Abstract
There is no doubt that genomes are organized nonrandomly in the nucleus of higher eukaryotes. But what is the functional relevance of this nonrandomness? In this Essay, we explore the biological meaning of spatial gene positioning by examining the functional link between the activity of a gene and its radial position in the nucleus.
Arguably one of the most important and tantalizing recent discoveries in the field of genome biology has been the realization that genomes are nonrandomly organized within the cell nucleus of higher eukaryotes (Misteli, 2007; Schneider and Grosschedl, 2007). Analysis of the location of genes and chromosomes in a number of cell types and tissues has revealed that genomic elements occupy preferential positions within the nucleus. The positions vary among tissues and cell types, and repositioning occurs during physiological processes such as differentiation and in pathological situations. Positioning patterns are also evolutionarily conserved pointing to a functional role for positioning in genome activity and homeostasis.
The preferential location of chromosomes and genes to particular nuclear locales has implications for all aspects of genome function. An emerging idea is that clustering of genes in transcription hot spots contributes to their efficient regulation and expression (Fraser and Bickmore, 2007; Lanctot et al., 2007) and that the relative positioning of chromosomes is important in the formation of translocations (Misteli, 2007). Furthermore, gene positioning has been linked to replication timing (Gilbert, 2001), and physical interactions between X chromosomes may play a role in X-inactivation (Erwin and Lee, 2008).
The idea of nonrandom positioning of chromosomes and genes has a long and often anecdotal history, much of which is based on correlative observations (Cremer and Cremer, 2006; Hochstrasser et al., 1986). A key obstacle in these studies was the difficulty in measuring the position of a chromosome or a gene in a quantitative manner. A breakthrough occurred when Bickmore and colleagues introduced the concept of radial position, that is, the position of a chromosome or a gene along the axis between the center of the nucleus and the periphery (Croft et al., 1999). In a landmark study, they demonstrated using quantitative analysis that in human lymphocytes chromosome 18 was preferentially located toward the periphery, whereas chromosome 19 was generally located toward the interior of the nucleus. At the time, there was little biological reason for analyzing the radial positions of chromosomes; it was simply a convenient parameter to measure. However, having established the concept of nonrandom radial positioning, the question arose as to whether the radial location of a locus was related to its function.
Correlating Gene Activity and Radial Gene Position: The Pros
The possibility that radial positioning is functionally relevant was hinted at in the original study by Bickmore and colleagues by the fact that internally localized chromosome 19 is the most gene-dense human chromosome, whereas peripheral chromosome 18 is one of the least gene-dense chromosomes. The case for a functional role of position was further strengthened by the finding that the GC-rich portion of the genome, which is also enriched in genes, tends to be more internally positioned than GC-poor, gene-poor DNA (Ferreira et al., 1997). Moreover, late-replicating regions of the genome, containing predominantly nongenic regions, are generally found at the nuclear periphery, whereas early-replicating regions, which are rich in active genes, are located closer to the center of the nucleus (Gilbert, 2001).
The strongest support for a functional link between radial position and gene activity thus far comes from the observation of movement of several genes from a peripheral position into the interior upon their activation. Prominent examples include β-globin during differentiation of mouse erythroid cells; IgH and IgK in murine B cell differentiation; GATA-3 and c-maf during murine T cell differentiation; and Mash1 during differentiation of mouse neurons (Hewitt et al., 2004; Kosak et al., 2002; Ragoczy et al., 2006; Williams et al., 2006). Although these genes tend to localize closely with the very edge of the nucleus when inactive, others such as GFAP in murine astrocytes or HoxB1 and HoxB9 in mouse embryos also undergo a shift toward a more internal location upon activation, but even in their inactive state they do not localize at the nuclear envelope (Chambeyron and Bickmore, 2004; Takizawa et al., 2008). Support for a link between radial positioning and gene activity also comes from analysis of the two alleles of the monoallelically expressed GFAP gene in single nuclei revealing that the active allele is generally found more internally compared to its inactive copy within the same nucleus (Takizawa et al., 2008). Changes in radial positions of genes coincidental with changes in their expression are not unique to mammalian cells. In yeast, silent genes are often associated with the periphery and reporter genes placed near telomeres, which cluster at the nuclear periphery, are efficiently silenced (Akhtar and Gasser, 2007). On the other hand, a number of yeast genes move toward the yeast periphery upon activation (Brown and Silver, 2007).
