Abstract
DNA repair and maintenance of genome stability are crucial to cellular and organismal function, and defects in these processes have been implicated in cancer and ageing. Detailed molecular, biochemical and genetic analyses have outlined the molecular framework involved in cellular DNA-repair pathways, but recent cell-biological approaches have revealed important roles for the spatial and temporal organization of the DNA-repair machinery during the recognition of DNA lesions and the assembly of repair complexes. It has also become clear that local higher-order chromatin structure, chromatin dynamics and non-random global genome organization are key factors in genome maintenance. These cell-biological features of DNA repair illustrate an emerging role for nuclear architecture in multiple aspects of genome maintenance.
Genomes are continuously assaulted and must be efficiently and accurately repaired to maintain their integrity. The most common challenges come from environmental stress, such as ultraviolet light, but also from endogenous events, such as the intracellular production of reactive oxygen species, recombination during the immune system’s response and stalled DNA-replication forks1. These insults lead to base damage, single-stranded DNA (ssDNA) breaks and double-stranded DNA breaks (DSBs). DSBs are particularly dangerous to cells as their inefficient or inaccurate repair can result in mutations and chromosomal translocations that may induce cancer2. Elaborate cellular repair and genome surveillance mechanisms counteract genomic damage from DSBs.
DSB repair is carried out by two main pathways: non- homologous end joining (NHEJ) and homologous recombination repair3 (HRR) (BOX 1). NHEJ is the major pathway for repairing non-replication-associated breaks and occurs predominantly in the G1 phase of the cell cycle. By contrast, HRR occurs mainly during late S–G2 phase. And, whereas DSB ends are simply joined in NHEJ, HRR uses a sister homologue as a template for repair3 (BOX 1). But DSB repair involves more than simply rejoining broken regions of chromosomes. DSBs result in the activation of a complex cellular DNA-damage response (DDR) cascade, which involves sensing the DNA damage and subsequent amplification and transmission of a damage signal to evoke a multitude of cellular responses — particularly activation of cell-cycle checkpoint kinases, which stall damaged cells in their cell cycle until resolution of the lesion. If unresolved DSBs persist, the DDR activates apoptotic pathways or directs damaged cells into senescence. DDR pathways are universal and the majority of the involved proteins are highly conserved from yeast to humans4.
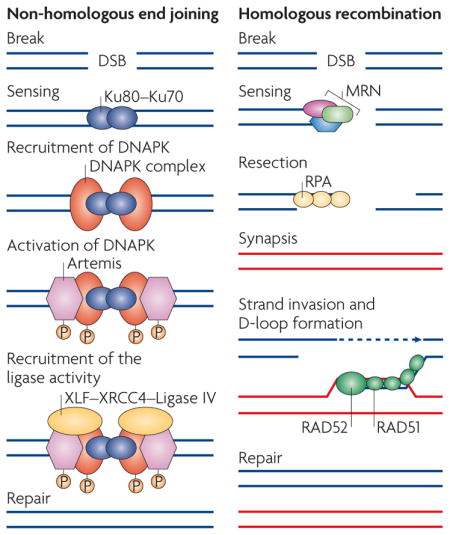
Box 1. The two main types of double-stranded DNA-break repair.
Non-homologous end joining
A DNA lesion (a double-stranded DNA break (DSB)) is sensed by the Ku80–Ku70 heterodimer, which in turn recruits the DNA-dependent protein kinase catalytic subunit DNAPKcs, resulting in assembly of the DNAPK complex and activation of its kinase activity (see the figure; left panel). Increasing evidence suggests that DNAPK functions as a regulatory component of non-homologous end joining (NHEJ), potentially facilitating and regulating the processing of DNA ends. DNAPK also increases the recruitment of XRCC4, DNA ligase IV, XLF and Artemis, which carry out the final rejoining reaction.
Homologous recombination repair
A DNA lesion is recognized by the MRN (MRE11–RAD50–NBS1) complex, which is recruited to the DSB to generate single-stranded DNA by resection (see the figure; right panel). The single-stranded ends are bound by replication protein A (RPA), RAD51 and RAD52 and can subsequently invade the homologous template, creating a D-loop and a Holliday junction, to prime DNA synthesis and to copy and ultimately restore genetic information that was disrupted by the DSB.
