Abstract
Multiple species of bacteria are able to sequester the host zymogen plasminogen to the cell surface. Once localised to the bacterial surface, plasminogen can act as a cofactor in adhesion, or, following activation to plasmin, provide a source of potent proteolytic activity. Numerous bacterial plasminogen receptors have been identified, and the mechanisms by which they interact with plasminogen are diverse. Here we provide an overview of bacterial plasminogen receptors and discuss the diverse role bacterial plasminogen acquisition plays in the relationship between bacteria and the host.
1. Introduction
Recruitment of plasminogen to the bacterial cell surface is emerging as a central theme in host/pathogen interactions. The glycoprotein plasminogen is found in plasma and extracellular fluids at concentrations of approximately 2 μM. Upon activation, plasminogen is converted to the serine protease plasmin [1]. Plasmin is able to degrade fibrin clots, connective tissue, extracellular matrix (ECM), and adhesion proteins. Plasmin itself contributes to a number of amplification loops which leads to increased plasminogen activation. Plasmin-mediated proteolysis of cell membrane proteins exposes cryptic plasminogen-binding sites within receptors, subsequently enhancing the recruitment of plasminogen to cell surfaces [2]. Similarly, cleavage of the inactive form of the urokinase plasminogen activator pro-uPA by cell bound plasmin generates the active two-chain uPA. This feedback activation results in a significant increase in plasmin activation within biological systems [3]. Additionally, activation of prometalloproteases by plasmin results in degradation of the collagen structural components of the ECM, leading to widespread tissue destruction. Recruitment of plasminogen to the surface of bacteria by specific plasminogen receptors was first reported over 20 years ago [4]. Since then, the importance of this interaction in bacterial virulence has become the focus of a large body of research. It is now clear that recruitment of plasminogen to bacterial cell surfaces is a feature common to both pathogenic and commensal bacteria. This paper provides an overview of known bacterial plasminogen receptors and examines the diverse roles they play in the host-bacteria interaction.
2. Plasminogen
Plasminogen is the inactive zymogen form of the enzyme plasmin [5, 6]. Posttranslational processing results in several different forms of plasminogen (Figure 1). The circulating mature form of plasminogen is known as Glu-plasminogen as a consequence of the glutamic acid residue at the N-terminus. Glu-plasminogen consists of the preactivation peptide followed by five characteristic kringle domains and then the serine protease active site in the C-terminal region [6] (Figure 1). The amino acid residues His603, Asp646 and Ser741, make up the catalytic triad of the serine protease domain. This domain catalyses the hydrolysis of peptide bonds, resulting in peptides with C-terminal arginine and lysine residues [6]. The kringle domains of plasmin(ogen) mediate interactions with multiple ligands, including fibrin(ogen) and mammalian cellular plasmin(ogen) receptors [7]. In particular Kringles 1, 2, 4, and 5 (K1-K5) contain lysine-binding sites (LBS) comprised of a hydrophobic cleft formed by aromatic residues that most commonly bind C-terminal lysine residues and internal lysine residues of receptors. As described in Figure 1, the kringles show differing affinities for free lysine and lysine-like compounds such as ω-aminocarboxylic ligands, in the following order of binding affinity K1 > K4 > K5 > K2 [8]. Kringle 3 shows no detectable binding to Lys or Lys-like compounds [9]. Intramolecular binding between lysine residues and the LBS of these Kringles maintains Glu-plasminogen in a closed conformation which is less susceptible to activation [6, 10]. Competitive binding interactions with fibrin(ogen) or plasminogen receptors allows Glu-plasminogen to adopt an open conformation, exposing the activation loop (Arg561-Val562) to cleavage by specific mammalian plasminogen activators thus forming Glu-plasmin [6, 7] (Figure 1). Alternatively, cleavage of the Lys77-Lys78 peptide bond may also occur leaving the plasminogen molecule with a Lys residue at the-N terminus (Lys-plasminogen) [6] (Figure 1). Lys-plasminogen has a more open, U-shaped conformation than Glu-plasminogen making it more readily activated to Lys-plasmin by the plasminogen activators [11, 12]. The resulting two-chain Glu- or Lys-plasmin molecule consists of the plasmin heavy chain A in the N-terminal region and the plasmin light chain B in the C-terminal region held together by interchain disulfide bonds (Figure 1).
Figure 1.
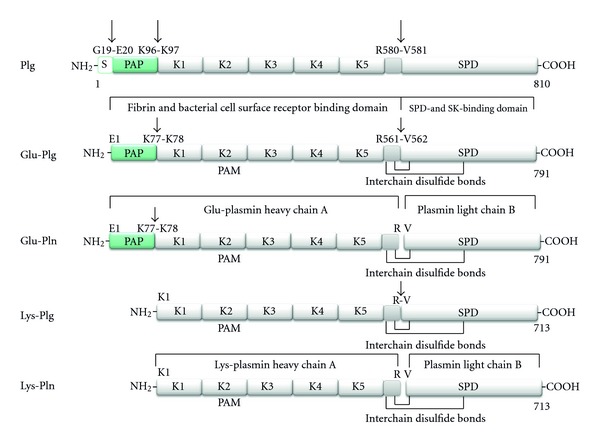
Structural domains of human plasmin(ogen) forms. Human plasminogen is synthesised as an 810 amino acid protein. The 19 amino acid residue signal sequence is removed resulting in the circulating mature form (791 amino acids, ~90,000 kDa) known as Glu-plasminogen (Glu-plg) as it contains an N-terminal glutamic acid. Glu-plg contains a hairpin-loop structure called the PAN domain encompassing the preactivation peptide (PAP), followed by 5 homologous kringle domains (K1–K5) containing three intradomain disulfide bridges, followed by a peptidase S1 domain (SPD). The preactivation peptide is generated by plasmin cleavage giving rise to Lys-plg (713 amino acids, ~80,000 kDa). The conversion of Glu-plg or Lys-plg to their respective plasmin forms occurs by hydrolysis of the Arg-Val peptide bond shown by either uPA or tPA, yielding chain A and the smaller chain B, which remain covalently associated by interchain disulfide bonds. Kringles 1, 2, 4, and 5 contain lysine-binding sites (LBS) with affinity for free lysine and lysine-like compounds such as ω-aminocarboxylic ligands in the following order of binding affinity K1 > K4 > K5 > K2 [8]. Kringle 3 shows no detectable binding to Lys or Lys-like compounds [9], related to a sequence variation in its LBS. Glu-plg thus binds to various lysine-containing proteins via Kringles 1, 2, 4, and 5. Streptokinase (SK) and staphylokinase (not shown) bind in a 1 : 1 complex with the SPD to generate an activator complex. Not shown: Mini plasminogen (K5 plus the SPD) can also be generated by stromelysin-19 cleavage of the Pro466-Val467 bond of Plg. Sequence data are derived from UniProt (swiss-prot entry P00747). Plasminogen (EC = 3.4.21.7) (http://www.uniprot.org/uniprot/P00747).
Two differentially glycosylated variants of human Glu-plasminogen exist. Both variant 1 and 2 contain O-linked glycosylation site, whereas variant 1 contains an additional N-linked glycosylation site (located at Asn289 within Kringle 3) [13–15]. These glycosylation patterns appear to affect both the stability and affinity of the protein to interact with lysine moieties as well as its subsequent activation rate [16]. Differences in glycosylation were recently shown to affect the positioning of Kringle 3 (a non-LBS containing Kringle) in the X-ray crystal structure of plasminogen, which has consequences for efficient Glu-plasminogen activation [10].
3. Plasminogen-Binding Bacteria
Commandeering the host plasminogen activation system is a common mechanism employed by a variety of bacteria [17–20]. The ability to acquire cell surface plasminogen is not host species restricted or limited to specific sites of infection. Rather, the ability to recruit plasminogen is emerging as a central theme in the interaction between host and bacteria. Early studies by Ullberg et al. showed that 5 out of 11 species of gram-negative bacteria tested and 9 out of 17 species of gram-positive bacteria tested displayed a specific and high affinity interaction with Glu-plasminogen [21, 22], although plasminogen acquisition by different strains within each species varied significantly. Many studies have since focused on the ability of highly pathogenic bacteria to interact with plasminogen, including Streptococcus pyogenes, S. pneumoniae, Staphylococcus aureus, Helicobacter pylori, Mycobacterium tuberculosis, Neisseria meningitides, and N. gonorrhoeae, [23–25]. There is also a growing body of evidence to indicate that animal pathogens sequester plasminogen. Examples of this include Mycoplasma hyopneumoniae and M. gallisepticum which bind porcine and chicken plasminogen, respectively, [26, 27] and the canine pathogen S. suis [28].
Interactions with plasminogen are not solely the domain of pathogenic bacteria, with a number of commensal species also reported to bind plasminogen with both high affinity and specificity, including several species of oral streptococci [29], bifidobacteria [30], and lactobacillus [31]. The role of bacterial-plasminogen recruitment in pathogenesis will be discussed in more detail later; however, given the above findings it appears that plasminogen recruitment by bacteria may have a multifaceted role in the interaction with the host. This may underlie the diversity of plasminogen receptors expressed by bacteria and the different mechanisms of interaction which have been described to date.
4. Bacterial Plasminogen Receptors
Recruitment of plasminogen to the bacterial cell surface is mediated directly by either specialised cell surface receptors or cytoplasmic and glycolytic pathway proteins localised to the bacterial cell surface or indirectly via interactions with host plasma proteins such as fibrinogen. Table 1 gives an overview of the most well-characterised bacterial plasminogen receptors.
