antibodies L11F11, L11G11, and L15F1cross-reacted with a disaccharide substructure of blood group A (Adi; GalNAcα1-3Gal-R), BG-B, and the Forssman disaccharide (Forssman Di; GalNAcα1-3GalNAc-R).
The BG-A glycan is best known as one of the antigens that defines a person’s blood type and contributes to compatibility for blood transfusions and organ transplants.44 BG-A can be present on red blood cells, epithelial cells, and secreted proteins, such as mucins. Red blood cells typically display the BG-A trisaccharide epitope on type 2 and type 4 chains, while epithelial cells and secreted mucins display BG-A on type 1 and type 3 chains. Altered expression of blood group antigens is often associated with malignancy, but BG-A and BG-A1 are not considered tumor-associated carbohydrate antigens due to their ubiquitous expression in other tissues.43, 44 Mice are capable of biosynthesizing blood group A glycans, but expression is very weak and restricted to a subset of tissues, such as the colon. In addition, blood group A epitopes may be displayed on different glycan chains in humans and mice, which could allow the mouse immune system to recognize certain forms as non-self. Therefore, this family of antibodies most likely represents a blood group incompatibility between mice and humans.
The next family of antibodies included 2 clones that bound best to BG-H2. Interestingly, these antibodies both cross-reacted with maltose (Glcα1-4Glc); however, no reactivity with blood group H1 (BG-H1; blood group H on a type 1 chain) was observed. The blood group H antigen (BG-H) is a biosynthetic precursor to the blood group A and blood group B antigens.44 Like blood group A, BG-H is displayed on type 2 and type 4 chains on red blood cells and type 1 and type 3 chains on epithelial cells and secreted proteins. BG-H expression is also altered on malignant cells, and BG-H2 can be over expressed on tumor tissue; however, BG-H2 is not generally considered a tumor-associated antigen.43, 44
These studies provide an extensive analysis of the antigens targeted by tumor cell binding antibodies present after immunization with whole cancer cells. While these antibodies are likely to be part of the induced immune response triggered by vaccination, tumor cell binding does not, in and of itself, prove that the antibodies are part of an anti-tumor response. It is possible that some or all of these antibodies are part of the natural repertoire of antibodies present in mice. Several lines of evidence suggest that this is not the case. First, the vast majority of antibodies in mammals do not bind carbohydrates. Therefore, one would produce very few carbohydrate-binding monoclonal antibodies from the natural B cell population. Second, in other work, we have profiled the natural repertoire of carbohydrate-binding antibodies present in mice.45 We found that they have no measureable antibodies to BG-A1, BG-H2, maltose, and SLeA. Therefore, the 12 antibodies that bind at least one of these glycans appear to be novel antibodies present after immunization, although we cannot rule out the possibility that they were present at a level below our detection limit. Mice occasionally have measurable antibody levels to the LeX and LeA antigens, but levels are very low. Therefore, one would not expect to obtain a high number of LeX or LeA binding monoclonal antibodies from the natural repertoire of B cells. Taken together, these factors indicate that the antibodies we evaluated are most likely induced as part of the response to whole cell immunization.
Our results highlight several key points. First, anti-glycan responses can be an important component of the immune response to whole cells. Nearly half of the antibodies with tumor-binding activity recognized glycan antigens (41%; 20/49). Interestingly, the antibodies primarily targeted a narrow group of 5 glycan antigens: Sialyl Lewis A (SLeA), Lewis A (LeA), Lewis X (LeX), blood group A (BG-A), and blood group H on a type 2 chain (BG-H2). Second, differences in blood group expression on the cells used for immunization and the host or patient can be a major focus of the anti-glycan immune response. This is an especially important consideration during pre-clinical development of cancer vaccines, where foreign or blood group incompatible glycans may dominant the response when immunizing an animal with a human cell vaccine. However, it is not yet clear if this would be beneficial or detrimental for a whole cell cancer vaccine. For example, it is possible that blood group incompatibility could help break tolerance and permit a more vigorous immune response in the same way the expression of the alpha-Gal antigen enhances immune responses when artificially expressed on human tumor cell lines.46, 47 Alternatively, it might focus the immune response on a subset of antigens that will not lead to a productive anti-tumor response. Although additional studies will be required to determine if these results are general for other types of cells, other routes of injection, and other organisms, our study highlights the potential of glycan array technology for high-throughput antigen identification.
Supplementary Material
Acknowledgments
We thank Jack Simpson (Protein Chemistry Laboratory, SAIC/NCI-Frederick) for MALDI-MS analysis of BSA conjugates. This research was supported in part by the Intramural Research Program of the NIH, NCI.