The molecular identity of DDR pathways has been outlined over the past decades using mostly biochemical, genetic and molecular methods. However, these approaches do not fully address the key issue of how repair occurs in the context of the genome. Understanding the cell biology of DNA repair and genome stability is particularly relevant, as many steps in the DNA-damage repair process rely on the spatial and temporal coordination of events. In addition, many key features of the DNA-repair machinery and the DDR are directly linked to the complex and highly organized structure of the cell nucleus5. Like many nuclear proteins, DNA-repair factors diffuse rapidly in the nuclear space and are then recruited to DNA lesions. Furthermore, the elaborate higher-order organization of chromatin is important in assembling the repair machinery, making DNA lesions accessible to the repair complexes, and might influence how readily a lesion is detected and how the repair machinery responds to it. The fact that the cell nucleus is not a homogeneous structure, but rather contains a large number of functionally distinct subcompartments, raises the question of whether DNA repair occurs everywhere in the nucleus with equal efficiency or whether specialized repair centre sites exist. Of particular importance for genome stability is the global organization of genomes in the nucleus, as the non-random arrangement of genes and chromosomes in the nucleus may predispose particular genome regions to rearrangements.
In this Review, we discuss how DNA repair occurs in the context of nuclear architecture. We will focus on DSB repair through the NHEJ and HRR pathways, as those are most relevant for human disease and cancer. In particular, we will summarize the role of spatial and temporal coordination in DSB-repair complex assembly, the influence of higher-order chromatin structure on the repair process and the effect of nuclear compartmentalization and non-random genome organization on maintenance of genome integrity.
Spatio-temporal organization of repair
The assembly of the cellular machinery, which recognizes DNA damage, triggers cellular signalling pathways to combat DNA lesions and eventually repairs them, involves many protein components. Their assembly at sites of DNA damage involves complex spatial and temporal coordination of many dynamic interactions among repair proteins and with the chromatin fibre.
Repair complex assembly
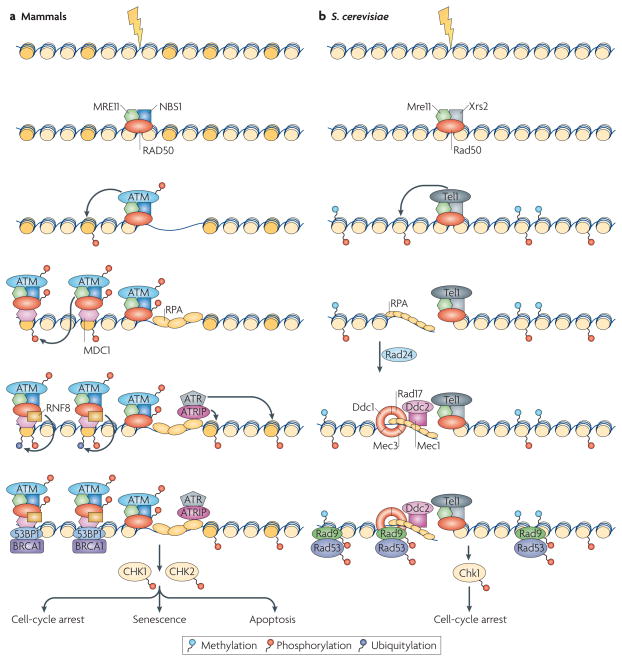
The assembly of the repair machinery on a DNA lesion occurs in a highly ordered, spatially and temporally coordinated manner, according to a strict hierarchy. The cascade of events starts in mammalian cells with the rapid accumulation of the primary damage sensor — the MRN (MRE11–RAD50–NBS1) complex6–8 — which detects the lesion. This complex is crucial for the recruitment and activation of the key DDR signalling kinase ATM6,9,10, which phosphorylates the histone variant H2AX at nucleosomes around the DSBs. This is followed by binding to the DSB-flanking chromatin of the mediator protein MDC1, which has affinity for phospho-H2AX11 and acts as a platform for the association of upstream12–15 and downstream effectors16–21. MDC1 triggers the recruitment of chromatin remodelling and modification complexes, which allow the association of downstream factors, such as 53BP1 and BRCA1 (FIG. 1)16–21. The resulting ssDNA is recognized by replication protein A (RPA), which subsequently recruits ATR, another downstream transducing kinase2 (FIG. 1). The key components and sequence of events of the DSB response are largely conserved in Saccharomyces cerevisiae22,23 (FIG. 1).