Lack of Correlation between Gene Activity and Radial Position: The Cons
Despite this list of correlations, we now know that the notion of localization of inactive genes at the periphery and active ones in the nuclear interior is an oversimplification and is not a universal hallmark of gene activation. For most biallelically expressed genes the two alleles are often in vastly different radial positions in the same nucleus, yet their activity status appears similar based on the strength of fluorescence in situ hybridization signals (Figure 1A). Additionally, a recent study of the monoallelically expressed GFAP gene demonstrated that although the inactive locus is generally more peripheral than the active one, in a fraction of nuclei the inactive allele was more internally localized than the active allele (Takizawa et al., 2008). Another general observation argues against a strong link between radial location and gene activity: if radial positioning were directly linked to expression, it would follow that transcription should occur predominantly in the interior of the nucleus. Yet, active sites of RNA polymerase II transcription are distributed uniformly throughout the nucleus (except for the nucleoli) with no apparent radial preference (Wansink et al., 1993), although preferential internal transcription zones might exist in specialized cells (Kosak et al., 2007). Similarly, heterochromatin, which is largely transcriptionally silent, is not restricted to a specific radial position, and large blocks of heterochromatin can be found throughout the nucleus (Figure 1B).
Figure 1. Radial Positioning of Genes.
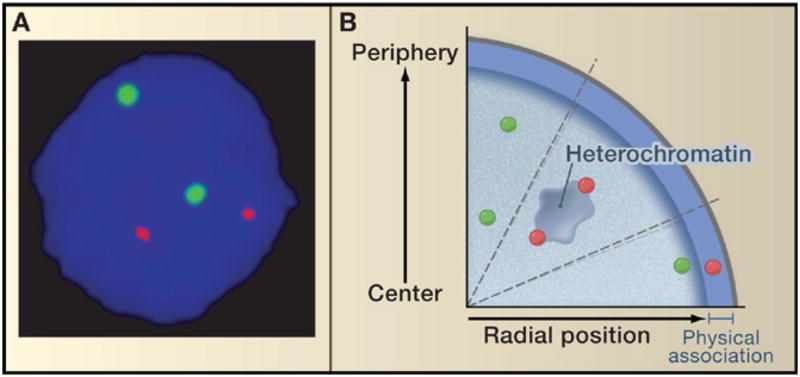
(A) Active genes can be anywhere in the nucleus. The radial positions of biallelically expressed genes often vary between the two homologous alleles in the same nucleus. Shown are the locations of the two alleles of the IGH (green) and MYC (red) genes in human lymphocytes.
(B) Functional significance of radial positioning. (Top) Active genes (green) exhibit a large range of radial positions; the precise radial position of a locus does not correlate with its activity level. (Middle) Inactive genes (red) may associate with heterochromatin blocks at various radial positions. (Bottom) In contrast to radial positioning, physical association with the nuclear periphery is often linked to silencing. Genes that are in close proximity to the nuclear envelope but do not physically interact with it may be active.
A general link between gene activity and radial position is even more strongly challenged by observations on single genes. Many gene loci remain in the same radial positions when their expression changes (Hewitt et al., 2004; Meaburn and Misteli, 2008; Zink et al., 2004). A lack of direct causality between gene expression and radial position is also highlighted by the fact that genes can become repositioned radially in the absence of detectable changes to their transcriptional output. For example, the Pah gene becomes more internally localized during differentiation of mouse neurons, and VEGF becomes more peripherally localized during the induction of tumor formation in breast epithelia, despite no change in expression (Meaburn and Misteli, 2008; Williams et al., 2006). In a recent study of 11 randomly selected genes analyzed under various growth and differentiation conditions, no general correlation between activity and radial position was found (Meaburn and Misteli, 2008). Finally, even observations on a peripherally silenced gene undermine the notion of a close link between repression and radial positioning. The β-globin gene, which is peripheral in its inactive form, remains at the periphery during early stages of activation and only then undergoes internalization (Ragoczy et al., 2006). This latter observation suggests that internal positioning is not a requirement for activity and that transcription alone does not drive the position of a gene. Taken together, the fact that genes can alter radial position without changes in expression, and that many genes do not undergo positional changes when their expression levels are modulated, indicates that radial positioning is functionally not tightly linked to gene activity.