Table 1.
Bacterial plasminogen receptors and their interactions with plasminogen.
| Plasminogen receptor | Bacterial species | Cell surface attachment | Binding affinity (KD) | Binding interactions and characteristics | References |
|---|---|---|---|---|---|
| Bfp60 | Bacteroides fragilis | Anchored | ND | ND | [42] |
| Choline-binding protein E (CBPE) | Streptococcus pneumoniae | Anchored | ND | Binds plg via internal lysine residues K259, K267, and K319 present in the phosphorylcholine esterase domain. | [43, 44] |
| CRASP-1, 3, 4, and 5 | Borrelia burgdorferi | Anchored | ND | ND | [45] |
| ErpP, ErpC, and ErpA | Borrelia burgdorferi | Anchored | Glu-plg: KD = 25 nM | Plg binding is associated with C-terminal lysine residues. Bound plg can be activated by uPA. | [46] |
| Erp63 | Borrelia spielmanii | Anchored | ND | ND | [47] |
| Flagella | Escherichia coli | Anchored | ND | ND | [48] |
| GlnA1 | Mycobacterium tuberculosis | Anchored | ND | Interact with LBS within plg | [49] |
| LenA | Leptospira interogans | Anchored | ND | Interacts with the K1–K3 plg fragment | [46, 50] |
| Leptospiral surface adhesin Lsa66 | Leptospira interrogans | Anchored | Plg: KD = 68.8 nM | ND. Bound plg can be activated by uPA | [51] |
| Lp30 | Leptospira interrogans | Anchored | Plg: KD = 167.39 nM | ND. Bound plg can be activated by uPA | [51] |
| LIC12238 LIC10494 LIC12730 |
Leptospira interrogans | Anchored | Plg: KD = 11.97 nM Plg: KD = 10.98 nM ND |
ND. Bound plg can be activated by uPA | [52] |
| LipL32, LipL40 | Leptospira interrogans | Anchored | ND | [52] | |
| Lp29, Lp49 | Leptospira interrogans | Anchored | ND | [52] | |
| Lsa20 Lsa66 |
Leptospira interrogans | Anchored | ND Plg: KD = 68.8 nM |
ND | [53] [51] |
| Streptococcus pyogenes | Glu-plg: KD = 1.6 nM–7.6 nM | K2 | [34, 35] | ||
| M and M-like protein | Streptococcus canis | Anchored | Mini-plg: KD = 2.7 nM | High affinity for plg K5 | [28] |
| Streptococcus equi | Plg: KD = 18 nM | Plg binding not competed out by excess K1–3, but inhibited by EACA, suggesting a role for K4 or K5 | [54] | ||
| Mhp 107 | Mycoplasma hyopneumoniae | Anchored | Plg (porcine): KD = ND | ND | [26] |
| MPL36 | Leptospira interrogans | Anchored | ND | [52] | |
| Outer surface protein A (OspA) | Borrelia burgdorferi | Anchored | Glu-plg: KD = 260 μM | Interacts with LBS within plg and pln. Bound plg can be activated by both uPA and tPA | [55] |
| 70 kDa surface protein (OppA) | Borrelia burgdoferi | Anchored | ND | ND | [55, 56] |
| PavB | Streptococcus pneumonia | Anchored | ND | ND | [57] |
| PfbB | Streptococcus pneumonia | Anchored | ND | ND | [58] |
| PfbA | Streptococcus pneumonia | Anchored | ND | ND | [59] |
| Plasminogen-binding protein (Pbp) | Bacteroides fragilis | Anchored | ND | ND | [60] |
| PbbA and pgbB | Helicobacter pylori | Anchored | ND | Interacts with LBS of plg | [61] |
| P116 | Mycoplasma hyopneumoniae | Anchored | Asp-plg (porcine): KD = 44 nM | ND | [26] |
| Protein E | Haemophilus influenzae | Anchored | ND | Interacts with LBS of plg | [62] |
| Type 1fimbriae | Escherichia coli | Anchored | Glu-plg: KD = 200 nM | ND | [63, 64] |
| Streptococcus pneumoniae | Plg: KD1 = 0.55 nM; KD2 = 86.2 nM | Residues 248–256; C-terminal lysyl residues LL433 and LL434. Interacts with LBS within Plg | [65–67] | ||
| Streptococcus pyogenes | Glu-Plg: KD = 1.6 nM; Lys-plg: KD = 127 nM | C-terminal lysine residues K434 and K434; Residues 252–255. Interacts with LBS within Plg | [68, 69] | ||
| Streptococcus suis | Plg: KD = 14 nM | Contains internal nonapeptide motif | [70] | ||
| Bifidobacterium lactis | Plg: KD = 42 nM | Lysine and glutamic acid residues K251, K251, and E252 | [30] | ||
| Bacillus anthracosis | ND | Plg binding partially mediated by C-terminal lysine. Interacts with LBS within Plg | [71, 72] | ||
| α-enolase | Neisseria meningitidis | Nonanchored | ND | Undefined internal plg-binding motif | [73] |
| Streptococcus mutans | ND | Binds plg via C-terminal lysine | [74] | ||
| Streptococcus agalactiae | Glu-Plg: ND; Lys-Plg: ND | ND | [75] | ||
| Mycoplasma gallisepticum | ND | ND | [27] | ||
| Mycoplasma fermentas | ND | ND | [76] | ||
| Borrelia burgdorferi | Glu-plg: KD = 125 nM | Interacts with LBS within Plg | [77] | ||
| Ag85B | Mycobacterium tuberculosis | Nonanchored | ND | Interacts with LBS within plg | [49] |
| Aspartase | Haemophilus influenzae | Nonanchored | ND | K4. Potent stimulator of tPA but not uPA | [78] |
| Bifidobacterium animalis | Plg: KD = 11.97 nM | Interacts with LBS within plg | [79] | ||
| DNaK | Neisseria meningitidis | Nonanchored | ND | Undefined internal plg-binding motif | [73] |
| Mycobacterium tuberculosis | ND | Interacts with LBS within plg | [49] | ||
| Elongation factor-tu (EF-tu) | Bacillus anthracosis | Nonanchored | ND | Interacts with LBS within plg | [71] |
| Fructose-1,6-bisphosphate aldolase | Mycoplasma tuberculosis | Nonanchored | Plg: KD = 6.73 nM | ND | [80] |
| Streptococcus pneumoniae | Pln: KD1 = 28 nM; KD2 = 52 nM Plg: KD1 = 0.43 μM; KD2 = 0.16 nM |
Binds plg via two C-terminal lysine residues separated by isoleucine and alanine | [67, 81] | ||
| Glyceraldehyde 3-phphate dehydrogenase (GAPDH; GAPC; SDH; Plr) | Streptococcus pyogenes | Nonanchored | ND | ND | [82] |
| Bacillus anthracis | Plg: KD = 78.5 nM; 572 nM | ND | [83] | ||
| Streptococcus equisimilis | Plg: KD = 220 nM; Pln: KD = 25 nM | ND | [84] | ||
| Peroxiredoxin | Neisseria meningitidis | Nonanchored | ND | Undefined internal plg-binding motif | [73] |
| Phosphoglycerate kinase | Streptococcus equisimilis | Nonanchored | ND | Shown to bind both plg and pln | [85] |
| SkzL | Streptococcus agalactiae | Nonanchored | Glu-plg: KD = 3−16 nM; Lys-plg: KD = 80 nM; Pln KD = 50 nM | ND | [86] |
ND: not determined, plg: plasminogen, pln: plasmin, LBS: lysine binding site, K1–5: kringle 1–5.
4.1. Specialised Cell Surface Receptors
Cell surface expressed receptors can be defined as those proteins which have a recognisable N-terminal signal sequence and membrane anchor motif. Several cell surface expressed plasminogen receptors have been well characterised, and it is interesting to note that many of these appear to have internal plasminogen-binding sites. Among the best characterised of these is the group A streptococcal plasminogen binding M protein. This coiled-coil alpha helical protein extends from the streptococcal cell surface and binds Glu-plasminogen with an affinity of Kd 1-2 nM [32, 33]. A combination of bacterial mutants, synthetic peptides and amino-acid substitution in recombinant proteins has been utilised to demonstrate that plasminogen binding to group A streptococcal M proteins is dependent on the presence of an internal plasminogen-binding repeat domain, consisting of positively charged arginine and histidine residues [34–36]. X-ray crystallography studies of the interaction between a 30-amino acid peptide comprising the plasminogen binding domain of streptococcal M protein (VEK-30) and a modified version of K2 of plasminogen indicate that Arg17 and His18 of VEK-30 form a pseudolysine structure that interacts with the LBS of this kringle [36]. This work supports earlier studies which showed that group A streptococcal plasminogen-binding M proteins interact with K2 of plasminogen, which contains a low affinity lysine-binding site [37]. Despite the fact that plasminogen binding by M proteins is readily inhibited by the lysine analogue EACA [32], mutation of the lysine residues within the bacterial interaction motif is not sufficient to fully abrogate plasminogen binding [34]. This highlights the important point that EACA competition alone is insufficient to demonstrate the role of lysine residues in interactions with plasminogen and its many receptors. Rather, the ability of lysine analogues to compete out plasminogen binding can be interpreted as demonstrating a role for the LBS within the kringle domains of plasminogen.