Figure 1. Assembly of DNA-damage response complexes.
a | In mammalian cells, the presence of a double-stranded DNA break (DSB) is originally sensed by the MRN (MRE11–RAD50–NBS1) sensor complex. The recruitment of MRN activates the transducer kinase ATM, which associates with DSBs and phosphorylates the histone variant H2AX (dark yellow) in DSB-flanking nucleosomes. The mediator MDC1 protein then binds to γ-H2AX and recruits additional copies of MRN and ATM, leading to spreading of the repair machinery along the chromosome. MDC1 also recruits ubiquitin ligase activities (for example, RNF8) that are responsible for the recruitment of downstream factors such as 53BP1 and BRAC1. DNA is then resected to single-stranded DNA (ssDNA) and recognized by replication protein A (RPA), which results in the recruitment of ATR through its interacting partner ATRIP. Both the ATM- and the ATR-dependent branches of the pathway, independently or in concert, lead to the activation of the checkpoint kinases CHK1 and CHK2. b | In Saccharomyces cerevisae, the DNA-damage response (DDR) cascade starts with the binding of the MRX (Mre11–Rad50–Xrs2) complex to the DSB. MRX recruits the Tel1 kinase, which phosphorylates histone H2A. After resection, ssDNA is sensed by RPA, which recruits the Ddc1–Mec3–Rad17 (known as 9–1–1 complex in mammals) sensor complex (Rad24 acts as a clamp loader) and the Ddc2–Mec1 complex independently. Next, Rad9 is recruited through its affinity for phosphorylated H2A and methylated H3, and the checkpoint kinases Chk1 and Rad53 are activated.
The hierarchical nature of assembly of repair complex components is powerfully demonstrated both in yeast and mammalian cells by protein-tethering experiments. In these studies, repair factors and checkpoint proteins are permanently immobilized on chromatin as fusion proteins with the Lac repressor, which binds to a Lac operator sequence placed in the genome, thus mimicking the effect of their immobilization during the repair process24–26. In support of a top–down hierarchy of assembly, immobilization of the upstream mammalian sensors NBS1 and MRE11 triggers the entire DDR downstream cascade by attracting the ATM kinase and several downstream factors25,26. Similarly in yeast, co-tethering of components of the downstream Ddc2–Mec1 and Ddc1–Mec3–Rad17 (known as 9–1–1 complex in mammals) sensor complexes to chromatin bypasses the requirement for the recruitment of the Rad24 clamp loader to load the 9–1–1 clamp complex at the sites of damage24. This hierarchical organization of the repair machinery is, however, complemented by crosstalk between the factors. In mammalian cells, tethering of the mediator MDC1 leads to recruitment of the upstream factors NBS1 and MRE11 in an H2AX-independent manner26. The ability of downstream components to recruit upstream factors is probably crucial in the amplification of the damage signal by a single DSB25,26.