A Key Experiment
The pros and cons in the long-standing debate on the role of radial positioning in gene activity are entirely based on correlative observations, often in the absence of precise measurements of gene activity. A much needed key experiment was to artificially change the position of a gene and test the transcriptional consequences. This has recently been done in three laboratories by artificially tethering reporter genes to the nuclear periphery of mammalian cells using various nuclear envelope and lamina proteins. The results were more ambiguous than hoped for. In one system, transcription of a reporter gene was significantly repressed upon association with the nuclear periphery via tethering to the inner nuclear membrane protein emerin (Reddy et al., 2008). A second system looked at the expression of multiple endogenous genes in domains tethered to the periphery by the lamin-associated protein LAP2β. Although expression of some genes was negatively affected, that of others was not (Finlan et al., 2008). Finally, in a third approach, an inducible reporter was placed at the nuclear periphery by interaction with lamin B. Location of the reporter at the nuclear periphery did not prevent its activation upon stimulation and the locus retained its full transcriptional competence (Kumaran and Spector, 2008). The apparent discrepancies in these results likely reflect experimental differences between the approaches. For example, it is not clear whether the induction of transcription after tethering to the periphery involves the same regulatory mechanisms as ongoing transcription. Additionally, although the reporter gene in the study by Reddy et al. was repressed upon relocation to the periphery, the reduction in expression was ~2-fold but was not complete unlike the case for endogenous genes in the study by Finlan et al. This suggests that despite the repressive effect of the nuclear periphery, association with the periphery alone does not totally silence the locus but merely reduces its transcription. Finally, the discrepancies might reflect the presence of the various reporter genes in different microenvironments, either as part of the endogenous heterogeneity of peripheral chromatin or created by the various experimental approaches. Although these important experiments do not unequivocally resolve the role of radial positioning in gene expression, they do mirror the findings from correlation-based experiments. So, why then is it that radial positioning seems to have different effects on different genes?
Peripheral Location Is Not Equal to Peripheral Association
A likely reason for the difficulties in interpreting correlative experiments between positioning and gene activity is that it is important to make a distinction between radial positioning toward the periphery and direct physical association with the nuclear envelope (Figure 1B). Although the tethering experiments suggest that physical association with the nuclear envelope can contribute to transcriptional repression, there is no reason to think that being near the periphery without physically associating with the nuclear lamina leads to repression. A good example is the human CFTR gene—it is located at the nuclear periphery when inactive and, although it becomes more internally localized when active, it remains in the very peripheral region of the nucleus (Zink et al., 2004). In addition, there is very little evidence to suggest that the precise location of a locus along the radial axis matters (Figure 1B). A locus halfway between the nuclear center and the periphery does not seem to have a lower probability of being active than a locus at the very center. So, we should not think about a correlation between the radial position of a gene and its activity, but about the functional role of physical association with the nuclear periphery. This seems a particularly critical point given that the light microscopy methods used in many positioning studies have resolution limits of ~250 nm and cannot distinguish between physical association and mere spatial proximity.
How the physical association of genes with the nuclear periphery affects their function is still largely unclear (Akhtar and Gasser, 2007; Brown and Silver, 2007; Misteli, 2004). The nuclear edge has traditionally been thought to provide a repressive milieu. This assumption came from microscopy images where the nuclear envelope abuts regions of dense chromatin or heterochromatin. This is supported at the molecular level by the fact that the major structural component of the nuclear periphery, the lamina, interacts with heterochromatin proteins such as heterochromatin protein 1, which is essential for the organization of heterochromatin. In support of a repressive role of the nuclear envelope, analysis of lamina-associated domains in human and fly cells revealed an enrichment of gene-poor and transcriptionally inactive regions in the lamin-associated domains (Guelen et al., 2008; Pickersgill et al., 2006). In addition, ChIP-microarray analysis of human cells has detected extensive interactions between the nuclear pore complex (NPC) and gene-poor regions (Brown et al., 2008a). These findings in mammalian cells are closely mirrored by those in the budding yeast Saccharomyces cerevisiae where extensive evidence exists for silencing, particularly of telomeres and mating-type loci, at the periphery (Akhtar and Gasser, 2007; Brown and Silver, 2007; Misteli, 2004).
However, things are more complicated. It increasingly appears that different parts of the nuclear periphery have distinct roles in transcriptional regulation. ChIP studies in yeast and mammalian cells suggest a strong correlation between association with the NPC and gene activation (Brown et al., 2008a; Brown and Silver, 2007). This idea is exemplified in flies where the dosage compensation complex mediating global upregulation of gene expression on the male X chromosome (which is localized at the nuclear periphery) directly interacts with NPCs (Mendjan et al., 2006). In contrast, genome-wide mapping studies in flies and mammalian cells indicate that genome regions associated with stretches of the lamina between the NPCs are predominantly transcriptionally repressed (Guelen et al., 2008; Pickersgill et al., 2006). Thus, different regions of the nuclear periphery may exert different regulatory effects on a gene, further confounding the analysis of radial gene position and gene activity.