Plasminogen-binding M proteins are expressed by approximately 15% of group A streptococcal isolates, and similar proteins have been identified in a variety of group C and G streptococcal strains [38, 39]. Recently, a plasminogen-binding M protein expressed by the group G streptococci S. canis was reported to bind to miniplasminogen, a plasminogen variant consisting of only K5 and the serine protease domain [28]. Similarly, the M-like protein of group C streptococcus GCS3 likely interacts with K4 or K5 of plasminogen [40]. K4 and K5 show a high affinity for lysine-based ligands when compared with K2 [8], so, whilst specific plasminogen-binding sites within the M proteins of group C streptococcus and S. canis are yet to be defined, it is likely that they display markedly different properties to the internal motif described for the group A streptococcal plasminogen-binding M proteins. It is possible that these receptors mediate plasminogen binding at different sites or stages of infection; however, this hypothesis has yet to be fully explored. Based on the crystal structure of plasminogen, it has been suggested that the interaction of K5 with lysine residues is key to the structural change of plasminogen from its closed to open form [41]. It is tempting to hypothesise that bacteria which do not express their own plasminogen activators, such as S. canis may have evolved plasminogen interaction mechanisms that allow more efficient activation by host activators.
Internal plasminogen-binding sites have also been proposed for several bacterial lipoproteins identified as plasminogen receptors. B. burgdorferi binds plasminogen via an array of lipoproteins, including ErpP, ErpC, Erp, and OspA [46], while B. recurrentis and B. hermsii mediate plasminogen binding by the lipoproteins HcPA and BhCRASP1 [87, 88]. Similarly, several as yet uncharacterised lipoproteins of Francisella tularensis have been found to interact with plasminogen in human plasma via ligand blot analysis [89]. Whilst specific plasminogen binding sites within all these proteins have not been fully defined, the role of C-terminal lysines appears limited for those that have been characterised. Truncated Erp proteins lacking three native C-terminal lysine residues show only a partial reduction in plasminogen binding, supporting a role for both C-terminal lysine residues and an unidentified internal binding site in the interaction with plasminogen [46]. Similarly, mutation of residues Lys259, Lys267, and Lys319 within the choline-binding protein E (CBPE) of S. pneumoniae results in a 70% reduction in plasminogen when compared to the wild-type protein [43]. A number of other receptors with less well-defined plasminogen-binding sites are listed in Table 1.
4.2. Cytoplasmic and Glycolytic Pathway Proteins
In addition to specialised cell surface expressed plasminogen receptors, a number of proteins, usually considered to be restricted to the cytoplasm, have been found on the bacterial cell surface and are involved in interactions with plasminogen. Examples include the glycolytic pathway enzymes α-enolase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), DNaK, and elongation factor Tu (efTu) [30, 71, 79]. The mechanisms underlying the cell surface localisation of these proteins are not defined; however, their cell surface location has been confirmed in multiple species of bacteria [73, 82, 83, 85]. Unlike traditional cell wall anchored proteins, the contribution of glycolytic pathway enzymes to whole cell binding can typically only be shown indirectly, using blocking antibodies or competing concentrations of soluble recombinant proteins. This stems from the fact that these proteins are often metabolically essential for bacterial survival which prevents the construction of isogenic knockout mutant strains.
Interactions between the glycolytic pathway enzyme enolase and plasminogen have been characterised for several species of bacteria (Table 1). The most extensively studied of the bacterial enolases include those expressed by S. pneumoniae and S. pyogenes. Both have been shown to have a higher affinity for Lys-plasminogen than the circulating Glu-plasminogen or plasmin [90, 91].
Reports on the mechanism of plasminogen binding by bacterial enolases have been conflicting. Pneumococcal enolase contains an internal nonapeptide motif (FYDKERKVY), with the C-terminal lysine residues playing only a minor role in the interaction with plasminogen [65]. Like S. pneumoniae, several other bacterial enolases also appear to mediate plasminogen binding via internal lysine residues, including the enolase from Bifidobacterium lactis, for which internal residues Lys251 and Lys255, as well as the negatively charged Glu252 are responsible for plasminogen binding [30]. However a recent study of the plasminogen-binding of oral Streptococcal enolase variants showed that plasminogen binding activity is conserved despite the loss of lysine residues within the internal nonapeptide, with the authors suggesting that the role of the first lysine in the internal nonapeptide in plasminogen binding may not be as critical as first thought [29]. For the S. pyogenes enolase (SEN), internal lysines Lys252 and Lys255 contribute significantly to plasminogen binding. However, the high affinity of SEN for plasminogen is also mediated in part by two lysine residues at the C-terminus (Lys434 and Lys435) which are thought to stabilise the conformation of SEN's plasminogen binding site [68]. Site-directed mutagenesis of either the C-terminal or internal lysine motifs abrogate binding of plasminogen by SEN [69]. In contrast, the enolase of S. mutans does not have a functional internal plasminogen-binding site and may mediate plasminogen binding by C-terminal lysine residues only [74]. In all reported cases, bacterial plasminogen binding by enolase is inhibited by the lysine analogue EACA, indicating a role for the lysine binding sites within plasminogen in this interaction. However, it has yet to be established which LBS within plasminogen mediate interactions with enolase. It is possible that the different motifs responsible for plasminogen binding within diverse enolases interact with distinct LBS within plasminogen. One could hypothesise that C-terminal lysines interact with K1 of plasminogen, whilst internal lysines bind to K5, akin to the model proposed by Law et al. 2012 for the interaction of plasminogen and fibrin(ogen). Plasminogen binding is not conserved in all enolases, as evidenced by the finding that enolase from Bacteroides fragilis does not interact with plasminogen [92].
Similar to enolase, glyceraldehyde 3 phosphate dehydrogenase is a glycolytic pathway enzyme which has been shown to be located on the bacterial cell surface and to interact with plasminogen. GAPDH of S. pyogenes and S. pneumoniae binds preferentially to Lys-plasminogen and plasmin, and this interaction is mediated by the C-terminal lysine residue in GAPDH [93]. Interestingly, it has been shown that GAPDH of group B streptococcus interacts with both Glu-and Lys-plasminogen but not plasmin [94].
4.3. Indirect Plasminogen Binding
A number of bacterial pathogens possess the ability to interact with additional plasma proteins including IgG, α2-macroglobulin, albumin, numerous complement factors, and fibrinogen [95]. These interactions are involved in pathogenic processes such as cell adherence and colonisation, evasion of the immune system and dissemination [95–97]. For S. pyogenes, the interaction of bacterial cell surface receptors with fibrinogen has been shown to play a role in the acquisition of cell surface plasmin activity.
Fibrinogen is a large, 340 kDa protein made up of two identical subunits connected by numerous disulphide linkages. Each subunit consists of three nonidentical polypeptide chains denoted Aα, Bβ, and γ [98]. These polypeptide chains are folded into a number of structural domains. The central E domain consists of the N-termini of all six polypeptide chains, the two D domains (one in each subunit) consist of C-terminal regions of Bβ and γ chains and a portion of the Aα chain, while the remaining portions of the two Aα chains form 2 αC domains [99]. Cleavage of fibrinogen by thrombin is the last step in the coagulation pathway and leads to the formation of fibrin. After thrombin cleavage, previously unexposed (cryptic) sites are revealed in fibrin molecules which initiate fibrin polymerisation and clot formation [100]. Polymerisation results in the exposure of additional cryptic-binding sites for a range of cell types, growth factors, and proteins including those involved in fibrinolysis, such as tissue plasminogen activator, plasminogen, plasminogen activator inhibitor, and α2-antiplasmin [101–103]. This diverse range of ligand interactions allows fibrin to participate in a variety of processes involved in tissue regeneration and also facilitates the tight regulation of haemostasis.
S. pyogenes secretes streptokinase, a plasminogen activating protein. Streptokinase binds to plasminogen SPD (Figure 1) and induces conformational changes in the latent active site of plasminogen producing an enzymatically active complex which, in addition to plasmin activity, also displays plasminogen activation activity [104]. While the main physiological role of plasmin is the degradation of fibrin, plasmin can also cleave a variety of other substrates including fibrinogen. Cleavage of soluble fibrinogen exposes cryptic sites within the molecule which allow it to interact with ligands that were previously nonreactive with the intact protein. Plasminogen-binding sites have been identified in D domain fibrinogen fragments [105] and the binding of plasminogen to this fragment enhances streptokinase-mediated plasminogen activation [106]. Additionally, fragment D is sufficient for interaction with fibrinogen receptors on the GAS cell surface [107, 108]. Therefore, at the site of infection, Plg-SK activator complexes can cleave fibrinogen, producing D domain fragments. These D domain fragments are then able to interact with both plasmin(ogen) (present in the activator complex and/or as free plasmin) and bacterial cell surface fibrinogen receptors thereby mediating the acquisition of unregulated plasmin activity onto the bacterial cell surface. This mechanism of plasmin acquisition appears to be important for those GAS strains that do not possess high-affinity plasminogen-binding proteins but do express fibrinogen-binding proteins such as PrtF1 and PrtF2 variants [109], M protein variants [110] and the lipoprotein Spy_0591 [111]. It is currently not known if a similar mechanism of plasmin acquisition involving fibrinogen fragments and bacterial fibrinogen receptors is functioning in other bacterial species.