Footnotes
Full list of array components on the glycan array; full microarray data for each of the antibodies; array images for each glycan-binding antibody.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Chiang CLL, Benencia F, Coukos G. Sem Immunol. 2010;22:132–143. doi: 10.1016/j.smim.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eager R, Nemunaitis J. Molecular Therapy. 2005;12:18–27. doi: 10.1016/j.ymthe.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Hsueh EC, Morton DL. Sem Cancer Biol. 2003;13:401–407. doi: 10.1016/j.semcancer.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Copier J, Dalgleish A. Int Rev Immunol. 2006;25:297–319. doi: 10.1080/08830180600992472. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan AM, Wang R, Kappe SHI. Human Vaccines. 2010;6 doi: 10.4161/hv.6.1.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le DT, Pardoll DM, Jaffee EM. Cancer J. 2010;16:304–310. doi: 10.1097/PPO.0b013e3181eb33d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small EJ, Sacks N, Nemunaitis J, Urba WJ, Dula E, Centeno AS, Nelson WG, Ando D, Howard C, Borellini F, Nguyen M, Hege K, Simons JW. Clin Cancer Res. 2007;13:3883–3891. doi: 10.1158/1078-0432.CCR-06-2937. [DOI] [PubMed] [Google Scholar]
- 8.Vermorken JB, Claessen AME, Van Tinteren H, Gall HE, Ezinga R, Meijer S, Scheper RJ, Meijer CJLM, Bloemena E, Ransom JH, Hanna MG, Jr, Pinedo HM. Lancet. 1999;353:345–350. doi: 10.1016/S0140-6736(98)07186-4. [DOI] [PubMed] [Google Scholar]
- 9.Eric L, Yeo CJ, Lillemoe KD, Biedrzycki B, Kobrin B, Herman J, Sugar E, Piantadosi S, Cameron JL, Solt S, Onners B, Tartakovsky I, Choi M, Sharma R, Illei PB, Ralph HH, Abrams RA, Le D, Elizabeth J, Laheru D. Ann Surgery. 2011;253:328–335. doi: 10.1097/SLA.0b013e3181fd271c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dube DH, Bertozzi CR. Nat Rev Drug Discovery. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 11.Heimburg-Molinaro J, Lum M, Vijay G, Jain M, Almogren A, Rittenhouse-Olson K. Vaccine. 2011;29:8802–8826. doi: 10.1016/j.vaccine.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Z, Wang Q. Curr Opin Chem Biol. 2009;13:608–617. doi: 10.1016/j.cbpa.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buskas T, Thompson P, Boons GJ. Chem Comm. 2009:5335–5349. doi: 10.1039/b908664c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keding SJ, Danishefsky SJ. Proc Natl Acad Sci USA. 2004;101:11937–11942. doi: 10.1073/pnas.0401894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rettig WJ, Cordon-Cardo C, Ng JS, Oettgen HF, Old LJ, Lloyd KO. Cancer Res. 1985;45:815–21. [PubMed] [Google Scholar]
- 16.Sakamoto J, Furukawa K, Cordon-Cardo C, Yin BWT, Rettig WJ, Oettgen HF, Old LJ, Lloyd KO. Cancer Res. 1986;46:1553–1561. [PubMed] [Google Scholar]
- 17.Hellstrom I, Garrigues HJ, Garrigues U, Hellstom KE. Cancer Res. 1990;50:2183–2190. [PubMed] [Google Scholar]
- 18.King M, Parsons S, Wu A, Jones N. Transfusion. 1991;31:142–149. doi: 10.1046/j.1537-2995.1991.31291142945.x. [DOI] [PubMed] [Google Scholar]
- 19.Fyfe G, Cebra-Thomas JA, Mustain E, Davie JM, Alley CD, Nahm MH. J Immunol. 1987;139:2187–2194. [PubMed] [Google Scholar]
- 20.Xu M, Wang F, Gildersleeve JC, Achilefu S. Hybridoma. 2010;29:355–9. doi: 10.1089/hyb.2010.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pancino GF, Osinaga E, Vorauher W, Kakouche A, Mistro D, Charpin C, Roseto A. Hybridoma. 1990;9:389–395. doi: 10.1089/hyb.1990.9.389. [DOI] [PubMed] [Google Scholar]
- 22.Meichenin M, Rocher J, Galanina O, Bovin N, Nifant’ev N, Sherman A, Cassagnau E, Heymann MF, Bara J, Fraser RH, Le Pendu J. Cancer Res. 2000;60:5499–5507. [PubMed] [Google Scholar]
- 23.Magnani JL, Nilsson B, Brockhaus M. J Biol Chem. 1982;257:14365–14369. [PubMed] [Google Scholar]
- 24.Rillahan CD, Paulson JC. Annu Rev Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oyelaran O, Gildersleeve JC. Curr Opin Chem Biol. 2009;13:406–13. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Palma AS, Feizi T. Biol Chem. 2009;390:647–656. doi: 10.1515/BC.2009.071. [DOI] [PubMed] [Google Scholar]