Different Genes, Different Behavior
Another reason why some genes undergo repositioning whereas others do not when gene activity changes is revealed by closer analysis of the types of genes that become repositioned and the circumstances of their repositioning. Strikingly, most genes for which a correlation between positioning and activity has been reported are those whose activity is tightly linked to differentiation events—examples include IgH and β-globin during B cell and erythroid cell differentiation, respectively (Kosak et al., 2002; Ragoczy et al., 2006), and genes of the hoxB cluster during development (Chambeyron and Bickmore, 2004). These genes transition from a silenced state to an active one as part of the differentiation process. On the other hand, many of the genes for which movement does not correlate with expression are genes whose activity changes but are never induced or completely silenced during differentiation, such as BCL2, TP53, and ERBB2IP during mammary epithelial cell differentiation (Meaburn and Misteli, 2008). Thus, a key difference between these groups of genes might be the changes in chromatin status they undergo as part of their activation. Although differentiation-induced genes are generally present in permanently repressed chromatin regions when they are inactive, many of the genes that do not change positions are already in an active, or possibly poised, chromatin state. Interestingly, most genes, including IgH, β-globin, and HoxB, that change position during their activation are associated with heterochromatin blocks in their inactive states (Figure 1B), but genes that do not undergo radial repositioning generally are not (Francastel et al., 1999). It is then possible that radial gene repositioning reflects to a large extent the dissociation of a locus from, often peripheral, heterochromatin blocks.
Neighborhood Matters
A further factor in determining whether a locus changes its position or not is its chromosomal neighborhood. Although a gene itself may not have an altered expression pattern, changes in expression at a nearby locus might drive its repositioning, making the gene simply a passenger in the spatial movements of adjacent regions. For example, in murine embryonic stem cells, Pah and Igf1 are adjacent to the Mash1 locus and remain transcriptionally silent during neuronal differentiation yet become more internally positioned along with differentiation-induced Mash1 (Williams et al., 2006). A neighborhood effect is also suggested by the observation that radial positioning correlates with local gene density, with locally gene-dense regions preferentially having an internal position (Murmann et al., 2005). The influence of the chromosomal context of a gene regarding its propensity to become repositioned has also been suggested when comparing gene behavior between species (Brown et al., 2006, 2008b). In human erythroblasts, the α-globin gene becomes repositioned away from its chromosome territory and upon activation is often juxtaposed with β-globin. During this differentiation process in mice, however, these genes do not become juxtaposed and α-globin remains within its chromosome territory. Interestingly, the chromosomal contexts of these genes in the human and mouse genomes are quite different. In human cells, α-globin is close to a telomere in a gene-dense region enriched in housekeeping genes, whereas in the mouse it is proximal to a centromere and in a region of lower gene density (Brown et al., 2006). Having said that, neighborhood effects are not universal. CFTR and its neighboring genes become repositioned away from the nuclear periphery independently of each other when activated (Zink et al., 2004). One difference between the neighborhoods of the Mash1 and CFTR loci is the number and type of genes activated. The genes surrounding CFTR are not coordinately regulated, whereas those flanking one side of Mash1 are. Thus, movements of genomic neighborhoods may occur preferentially if multiple genes are activated. An extreme case of this is the dramatic expulsion of large loops of several micrometers containing active multigene clusters from the body of the chromosome (Misteli, 2007). It is highly likely that neighborhood effects also apply to gene repression although no examples have been reported to date.
Beyond the Tip of the Iceberg
Radial positioning is routinely used as a surrogate to determine whether a gene undergoes a change in its nuclear location, but radial positioning is merely the proverbial tip of the iceberg. It is important to point out that a lack of apparent change in radial position does not mean that a locus does not change its position within the nucleus. A gene may alter its location relative to other gene loci, to intranuclear compartments, or to heterochromatin domains. Such relative movements, not necessarily accompanied by radial movements, are increasingly being recognized as functionally relevant. The importance of relative positioning lies in its ability to bring distantly located genome regions into close spatial proximity, allowing their physical interaction. A striking example is the recently reported association of the active allele of the monoallelically expressed interferon-β gene with three regulatory sequences on distinct chromosomes (Apostolou and Thanos, 2008). The authors proposed that these sequences facilitate the assembly of the transcriptional machinery via their interaction with the interferon-β locus. How sequences on different chromosomes find each other in the nucleus and whether and how they move within the nucleus are key questions in the field. An intriguing possibility comes from the observation in living cells of linear actin/myosin-mediated motion of genome regions over long distances (Chuang et al., 2006; Dundr et al., 2007), perhaps pointing to the existence of directed gene transport mechanisms within the nucleus.