4.4. Physiological Significance of Plasminogen Acquisition by Bacteria
The broad proteolytic activity of plasmin necessitates tight in vivo regulation. Within the host, this is achieved by specific mechanisms that control the generation of plasmin from plasminogen and by mechanisms that restrict plasmin activity to specific locations as required. The major circulating inhibitor of plasmin is α2-antiplasmin. Lysine residues within α2-antiplasmin stabilise binding to the kringles of plasmin(ogen), resulting in rapid inhibition of plasmin in solution. However, once bound to surfaces such as fibrin, or cell surface receptors, plasmin is partially protected from inactivation by α2-antiplasmin [112–114]. Bacteria circumvent host regulatory mechanisms as cell surface bound plasminogen are more readily activated to plasmin, and, as in the host, this plasmin activity is not readily inhibited by host inhibitors [18, 19, 115]. Protection of plasmin from inhibition by binding to cell surface receptors appears to be central to the pathogenesis of several bacterial species and is utilised in a variety of pathogenic processes (summarised in Figure 2) [66, 73, 80, 116, 117].
Figure 2.
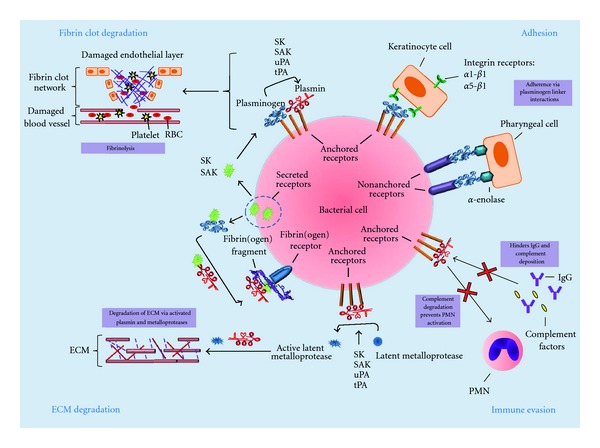
Mechanisms of bacterial cell surface plasmin(ogen) acquisition and its role in bacterial-host interactions. Plasmin(ogen) can be bound directly to the bacterial cell surface via cell-membrane-anchored receptors, nonanchored-cell-surface-associated receptors or indirectly through interactions with fibrinogen and cell surface fibrinogen receptors. Plasmin(ogen) localised on the bacterial cell surface is involved in four main processes; (1) ECM degradation via activated metalloproteases and plasmin; (2) fibrinolysis via plasmin; (3) immune evasion through plasmin-mediated degradation of immune effectors, including complement components and immunoglobulins; (4) adherence to host cells via plasminogen-linker interactions with host cell surface receptors. ECM: extracellular matrix; IgG: immunoglobulin G; RBC: red blood cell; SAK: staphylokinase; SEN: streptococcal α-enolase; Ska: streptokinase; tPA: tissue plasminogen activator; uPA; urokinase plasminogen activator.
Several bacterial species associated with highly invasive infections express receptors for plasminogen and plasmin, including S. pyogenes, S. pneumoniae, S. aureus, P. aeruginosa, Y. pestis, and S. enteritidis. Local thrombosis and microvascular occlusion during the early inflammatory response to bacterial infection can capture bacteria and prevent bacterial dissemination into deeper tissues. Surface-associated plasmin activity can facilitate fibrinolysis, preventing clot formation or promote the release of bacteria from a formed clot (Figure 2) [17, 107]. Furthermore, plasmin degradation of fibrinogen can initiate the release of products that affect blood vessel permeability and the accumulation of inflammatory cells [1, 118]. A major pathogenic consequence of bacterial plasminogen recruitment thus appears to be severe tissue destruction and overstimulation of the inflammatory response.
The direct degradation of ECM and basement membrane proteins and the activation of matrix metalloproteases by plasmin may enable bacteria to break down host tissue barriers (Figure 2). This is evidenced by the repeated demonstration that plasmin-coated bacteria are capable of penetrating ECM or basement membranes in vitro [119–121]. Plasminogen immobilised to the surface of E. coli, H. pylori, and N. meningitidis shows enhanced tPA-mediated plasminogen activation; whilst tPA- and uPA-activated plasmin at the surface of S. typhimurium, B. burgdorferi, S. pneumoniae, S. agalactiae, and M. fermentans facilitates the degradation of various ECM components, migration through endothelial and epithelial cell layers, or invasion of epithelial cells [18, 75, 122, 123]. The role of plasminogen acquisition in highly invasive infections is supported by a number of studies using animal models of infection, as well as several epidemiological studies. The ability to accumulate cell surface plasmin has been shown to be a prerequisite for systemic S. pyogenes infection in a humanised plasminogen mouse model [124–126]. Moreover, B. burgdorferi with active plasmin bound to their surface causes a more severe form of bacteraemia than their counterparts without active plasmin in a mouse model of spirochetemia [116]. Additionally, the abrogation of enolase-mediated plasminogen binding by S. pneumoniae significantly reduces the virulence of this pathogen in mice [65]. There are also epidemiological data to support the role of plasmin acquisition in bacterial pathogenesis. E. coli strains isolated from patients with colonic disease have been shown to bind significantly more plasminogen than E. coli isolates from healthy patients [127]. Similarly a study of S. pyogenes isolates from Northern Australia showed that isolates associated with invasive disease acquired significantly more cell surface plasminogen than noninvasive isolates [38]. However, oral streptococci display specific, high affinity plasminogen binding irrespective of their association with either benign dental plaque or severe inflammatory disease [128], and many commensal bacteria have been shown to recruit plasminogen to the bacterial cell surface (Table 1). This suggests that sequestration of plasminogen by bacteria may be important for bacterial survival in the host environment, with reports indicating a role for this interaction in both immune evasion and host colonisation [68, 129–131].
Plasmin plays an integral role in the recruitment of host immune cells to sites of bacterial infection and is able to degrade essential components of the innate immune response such as the complement factors C3b, C4b, and C5 (Figure 2) [132–134]. Plasmin at the bacterial cell surface therefore provides organisms with the capacity to degrade immunoglobulins and complement proteins, thereby inhibiting the host immune response. Specifically, plasminogen activation by the bacterial activators staphylokinase (of S. aureus) and PgtE (of S. typhimurium) results in degradation of C3b, thereby preventing complement-driven phagocytosis. PgtE-generated plasmin has also been shown to degrade complement factors C4b and C5 [129, 133]. Similarly, uPA activated plasminogen at the surface of L. interogans and B. anthracis prevents deposition of IgG and C3b on the bacterial surface [135] and leads to a subsequent decrease in macrophage phagocytosis [71]. Furthermore, the ability of certain bacteria to activate plasminogen has been shown to alter the response of inflammatory cells to infection. The expression of the plasminogen activator Pla by Yersinia pestis appears to decrease the level of neutrophil infiltration in a mouse model of infection [136]. Clearly, there is a role for bacterial plasminogen acquisition in protecting bacteria from the host immune response. Whilst this has obvious significance for the initiation of systemic bacterial disease, it also has implications in host colonisation by commensal and pathogenic organisms alike.
A further role for plasminogen recruitment in bacterial colonisation has been demonstrated by several studies of plasminogen recruitment by streptococci. Plasminogen has been shown in vitro to act as a linker molecule between enolase at the surface of pharyngeal cells, and SEN at the surface of S. pyogenes, thus facilitating the adhesion process (Figure 2) [137]. When this bridging plasminogen molecule is activated by tPA to plasmin, it can digest intercellular junctions and disrupt cell monolayers in ECM models [137]. Similarly, the streptococcal M protein GSC3 has been shown to mediate plasminogen-dependant adherence of streptococci to pharyngeal cells [40], implying a role for plasminogen binding in colonisation of the throat and oral cavity by bacteria. A role for plasminogen binding in colonisation has been further demonstrated for S. pyogenes interaction with keratinocytes. Plasminogen on the bacterial cell surface promoted the internalisation of streptococci by keratinocytes through the interaction with α1β1- and α5β1-integrins (Figure 2) [130]. In all the cases reported so far, the role of bacterially bound plasminogen in adherence/internalisation appears to function independently of the serine protease activity of plasmin.
5. Conclusions
The expression of receptors which enable localisation of plasminogen to the cell surface is a phenotype common to a multitude of bacteria. Since the initial identification of bacterial plasminogen receptors over 20 years ago, a myriad of receptor types have been identified, associated with both pathogenic and commensal bacterial species. The vast array of mechanisms via which different receptors interact with Glu-plasminogen, Lys-plasminogen, plasmin, and mini-plasmin suggests that these receptors may have evolved to mediate interactions with this abundant human protein under diverse physiological conditions. Indeed, recent studies show bacterial plasmin(ogen) acquisition is central to the onset of invasive pathogenesis via fibrin and ECM degradation; immune evasion via degradation of various immune effectors; and colonisation of the host (Figure 2). Much remains to be learned about how diverse plasminogen receptors interact with plasminogen. For many receptors, there is limited information on specificities of interaction with different forms of plasminogen and plasmin and on the location of binding within the plasminogen molecule. Recent structural studies suggest that the mechanism through which receptors interact with plasminogen can have different effects on the structure and activation of this protein which may ultimately influence the pathogenic process for many bacterial species [10]. Plasminogen receptors clearly play a central role in the relationship between bacteria and the host, and further elucidation of the nuances of how microbes interact with plasminogen will contribute significantly to our understanding of both the plasminogen molecule and bacterial pathogenesis in the future.