- 27.Wu CY, Liang PH, Wong CH. Org Biomol Chem. 2009;7:2247–2254. doi: 10.1039/b902510n. [DOI] [PubMed] [Google Scholar]
- 28.Xu M, Wright WD, Higashikubo R, Roti Roti JL. Int J Hyperthermia. 1996;12:645–660. doi: 10.3109/02656739609027672. [DOI] [PubMed] [Google Scholar]
- 29.Kohler G, Milstein C. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 30.Campbell CT, Zhang Y, Gildersleeve JC. Curr Protocols Chem Biol. 2010;2:37–53. doi: 10.1002/9780470559277.ch090228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oyelaran O, McShane LM, Dodd L, Gildersleeve JC. J Proteome Res. 2009;8:4301–10. doi: 10.1021/pr900515y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manimala J, Li Z, Jain A, VedBrat S, Gildersleeve JC. Chem Bio Chem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 33.Manimala JC, Roach TA, Li ZT, Gildersleeve JC. Angew Chem Int Ed. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 34.Manimala JC, Roach TA, Li Z, Gildersleeve JC. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Li Q, Rodriguez LG, Gildersleeve JC. J Am Chem Soc. 2010;132:9653–62. doi: 10.1021/ja100608w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Campbell CT, Li Q, Gildersleeve JC. Mol BIOS’s. 2010;6:1583–91. doi: 10.1039/c002259d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shorebah MG, Jackson CL, Price PW, Meager R, Schulz T, Godwin AK, CIA Q, Gildersleeve JC. Stem Cells Dev. 2011;20:515–25. doi: 10.1089/scd.2010.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabrication of arrays and assessment of antibody binding was carried out as reported previously with minor modifications. Briefly, the array was blocked with 3% bovine serum albumin (BSA)/PBS for 2 hr, incubated with antibody supernatants at a dilution of 1:5 for 2 hr at room temperature, washed with PBS, and then incubated with 50 μL of Cy3-labeled goat anti-mouse immunoglobulin Ig (detects IgG and IgM; Jackson ImmunoResearch Laboratories) in 3% BSA for 1.5 hr. After washing and drying, the slides were scanned on a GenePix scanner (GenePix 4000A Microarray Scanner, Molecular Devices Corporation, Union City, CA). The fluorescence was quantified by using Gene-Pix Pro 6.0 software with a GenePix Array List file. The value for each array component was obtained by averaging the background corrected median intensities of the two replicate spots. Full array data can be found in the Supporting Information.
- 39.The two antibodies with weak binding to a variety of glycans were M12E9 and 4F211B5. No additional studies were carried out to further characterize these antibodies.
- 40.Sawada R, Sun SM, Wu X, Hong F, Ragupathi G, Livingston PO, Scholz WW. Clin Cancer Res. 2011;17:1024–1032. doi: 10.1158/1078-0432.CCR-10-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magnani JL. Arch Biochem Biophys. 2004;426:122–131. doi: 10.1016/j.abb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Hakamori S, Nudelman E, Levery SB, Kannagi R. J Biol Chem. 1984;259:4672–4680. [PubMed] [Google Scholar]
- 43.Henry S, Oriol R, Samuelsson B. Vox Sang. 1995;69:166–182. doi: 10.1111/j.1423-0410.1995.tb02591.x. [DOI] [PubMed] [Google Scholar]
- 44.Nydegger UE, Tevaearai H, Berdat P, Rieben R, Carrel T, Mohacsi P, Flegel WA. Ann NY Acad Sci. 2005;1050:40–51. doi: 10.1196/annals.1313.006. [DOI] [PubMed] [Google Scholar]
- 45.Yin Z, Nguyen HG, Chowdhury S, Bentley P, Bruckman MA, Miermont A, Gildersleeve JC, Wang Q, Huang X. Bioconjugate Chem. 2012;23:1694–1703. doi: 10.1021/bc300244a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdel-Motal UM, Wigglesworth K, Galili U. Vaccine. 2009;27:3072–3082. doi: 10.1016/j.vaccine.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Rossi GR, Mautino MR, Unfer RC, Seregina TM, Vahanian N, Link CJ. Cancer Res. 2005;65:10555–10561. doi: 10.1158/0008-5472.CAN-05-0627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.