Another manifestation of the potentially important role of relative positioning is the clustering of coregulated genes in nuclear space. Such clustering is well established for ribosomal genes that aggregate to form the nucleolus. Similar clustering has been suggested for genes transcribed by RNA polymerase II, exemplified by the association of coregulated genes during erythrocyte differentiation in transcription centers enriched in RNA polymerase II (Fraser and Bickmore, 2007). How generally applicable clustering of coregulated genes is and how precisely clustered genes associate with each other is not known. One model suggests that gene clusters represent transcription factories, which consist of preassembled transcription complexes that serve multiple genes in a centralized fashion (Fraser and Bickmore, 2007). Alternatively, other observations favor the interpretation that clustered genes associate with intranuclear structures termed interchromatin granules, which are enriched in pre-mRNA splicing factors and roughly correspond to nuclear speckles (Brown et al., 2006, 2008b; Lawrence and Clemson, 2008). In addition to gene activation, clustering of genes and chromosomes has also been implicated in repression, imprinting, and X chromosome inactivation (Fraser and Bickmore, 2007; Misteli, 2007; Schneider and Grosschedl, 2007), pointing to a ubiquitous role of relative positioning in genome function. It appears from these observations that the relative positioning of genes and genomic regions to each other undergoes more dramatic changes during various events and might therefore be functionally more important than radial positioning.
Conclusions
The discovery of distinct radial positions of chromosomes and genes has changed the way we think about genome organization. It has highlighted the non-randomness of higher-order genome organization and it has inspired the pursuit of how spatial genome organization contributes to function. Ironically, despite its importance in uncovering this fundamental principle of nuclear organization, the functional relevance of radial gene positioning has remained elusive. Clearly, radial gene positioning (and probably relative gene positioning too) are affected by multiple components, and positional changes of a given gene locus are not determined by a single mechanism. Furthermore, it appears that different genes behave very differently and it is not easy to deduce universal rules. A complicating factor in unraveling the positioning-function relationship is that many studies to date have focused on probing the effect of single parameters on the positioning of single genes. More complex, systematic, and unbiased methods of analysis are required to begin to understand the rules and consequences of genome positioning events. Fortunately, such methods are now available. There is no reason why the combined use of genome-wide expression analysis, genome-wide interaction maps based on chromosome conformation capture analysis, and high-throughput imaging to analyze large numbers of genes should not eventually reveal the true meaning of gene positioning. It will likely be one of the most important revelations in our understanding of how genomes function.
References
- Akhtar A, Gasser SM. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Brown CR, Silver PA. Curr Opin Genet Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Genes Dev. 2008a;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Green J, das Neves RP, Wallace HAC, Smith AJM, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. J Cell Biol. 2008b;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Eur J Histochem. 2006;50:161–176. [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Lee JT. Curr Opin Cell Biol. 2008;20:349–355. doi: 10.1016/j.ceb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J, Paolella G, Ramos C, Lamond AI. J Cell Biol. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francastel C, Walters MC, Groudine M, Martin DI. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. J Cell Biol. 2001;152:F11–F15. doi: 10.1083/jcb.152.2.f11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. Eur J Immunol. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Scalzo D, Alworth SV, Li F, Palmer S, Enver T, Lee JS, Groudine M. PLoS Biol. 2007;5:e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Clemson CM. J Cell Biol. 2008;182:1035–1038. doi: 10.1083/jcb.200808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. J Cell Biol. 2008;180:39–50. doi: 10.1083/jcb.200708204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Misteli T. Cell. 2004;119:153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Misteli T. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Murmann AE, Gao J, Encinosa M, Gautier M, Peter ME, Eils R, Lichter P, Rowley JD. Exp Cell Res. 2005;311:14–26. doi: 10.1016/j.yexcr.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. J Cell Biol. 1993;122:282–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Land S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, et al. J Cell Biol. 2004;I166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]