References
- 1.Danø K, Andreasen PA, Grøndahl-Hansen J, Kristensen P, Nielsen LS, Skriver L. Plasminogen Activators, Tissue Degradation, and Cancer. Advances in Cancer Research. 1985;44(C):139–266. doi: 10.1016/s0065-230x(08)60028-7. [DOI] [PubMed] [Google Scholar]
- 2.Plow EF, Herren T, Redlitz A, Miles LA, Hoover-Plow JL. The cell biology of the plasminogen system. The FASEB Journal. 1995;9(10):939–945. doi: 10.1096/fasebj.9.10.7615163. [DOI] [PubMed] [Google Scholar]
- 3.Ellis V, Scully MF, Kakkar VV. Plasminogen activation initiated by single-chain urokinase-type plasminogen activator. Potentiation by U937 monocytes. The Journal of Biological Chemistry. 1989;264(4):2185–2188. [PubMed] [Google Scholar]
- 4.Lottenberg R, Broder CC, Boyle MD. Identification of a specific receptor for plasmin on a group A streptococcus. Infection and Immunity. 1987;55(8):1914–1918. doi: 10.1128/iai.55.8.1914-1918.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreasen PA, Kjoller L, Christensen L, Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: a review. International Journal of Cancer. 1997;72(1):1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Ponting CP, Marshall JM, Cederholm-Williams SA. Plasminogen: a structural review. Blood Coagulation and Fibrinolysis. 1992;3(5):605–614. [PubMed] [Google Scholar]
- 7.Ranson M, Andronicos NM. Plasminogen binding and cancer: promises and pitfalls. Frontiers in Bioscience. 2003;8:s294–s304. doi: 10.2741/1044. [DOI] [PubMed] [Google Scholar]
- 8.Marti DN, Hu CK, An SSA, Von Haller P, Schaller J, Llinás M. Ligand preferences of kringle 2 and homologous domains of human plasminogen: canvassing weak, intermediate, and high-affinity binding sites by 1H-NMR. Biochemistry. 1997;36(39):11591–11604. doi: 10.1021/bi971316v. [DOI] [PubMed] [Google Scholar]
- 9.Söhndel S, Hu CK, Marti D, et al. Recombinant gene expression and 1H NMR characteristics of the kringle (2 + 3) supermodule: spectroscopic/functional individuality of plasminogen kringle domains. Biochemistry. 1996;35(7):2357–2364. doi: 10.1021/bi9520949. [DOI] [PubMed] [Google Scholar]
- 10.Law RHP, Caradoc-Davies N, Coweison N, et al. The X-ray crystal structure of full-lngth human plasminogen. Cell Reports. 2012;1(3):p. 6. doi: 10.1016/j.celrep.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Weisel JW, Nagaswami C, Korsholm B, Petersen LC, Suenson E. Interactions of plasminogen with polymerizing fibrin and its derivatives, monitored with a photoaffinity cross-linker and electron microscopy. Journal of Molecular Biology. 1994;235(3):1117–1135. doi: 10.1006/jmbi.1994.1061. [DOI] [PubMed] [Google Scholar]
- 12.Claeys H, Vermylen J. Physico chemical and proenzyme properties of NH2 terminal glutamic acid and NH2 terminal lysine human plasminogen. Influence of 6 aminohexanoic acid. Biochimica et Biophysica Acta. 1974;342(2):351–359. doi: 10.1016/0005-2795(74)90090-7. [DOI] [PubMed] [Google Scholar]
- 13.Hayes ML, Castellino FJ. Carbohydrate of the human plasminogen variants. III. Structure of the θ-glycosidically linked oligosaccharide unit. The Journal of Biological Chemistry. 1979;254(18):8777–8780. [PubMed] [Google Scholar]
- 14.Hayes ML, Castellino FJ. Carbohydrate of the human plasminogen variants. II. Structure of the asparagine linked oligosaccharide unit. The Journal of Biological Chemistry. 1979;254(18):8772–8776. [PubMed] [Google Scholar]
- 15.Hayes ML, Castellino FJ. Carbohydrate of the human plasminogen variants. I. Carbohydrate composition, glycopeptide isolation, and characterization. The Journal of Biological Chemistry. 1979;254(18):8768–8771. [PubMed] [Google Scholar]
- 16.Mølgaard L, Ponting CP, Christensen U. Glycosylation at Asn-289 facilitates the ligand-induced conformational changes of human Glu-plasminogen. FEBS Letters. 1997;405(3):363–368. doi: 10.1016/s0014-5793(97)00221-4. [DOI] [PubMed] [Google Scholar]
- 17.Coleman JL, Benach JL. Use of the plasminogen activation system by microorganisms. Journal of Laboratory and Clinical Medicine. 1999;134(6):567–576. doi: 10.1016/s0022-2143(99)90095-1. [DOI] [PubMed] [Google Scholar]
- 18.Lähteenmäki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends in Microbiology. 2005;13(2):79–85. doi: 10.1016/j.tim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Walker MJ, McArthur JD, McKay F, Ranson M. Is plasminogen deployed as a Streptococcus pyogenes virulence factor? Trends in Microbiology. 2005;13(7):308–313. doi: 10.1016/j.tim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 20.McArthur JD, Cook SM, i C, Walker MJ. The role of streptokinase as a virulence determinant of Streptococcus pyogenes—potential for therapeutic targeting. Current Drug Targets. 2012;13(3):297–307. doi: 10.2174/138945012799424589. [DOI] [PubMed] [Google Scholar]
- 21.Ullberg M, Kronvall G, Karlsson I, Wiman B. Receptors for human plasminogen on gram-negative bacteria. Infection and Immunity. 1990;58(1):21–25. doi: 10.1128/iai.58.1.21-25.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullberg M, Kronvall G, Wiman B. New receptor for human plasminogen on gram positive cocci. APMIS. 1989;97(11):996–1002. doi: 10.1111/j.1699-0463.1989.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 23.Ullberg M, Kuusela P, Kristiansen BE, Kronvall G. Binding of plasminogen to Neisseria meningitidis and Neisseria gonorrhoeae and formation of surface-associated plasmin. Journal of Infectious Diseases. 1992;166(6):1329–1334. doi: 10.1093/infdis/166.6.1329. [DOI] [PubMed] [Google Scholar]
- 24.Ringner M, Valkonen KH, Wadstrom T. Binding of vitronectin and plasminogen to Helicobacter pylori. FEMS Immunology and Medical Microbiology. 1994;9(1):29–34. doi: 10.1111/j.1574-695X.1994.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 25.Kuusela P, Saksela O. Binding and activation of plasminogen at the surface of Staphylococcus aureus. Increase in affinity after conversion to the Lys form of the ligand. European Journal of Biochemistry. 1990;193(3):759–765. doi: 10.1111/j.1432-1033.1990.tb19397.x. [DOI] [PubMed] [Google Scholar]
- 26.Seymour LM, Deutscher AT, Jenkins C, et al. A processed multidomain Mycoplasma hyopneumoniae adhesin binds fibronectin, plasminogen, and swine respiratory cilia. The Journal of Biological Chemistry. 2010;285(44):33971–33978. doi: 10.1074/jbc.M110.104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Yu S, Shen X, et al. The Mycoplasma gallisepticum α-enolase is cell surface-exposed and mediates adherence by binding to chicken plasminogen. Microbial Pathogenesis. 2011;51(4):285–290. doi: 10.1016/j.micpath.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Fulde M, Rohde M, Hitzmann A, et al. SCM, a novel M-like protein from Streptococcus canis, binds (mini)-plasminogen with high affinity and facilitates bacterial transmigration. Biochemical Journal. 2011;434(3):523–535. doi: 10.1042/BJ20101121. [DOI] [PubMed] [Google Scholar]
- 29.Itzek A, Gillen CM, Fulde M, et al. Contribution of plasminogen activation towards the pathogenic potential of oral streptococci. PLoS ONE. 2010;5(11) doi: 10.1371/journal.pone.0013826.e13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Candela M, Biagi E, Centanni M, et al. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology. 2009;155(part 10):3294–3303. doi: 10.1099/mic.0.028795-0. [DOI] [PubMed] [Google Scholar]
- 31.Antikainen J, Kuparinen V, Lähteenmäki K, Korhonen TK. Enolases from Gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. FEMS Immunology and Medical Microbiology. 2007;51(3):526–534. doi: 10.1111/j.1574-695X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- 32.Berge A, Sjobring U. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. The Journal of Biological Chemistry. 1993;268(34):25417–25424. [PubMed] [Google Scholar]
- 33.Sanderson-Smith M, Batzloff M, Sriprakash KS, Dowton M, Ranson M, Walker MJ. Divergence in the plasminogen-binding group A streptococcal M protein family: functional conservation of binding site and potential role for immune selection of variants. The Journal of Biological Chemistry. 2006;281(6):3217–3226. doi: 10.1074/jbc.M508758200. [DOI] [PubMed] [Google Scholar]
- 34.Sanderson-Smith ML, Walker MJ, Ranson M. The maintenance of high affinity plasminogen binding by group A streptococcal plasminogen-binding M-like protein is mediated by arginine and histidine residues within the a1 and a2 repeat domains. The Journal of Biological Chemistry. 2006;281(36):25965–25971. doi: 10.1074/jbc.M603846200. [DOI] [PubMed] [Google Scholar]
- 35.Sanderson-Smith ML, Dowton M, Ranson M, Walker MJ. The plasminogen-binding group A streptococcal M protein-related protein Prp binds plasminogen via arginine and histidine residues. Journal of Bacteriology. 2007;189(4):1435–1440. doi: 10.1128/JB.01218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rios-Steiner JL, Schenone M, Mochalkin I, Tulinsky A, Castellino FJ. Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein. Journal of Molecular Biology. 2001;308(4):705–719. doi: 10.1006/jmbi.2001.4646. [DOI] [PubMed] [Google Scholar]