DNA-repair foci
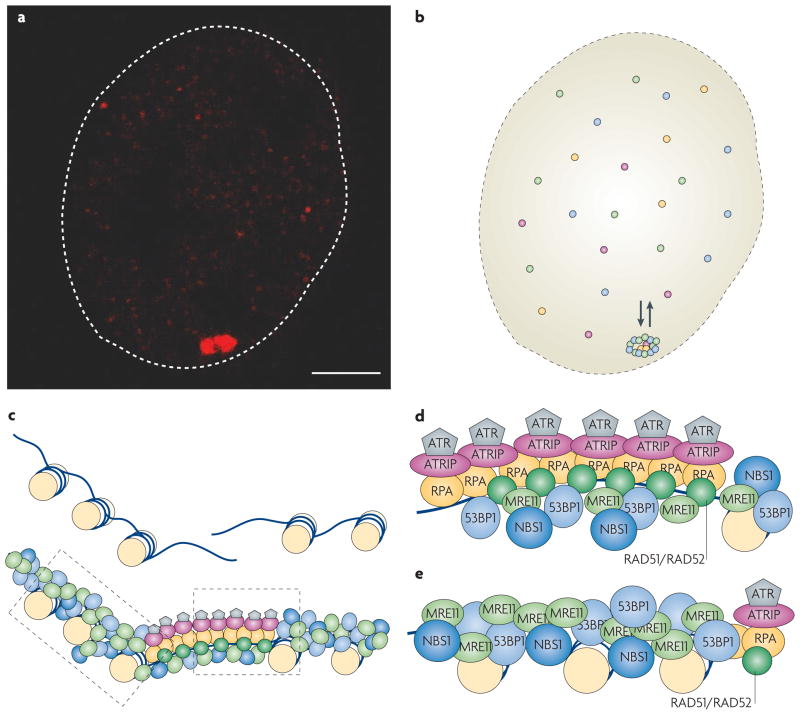
The DNA-repair process manifests itself cytologically in the form of DNA-repair foci (FIG. 2). These foci are structures that are formed by the recruitment and accumulation of DNA-repair factors at a site of DNA damage27. As a consequence, a single DSB nucleates a macroscopically discernible focus, which spreads up to one megabase from the break27 (FIG. 2a,b). Foci can be experimentally induced by γ-irradiation, radiometric drugs, laser microirradiation or sequence-specific endonucleases27. They are ubiquitous and form in organisms ranging from yeast to mammalian cells.
Figure 2. DNA-repair foci and their dynamics.
a | Repair foci are the cytological manifestations of the DNA-repair process. In this example, fluorescently tagged MDC1 (red) accumulates in NIH3-3T3 cells upon induction of a single double-stranded DNA break (DSB) by the ISceI endonuclease. The scale bar represents 2 μm. b | Repair foci are stable, yet highly dynamic structures. Although they persist for extended periods of time, their resident proteins are recruited from a freely diffusible nucleoplasmic pool that is in continuous dynamic exchange with repair foci. Species of repair factors are indicated in different colours. c | Schematic illustration of the ‘microstructure’ of repair foci. The centre of the repair focus contains resected single-stranded DNA (ssDNA). This region is occupied by a specific set of factors that generate and have affinity for ssDNA regions (grey, purple, yellow and dark green), including signalling molecules and components that are involved in restoring normal chromatin structure. The regions flanking the actual break are occupied by a distinct set of proteins (blue and light green) that is involved in spreading and amplifying the DNA-damage response (DDR) signal. d | Hypothetical organization of a ssDNA microcompartment. Factors that directly or indirectly associate with the resected ssDNA, such as replication protein A (RPA), the recombination factors RAD51 and RAD52, the ATR kinase and its interacting partner ATRIP, accumulate in a central ssDNA microcompartment. e | Hypothetical organization of a region that flanks a DSB. Factors such as the sensor complex MRN (MRE11–RAD50–NBS1), the mediator MDC1 and downstream factors 53BP1 and BRCA1 are found in the central region of the damage site (see panel d), but also spread up to a megabase away from the physical break. (Note that not all factors are shown.)
The structural organization of repair foci has not been elucidated; however, it appears that they are not merely random aggregates of repair components, as different factors exhibit distinct patterns of association with the DNA lesion28 (FIG. 2c–e). The recombination factors RAD51 and RAD52, the ATR kinase and its interacting partners ATRIP and RPA, and the DNA-clamp proteins RAD17 and RAD9 are restricted to a small area around the DSB, which corresponds mainly to stretches of ssDNA to which these proteins bind (FIG. 2d). By contrast, several factors, including the sensor complex MRN, the DDR mediator MDC1, the ATM kinase and the downstream factors 53BP1 and BRCA1, spread up to a megabase away from the break28 (FIG. 2e). Interestingly, the downstream effector kinases CHK1 and CHK2 and other effectors of DDR such as p53 and CDC25A do not visibly accumulate at damage sites28, suggesting that these proteins only interact transiently with the sites of damage, where they presumably become activated, and then rapidly spread to the entire nucleus. Members of the NHEJ pathway, such as the Ku80–Ku70 heterodimer and the DNA protein kinase DNAPK28, do not accumulate in foci either, presumably because they are only required at low copy numbers at sites of damage. Repair foci can form at all stages of the cell cycle, but their appearance is dictated by the cell-cycle dependence of the involved repair process.