- 37.Wistedt AC, Kotarsky H, Marti D, et al. Kringle 2 mediates high affinity binding of plasminogen to an internal sequence in streptococcal surface protein PAM. The Journal of Biological Chemistry. 1998;273(38):24420–24424. doi: 10.1074/jbc.273.38.24420. [DOI] [PubMed] [Google Scholar]
- 38.McKay FC, McArthur JD, Sanderson-Smith ML, et al. Plasminogen binding by group A streptococcal isolates from a region of hyperendemicity for streptococcal skin infection and a high incidence of invasive infection. Infection and Immunity. 2004;72(1):364–370. doi: 10.1128/IAI.72.1.364-370.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben Nasr A, Wistedt A, Ringdahl U, Sjobring U. Streptokinase activates plasminogen bound to human group C and G streptococci through M-like proteins. European Journal of Biochemistry. 1994;222(2):267–276. doi: 10.1111/j.1432-1033.1994.tb18865.x. [DOI] [PubMed] [Google Scholar]
- 40.Bergmann R, Dinkla K, Patric Nitsche-Schmitz D, et al. Biological functions of GCS3, a novel plasminogen-binding protein of Streptococcus dysgalactiae ssp. equisimilis. International Journal of Medical Microbiology. 2011;301(2):157–164. doi: 10.1016/j.ijmm.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Yang D, Sun YY, Nemkul N, et al. Plasminogen Activator Inhibitor-1 Mitigates Brain Injury in a Rat Model of Infection-Sensitized Neonatal Hypoxia-Ischemia. doi: 10.1093/cercor/bhs115. Cerebral Cortex. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira EDO, De Carvalho JB, Peixoto RJM, et al. The interaction of Bacteroides fragilis with components of the human fibrinolytic system. FEMS Immunology and Medical Microbiology. 2009;56(1):48–55. doi: 10.1111/j.1574-695X.2009.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Attali C, Frolet C, Durmort C, Offant J, Vernet T, Di Guilmi AM. Streptococcus pneumoniae choline-binding protein E interaction with plasminogen/plasmin stimulates migration across the extracellular matrix. Infection and Immunity. 2008;76(2):466–476. doi: 10.1128/IAI.01261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garau G, Lemaire D, Vernet T, Dideberg O, Di Guilmi AM. Crystal structure of phosphorylcholine esterase domain of the virulence factor choline-binding protein E from Streptococcus pneumoniae: new structural features among the metallo-β-lactamase superfamily. The Journal of Biological Chemistry. 2005;280(31):28591–28600. doi: 10.1074/jbc.M502744200. [DOI] [PubMed] [Google Scholar]
- 45.Hallström T, Haupt K, Kraiczy P, et al. Complement regulator-acquiring surface protein 1 of Borrelia burgdorferi binds to human bone morphogenic protein 2, several extracellular matrix proteins, and plasminogen. Journal of Infectious Diseases. 2010;202(3):490–498. doi: 10.1086/653825. [DOI] [PubMed] [Google Scholar]
- 46.Brissette CA, Haupt K, Barthel D, et al. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infection and Immunity. 2009;77(1):300–306. doi: 10.1128/IAI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seling A, Siegel C, Fingerle V, et al. Functional characterization of Borrelia spielmanii outer surface proteins that interact with distinct members of the human factor H protein family and with plasminogen. Infection and Immunity. 2010;78(1):39–48. doi: 10.1128/IAI.00691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lahteenmaki K, Westerlund B, Kuusela P, Korhonen TK. Immobilization of plasminogen on Escherichia coli flagella. FEMS Microbiology Letters. 1993;106(3):309–314. doi: 10.1111/j.1574-6968.1993.tb05981.x. [DOI] [PubMed] [Google Scholar]
- 49.Xolalpa W, Vallecillo AJ, Lara M, et al. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics. 2007;7(18):3332–3341. doi: 10.1002/pmic.200600876. [DOI] [PubMed] [Google Scholar]
- 50.Verma A, Brissette CA, Bowman AA, Shah ST, Zipfel PF, Stevenson B. Leptospiral endostatin-like protein a is a bacterial cell surface receptor for human plasminogen. Infection and Immunity. 2010;78(5):2053–2059. doi: 10.1128/IAI.01282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira R, de Morais ZM, Gonçales AP, Romero EC, Vasconcellos SA, Nascimento ALTO. Characterization of novel OmpA-like protein of Leptospira interrogans that binds extracellular matrix molecules and plasminogen. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0021962.e21962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vieira ML, Atzingen MV, Oliveira TR, et al. In vitro identification of novel plasminogen-binding receptors of the pathogen Leptospira interrogans. PloS one. 2010;5(6):p. e11259. doi: 10.1371/journal.pone.0011259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendes RS, Von Atzingen M, de Morais ZM. The novel leptospiral surface adhesin Lsa20 binds laminin and human plasminogen and is probably expressed during infection. Infection and Immunity. 2011;79(11):4657–4667. doi: 10.1128/IAI.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergmann R, Dinkla K, Patric Nitsche-Schmitz D, et al. Biological functions of GCS3, a novel plasminogen-binding protein of Streptococcus dysgalactiae ssp. equisimilis. International Journal of Medical Microbiology. 2011;301(2):157–164. doi: 10.1016/j.ijmm.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 55.Hu LT, Perides G, Noring R, Klempner MS. Binding of human plasminogen to Borrelia burgdorferi. Infection and Immunity. 1995;63(9):3491–3496. doi: 10.1128/iai.63.9.3491-3496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linden THU, Pratt SD, Perides G, Katz L, Rogers RA, Klempner MS. Isolation, cloning, and expression of a 70-kilodalton plasminogen binding protein of Borrelia burgdorferi. Infection and Immunity. 1997;65(12):4989–4995. doi: 10.1128/iai.65.12.4989-4995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensch I, Gámez G, Rothe M, et al. PavB is a surface-exposed adhesin of Streptococcus pneumoniae contributing to nasopharyngeal colonization and airways infections. Molecular Microbiology. 2010;77(1):22–43. doi: 10.1111/j.1365-2958.2010.07189.x. [DOI] [PubMed] [Google Scholar]
- 58.Papasergi S, Garibaldi M, Tuscano G, et al. Plasminogen- and fibronectin-binding protein B is involved in the adherence of Streptococcus pneumoniae to human epithelial cells. The Journal of Biological Chemistry. 2010;285(10):7517–7524. doi: 10.1074/jbc.M109.062075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi M, Terao Y, Mori Y, Hamada S, Kawabata S. PfbA, a novel plasmin- and fibronectin-binding protein of Streptococcus pneumoniae, contributes to fibronectin-dependent adhesion and antiphagocytosis. The Journal of Biological Chemistry. 2008;283(52):36272–36279. doi: 10.1074/jbc.M807087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sijbrandi R, Stork M, Luirink J, Otto BR. Pbp, a cell-surface exposed plasminogen binding protein of Bacteroides fragilis. Microbes and Infection. 2008;10(5):514–521. doi: 10.1016/j.micinf.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 61.Jönsson K, Guo BP, Monstein HJ, Mekalanos JJ, Kronvall G. Molecular cloning and characterization of two Helicobacter pylori genes coding for plasminogen-binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(7):1852–1857. doi: 10.1073/pnas.0307329101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barthel D, Singh B, Riesbeck K, Zipfel PF. Haemophilus influenzae uses the surface protein E to acquire human plasminogen and to evade innate immunity. The Journal of Immunology. 2012;188(1):379–385. doi: 10.4049/jimmunol.1101927. [DOI] [PubMed] [Google Scholar]
- 63.Parkkinen J, Korhonen TK. Binding of plasminogen to Escherichia coli adhesion proteins. FEBS Letters. 1989;250(2):437–440. doi: 10.1016/0014-5793(89)80772-0. [DOI] [PubMed] [Google Scholar]
- 64.Kukkonen M, Saarela S, Lähteenmäki K, et al. Identification of two laminin-binding fimbriae, the type 1 fimbria of Salmonella enterica serovar typhimurium and the G fimbria of Escherichia coli, as plasminogen receptors. Infection and Immunity. 1998;66(10):4965–4970. doi: 10.1128/iai.66.10.4965-4970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bergmann S, Wild D, Diekmann O, et al. Identification of a novel plasmin(ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Molecular Microbiology. 2003;49(2):411–423. doi: 10.1046/j.1365-2958.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 66.Bergmann S, Rohde M, Preissner KT, Hammerschmidt S. The nine residue plasminogen-binding motif of the pneumococcal enolase is the major cofactor of plasmin-mediated degradation of extracellular matrix, dissolution of fibrin and transmigration. Thrombosis and Haemostasis. 2005;94(2):304–311. doi: 10.1160/TH05-05-0369. [DOI] [PubMed] [Google Scholar]
- 67.Ehinger S, Schubert WD, Bergmann S, Hammerschmidt S, Heinz DW. Plasmin(ogen)-binding α-Enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. Journal of Molecular Biology. 2004;343(4):997–1005. doi: 10.1016/j.jmb.2004.08.088. [DOI] [PubMed] [Google Scholar]
- 68.Derbise A, Song YP, Parikh S, Fischetti VA, Pancholi V. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infection and Immunity. 2004;72(1):94–105. doi: 10.1128/IAI.72.1.94-105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cork AJ, Jergic S, Hammerschmidt S, et al. Defining the structural basis of human plasminogen binding by streptococcal surface enolase. The Journal of Biological Chemistry. 2009;284(25):17129–17137. doi: 10.1074/jbc.M109.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Esgleas M, Li Y, Hancock MA, Harel J, Dubreuil JD, Gottschalk M. Isolation and characterization of α-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology. 2008;154(part 9):2668–2679. doi: 10.1099/mic.0.2008/017145-0. [DOI] [PubMed] [Google Scholar]
- 71.Chung MC, Tonry JH, Narayanan A, et al. Bacillus anthracis interacts with plasmin(ogen) to evade C3b-dependent innate immunity. PLoS ONE. 2011;6(3) doi: 10.1371/journal.pone.0018119.e18119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agarwal S, Kulshreshtha P, Bambah Mukku D, Bhatnagar R. α-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochimica et Biophysica Acta. 2008;1784(7-8):986–994. doi: 10.1016/j.bbapap.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 73.Knaust A, Weber MVR, Hammerschmidt S, Bergmann S, Frosch M, Kurzai O. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. Journal of Bacteriology. 2007;189(8):3246–3255. doi: 10.1128/JB.01966-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones MN, Holt RG. Cloning and characterization of an α-enolase of the oral pathogen Streptococcus mutans that binds human plasminogen. Biochemical and Biophysical Research Communications. 2007;364(4):924–929. doi: 10.1016/j.bbrc.2007.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magalhães V, Veiga-Malta I, Almeida MR, et al. Interaction with human plasminogen system turns on proteolytic activity in Streptococcus agalactiae and enhances its virulence in a mouse model. Microbes and Infection. 2007;9(11):1276–1284. doi: 10.1016/j.micinf.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 76.Yavlovich A, Higazi AR, Rottem S. Plasminogen binding and activation by Mycoplasma fermentans. Infection and Immunity. 2001;69(4):1977–1982. doi: 10.1128/IAI.69.4.1977-1982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Floden AM, Watt JA, Brissette CA. Borrelia burgdorferi enolase is a surface-exposed plasminogen binding protein. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027502.e27502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sjöström I, Gröndahl H, Falk G, Kronvall G, Ullberg M. Purification and characterisation of a plasminogen-binding protein from Haemophilus influenzae. Sequence determination reveals identity with aspartase. Biochimica et Biophysica Acta. 1997;1324(2):182–190. doi: 10.1016/s0005-2736(96)00218-0. [DOI] [PubMed] [Google Scholar]
- 79.Candela M, Centanni M, Fiori J, et al. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology. 2010;156(6):1609–1618. doi: 10.1099/mic.0.038307-0. [DOI] [PubMed] [Google Scholar]
- 80.de la Paz Santangelo M, Gest P, Guerin E, et al. Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1, 6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli. The Journal of Biological Chemistry. 2011;286(46):40219–40231. doi: 10.1074/jbc.M111.259440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kolberg J, Sletten K. Monoclonal antibodies that recognize a common pneumococcal protein with similarities to streptococcal group A surface glyceraldehyde-3-phosphate dehydrogenase. Infection and Immunity. 1996;64(9):3544–3547. doi: 10.1128/iai.64.9.3544-3547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. Journal of Experimental Medicine. 1992;176(2):415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matta SK, Agarwal S, Bhatnagar R. Surface localized and extracellular Glyceraldehyde-3-phosphate dehydrogenase of Bacillus anthracis is a plasminogen binding protein. Biochimica et Biophysica Acta. 2010;1804(11):2111–2120. doi: 10.1016/j.bbapap.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 84.Gase K, Gase A, Schirmer H, Malke H. Cloning, sequencing and functional overexpression of the Streptococcus equisimilis H46A gapC gene encoding a glyceraldehyde-3-phosphate dehydrogenase that also functions as a plasmin(ogen)-binding protein. Purification and biochemical characterization of-the protein. European Journal of Biochemistry. 1996;239(1):42–51. doi: 10.1111/j.1432-1033.1996.0042u.x. [DOI] [PubMed] [Google Scholar]
- 85.Boone TJ, Burnham CAD, Tyrrell GJ. Binding of group B streptococcal phosphoglycerate kinase to plasminogen and actin. Microbial Pathogenesis. 2011;51(4):255–261. doi: 10.1016/j.micpath.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Wiles KG, Panizzi P, Kroh HK, Bock PE. Skizzle is a novel plasminogen- and plasmin-binding protein from Streptococcus agalactiae that targets proteins of human fibrinolysis to promote plasmin generation. The Journal of Biological Chemistry. 2010;285(27):21153–21164. doi: 10.1074/jbc.M110.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grosskinsky S, Schott M, Brenner C, et al. Borrelia recurrentis employs a novel multifunctional surface protein with anti-complement, anti-opsonic and invasive potential to escape innate immunity. PLoS ONE. 2009;4(3) doi: 10.1371/journal.pone.0004858.e4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rossmann E, Kraiczy P, Herzberger P, et al. Dual binding specificity of a Borrelia hermsii-associated complement regulator-acquiring surface protein for factor II and plasminogen discloses a putative virulence factor of relapsing fever spirochetes. Journal of Immunology. 2007;178(11):7292–7301. doi: 10.4049/jimmunol.178.11.7292. [DOI] [PubMed] [Google Scholar]
- 89.Clinton SR, Bina JE, Hatch TP, Whitt MA, Miller MA. Binding and activation of host plasminogen on the surface of Francisella tularensis. BMC Microbiology. 2010;10, article no. 76 doi: 10.1186/1471-2180-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pancholi V, Fischetti VA. α-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. The Journal of Biological Chemistry. 1998;273(23):14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 91.Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Molecular Microbiology. 2001;40(6):1273–1287. doi: 10.1046/j.1365-2958.2001.02448.x. [DOI] [PubMed] [Google Scholar]
- 92.Sijbrandi R, Den Blaauwen T, Tame JRH, Oudega B, Luirink J, Otto BR. Characterization of an iron-regulated alpha-enolase of Bacteroides fragilis. Microbes and Infection. 2005;7(1):9–18. doi: 10.1016/j.micinf.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 93.Winram SB, Lottenberg R. Site-directed mutagenesis of streptococcal plasmin receptor protein (Plr) identifies the C-terminal Lys334 as essential for plasmin binding, but mutation of the plr gene does not reduce plasmin binding to group A streptococci. Microbiology. 1998;144(part 8):2025–2035. doi: 10.1099/00221287-144-8-2025. [DOI] [PubMed] [Google Scholar]
- 94.Seifert KN, McArthur WP, Bleiweis AS, Brady LJ. Characterization of group B streptococcal glyceraldehyde-3-phosphate dehydrogenase: surface localization, enzymatic activity, and protein-protein interactions. Canadian journal of microbiology. 2003;49(5):350–356. doi: 10.1139/w03-042. [DOI] [PubMed] [Google Scholar]
- 95.Olsen RJ, Shelburne SA, Musser JM. Molecular mechanisms underlying group A streptococcal pathogenesis. Cellular Microbiology. 2009;11(1):1–12. doi: 10.1111/j.1462-5822.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 96.Kwinn LA, Nizet V. How group A Streptococcus circumvents host phagocyte defenses. Future Microbiology. 2007;2(1):75–84. doi: 10.2217/17460913.2.1.75. [DOI] [PubMed] [Google Scholar]
- 97.Tart AH, Walker MJ, Musser JM. New understanding of the group A Streptococcus pathogenesis cycle. Trends in Microbiology. 2007;15(7):318–325. doi: 10.1016/j.tim.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 98.Doolittle RF. Fibrinogen and fibrin. Annual Review of Biochemistry. 1983;53:195–229. doi: 10.1146/annurev.bi.53.070184.001211. [DOI] [PubMed] [Google Scholar]
- 99.Medved L, Weisel JW. Recommendations for nomenclature on fibrinogen and fibrin. Journal of Thrombosis and Haemostasis. 2009;7(2):355–359. doi: 10.1111/j.1538-7836.2008.03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mosesson MW, Siebenlist KR, Meh DA. The structure and biological features of fibrinogen and fibrin. Annals of the New York Academy of Sciences. 2001;936:11–30. doi: 10.1111/j.1749-6632.2001.tb03491.x. [DOI] [PubMed] [Google Scholar]
- 101.Tsurupa G, Hantgan RR, Burton RA, Pechik I, Tjandra N, Medved L. Structure, stability, and interaction of the fibrin(ogen) αC-domains. Biochemistry. 2009;48(51):12191–12201. doi: 10.1021/bi901640e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wagner OF, De Vries C, Hohmann C, Veerman H, Pannekoek H. Interaction between plasminogen activator inhibitor type 1 (PAI-1) bound to fibrin and either tissue-type plasminogen activator (t-PA) or urokinase-type plasminogen activator (u-PA). Binding of t-PA/PAI-1 complexes to fibrin mediated by both the finger and the kringle-2 domain of t-PA. Journal of Clinical Investigation. 1989;84(2):647–655. doi: 10.1172/JCI114211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sakata Y, Aoki N. Molecular abnormality of plasminogen. The Journal of Biological Chemistry. 1980;255(11):5442–5447. [PubMed] [Google Scholar]
- 104.Bajaj SP, Castellino FJ. Activation of human plasminogen by equimolar levels of streptokinase. The Journal of Biological Chemistry. 1977;252(2):492–498. [PubMed] [Google Scholar]
- 105.Váradi A, Patthy L. Location of plasminogen-binding sites in human fibrin(ogen) Biochemistry. 1983;22(10):2440–2446. doi: 10.1021/bi00279a021. [DOI] [PubMed] [Google Scholar]
- 106.Strickland DK, Morris JP, Castellino FJ. Enhancement of the streptokinase-catalyzed activation of human plasminogen by human fibrinogen and its plasminolysis products. Biochemistry. 1982;21(4):721–728. doi: 10.1021/bi00533a021. [DOI] [PubMed] [Google Scholar]
- 107.Wang H, Lottenberg R, Boyle MDP. Analysis of the interaction of group A streptococci with fibrinogen, streptokinase and plasminogen. Microbial Pathogenesis. 1995;18(3):153–166. doi: 10.1016/s0882-4010(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 108.Hess JL, Boyle MDP. Fibrinogen fragment D is necessary and sufficient to anchor a surface plasminogen-activating complex in Streptococcus pyogenes. Proteomics. 2006;6(1):375–378. doi: 10.1002/pmic.200500189. [DOI] [PubMed] [Google Scholar]
- 109.Katerov V, Andreev A, Schalén C, Totolian AA. Protein F, a fibronectin-binding protein of Streptococcus pyogenes, also binds human fibrinogen: isolation of the protein and mapping of the binding region. Microbiology. 1998;144(part 1):119–126. doi: 10.1099/00221287-144-1-119. [DOI] [PubMed] [Google Scholar]
- 110.Schmidt KH, Mann K, Cooney J, Kohler W. Multiple binding of type 3 streptococcal M protein to human fibrinogen, albumin and fibronectin. FEMS Immunology and Medical Microbiology. 1993;7(2):135–143. doi: 10.1111/j.1574-695X.1993.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 111.Margarit I, Bonacci S, Pietrocola G, et al. Capturing host-pathogen interactions by protein microarrays: identification of novel streptococcal proteins binding to human fibronectin, fibrinogen, and C4BP. The FASEB Journal. 2009;23(9):3100–3112. doi: 10.1096/fj.09-131458. [DOI] [PubMed] [Google Scholar]
- 112.Hall SW, Humphries JE, Gonias SL. Inhibition of cell surface receptor-bound plasmin by α2-antiplasmin and α2-macroglobulin. The Journal of Biological Chemistry. 1991;266(19):12329–12336. [PubMed] [Google Scholar]
- 113.Wiman B, Collen D. On the kinetics of the reaction between human antiplasmin and plasmin. European Journal of Biochemistry. 1978;84(2):573–578. doi: 10.1111/j.1432-1033.1978.tb12200.x. [DOI] [PubMed] [Google Scholar]
- 114.Plow EF, Freaney DE, Plescia J, Miles LA. The plasminogen system and cell surfaces: evidence for plasminogen and urokinase receptors on the same cell type. Journal of Cell Biology. 1986;103(6, part 1):2411–2420. doi: 10.1083/jcb.103.6.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kuusela P, Ullberg M, Saksela O, Kronvall G. Tissue-type plasminogen activator-mediated activation of plasminogen on the surface of group A, C, and G streptococci. Infection and Immunity. 1992;60(1):196–201. doi: 10.1128/iai.60.1.196-201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Coleman JL, Gebbia JA, Piesman J, Degen JL, Bugge TH, Benach JL. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell. 1997;89(7):1111–1119. doi: 10.1016/s0092-8674(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 117.Lottenberg R, Minning-Wenz D, Boyle MDP. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends in Microbiology. 1994;2(1):20–24. doi: 10.1016/0966-842x(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 118.Castellino FJ, Ploplis VA. Structure and function of the plasminogen/plasmin system. Thrombosis and Haemostasis. 2005;93(4):647–654. doi: 10.1160/TH04-12-0842. [DOI] [PubMed] [Google Scholar]
- 119.Lahteenmaki K, Virkola R, Pouttu R, Kuusela P, Kukkonen M, Korhonen TK. Bacterial plasminogen receptors: in vitro evidence for a role in degradation of the mammalian extracellular matrix. Infection and Immunity. 1995;63(9):3659–3664. doi: 10.1128/iai.63.9.3659-3664.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Korhonen TK, Virkola R, Lahteenmaki K, et al. Penetration of fimbriate enteric bacteria through basement membranes: a hypothesis. FEMS Microbiology Letters. 1992;100(1-3):307–312. doi: 10.1111/j.1574-6968.1992.tb14057.x. [DOI] [PubMed] [Google Scholar]
- 121.Eberhard T, Kronvall G, Ullberg M. Surface bound plasmin promotes migration of Streptococcus pneumoniae through reconstituted basement membranes. Microbial Pathogenesis. 1999;26(3):175–181. doi: 10.1006/mpat.1998.0262. [DOI] [PubMed] [Google Scholar]
- 122.Yavlovich A, Katzenell A, Tarshis M, Higazi AAR, Rottem S. Mycoplasma fermentans binds to and invades HeLa cells: involvement of plasminogen and urokinase. Infection and Immunity. 2004;72(9):5004–5011. doi: 10.1128/IAI.72.9.5004-5011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Coleman JL, Sellati TJ, Testa JE, Kew RR, Furie MB, Benach JL. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infection and Immunity. 1995;63(7):2478–2484. doi: 10.1128/iai.63.7.2478-2484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun H, Ringdahl U, Momeister JW, et al. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305(5688):1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- 125.Cole JN, McArthur JD, McKay FC, et al. Trigger for group A streptococcal M1T1 invasive disease. The FASEB journal. 2006;20(10):1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- 126.Sanderson-Smith ML, Dinkla K, Cole JN, et al. M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. The FASEB Journal. 2008;22(8):2715–2722. doi: 10.1096/fj.07-105643. [DOI] [PubMed] [Google Scholar]
- 127.Shen W, Steinruck H, Ljungh A. Expression of binding of plasminogen, thrombospondin, vitronectin, and fibrinogen, and adhesive properties by Escherichia coli strains isolated from patients with colonic diseases. Gut. 1995;36(3):401–406. doi: 10.1136/gut.36.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kinnby B, Booth NA, Svensäter G. Plasminogen binding by oral streptococci from dental plaque and inflammatory lesions. Microbiology. 2008;154(part 3):924–931. doi: 10.1099/mic.0.2007/013235-0. [DOI] [PubMed] [Google Scholar]
- 129.Rooijakkers SHM, Van Wamel WJB, Ruyken M, Van Kessel KPM, Van Strijp JAG. Anti-opsonic properties of staphylokinase. Microbes and Infection. 2005;7(3):476–484. doi: 10.1016/j.micinf.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 130.Siemens N, Patenge N, Otto J, Fiedler T, Kreikemeyer B. Streptococcus pyogenes M49 plasminogen/plasmin binding facilitates keratinocyte invasion via integrin-integrin-linked kinase (ILK) pathways and protects from macrophage killing. The Journal of Biological Chemistry. 2011;286(24):21612–21622. doi: 10.1074/jbc.M110.202671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Blom AM, Hallström T, Riesbeck K. Complement evasion strategies of pathogens-Acquisition of inhibitors and beyond. Molecular Immunology. 2009;46(14):2808–2817. doi: 10.1016/j.molimm.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 132.Ploplis VA, French EL, Carmeliet P, Collen D, Plow EF. Plasminogen deficiency differentially affects recruitment of inflammatory cell populations in mice. Blood. 1998;91(6):2005–2009. [PubMed] [Google Scholar]
- 133.Ramu P, Tanskanen R, Holmberg M, Lähteenmäki K, Korhonen TK, Meri S. The surface protease PgtE of Salmonella enterica affects complement activity by proteolytically cleaving C3b, C4b and C5. FEBS Letters. 2007;581(9):1716–1720. doi: 10.1016/j.febslet.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 134.Seya T, Nagasawa S. Limited proteolysis of complement protein C3b by regulatory enzyme C3b inactivator: isolation and characterization of a biologically active fragment, C3d,g. Journal of Biochemistry. 1985;97(1):373–382. doi: 10.1093/oxfordjournals.jbchem.a135064. [DOI] [PubMed] [Google Scholar]
- 135.Vieira ML, de Morais ZM, Vasconcellos SA, Romero EC, Nascimento ALTO. In vitro evidence for immune evasion activity by human plasmin associated to pathogenic Leptospira interrogans. Microbial Pathogenesis. 2011;51(5):360–365. doi: 10.1016/j.micpath.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 136.Degen JL, Bugge TH, Goguen JD. Fibrin and fibrinolysis in infection and host defense. Journal of Thrombosis and Haemostasis. 2007;5(supplement 1):24–31. doi: 10.1111/j.1538-7836.2007.02519.x. [DOI] [PubMed] [Google Scholar]
- 137.Pancholi V, Fontan P, Jin H. Plasminogen-mediated group A streptococcal adherence to and pericellular invasion of human pharyngeal cells. Microbial Pathogenesis. 2003;35(6):293–303. doi: 10.1016/j.micpath.2003.08.004. [DOI] [PubMed] [Google Scholar]