Whether the accumulation of repair factors in foci is essential for efficient repair is unclear. The accumulation of factors could play a structural role by keeping the broken ends in spatial proximity, enhancing the efficiency of repair. Another likely, and potentially essential, function of foci is the amplification of the DNA-damage signal by the recruitment of multiple copies of signalling kinases to sites of damage. Consistent with this idea, artificial foci formation by immobilization of a single or a combination of checkpoint proteins to chromatin triggers the DDR and delays the cell cycle in the absence of DNA lesions in both yeast and mammalian cells24,26 (see below).
The dynamic nature of repair foci
Although repair foci are stable cytological structures once they are formed, they are intrinsically highly dynamic29,30. Most of the factors involved are in constant dynamic exchange between the chromatin-bound pool and the freely diffusing nucleoplasmic fraction, and their residence time in the foci depends on the nature of their interaction with damaged chromatin as well as on their affinity for other components of the repair machinery. For example, the sensor NBS1 has a residence time of less than one second, whereas the mediator MDC1 dwells in repair foci for ~10 times longer30. It is tempting to speculate that, as has been demonstrated for RNA polymerase assembly during transcription31, modulation of the dynamic exchange of repair factors with their chromatin or protein targets by post-translational modifications might serve a regulatory function to rapidly adjust the DDR to environmental conditions31.
Analysis of dynamic properties of factors in repair foci using combined imaging and kinetic computational modelling approaches has uncovered fundamental features of repair-complex assembly32. First, they suggest, in agreement with a hierarchical assembly model, that the repair machinery assembles in a sequential manner rather than by random assembly at the site of damage. Surprisingly, although several repair factors, including NBS1, MDC1 and the RAD52 group of homologous recombination proteins, form a complex and co-localize in DNA-damage-induced foci, they do not exist as a pre-formed holocomplex in the nucleoplasm; they assemble ‘on the spot’ when they encounter damage29,30. Sequential assembly from single subunits also occurs during nucleotide-excision repair (NER) and appears more beneficial in terms of repair efficiency and timing than alternative mechanisms, such as random recruitment or pre-assembly of a holocomplex32.
The role of damaged DNA in DDR
The assembly of the DNA-repair machinery in repair foci and the activation of the DDR has generally been assumed to require the presence of DNA lesions. However, recent experiments in yeast and mammalian cells have challenged this seemingly obvious assumption. When repair components were immobilized on chromatin as Lac repressor fusion proteins, a full DDR response, including cell-cycle arrest, was triggered even in the absence of DNA breaks24,26. These observations show that it is not the DNA lesion per se, but rather the nucleation of repair foci upon tethering, that activates transducer kinases. Moreover, the lesion itself is not essential for the integrity of the repair complex24,26. Consistent with the idea of lesion-independent activation of transducer kinases, overexpression of a small domain of the ATR-stimulating TopBP1 protein leads to activation of ATR, cell-cycle arrest and senescence in the absence of DNA breaks33.
The possibility of activation of DDR in the apparent absence of DNA damage is also suggested by the finding that ATM becomes rapidly activated in response to changes in chromatin structure upon exposure of cells to mild hypotonic buffers or treatment with the histone deacetylase (HDAC) inhibitor TSA, which decondenses chromatin globally34. Furthermore, highly hyper-condensed, compacted mitotic chromosomes activate ATM and DNA-damage checkpoints in the absence of apparent DNA lesions35. DNA-damage signalling pathways and ATM are also activated by hypoxia, presumably in the absence of lesions36. Together, these observations raise the intriguing possibility that repair proteins do not probe the genome solely for DNA breaks, but possibly also for changes in chromatin structure.
DNA repair in the context of chromatin
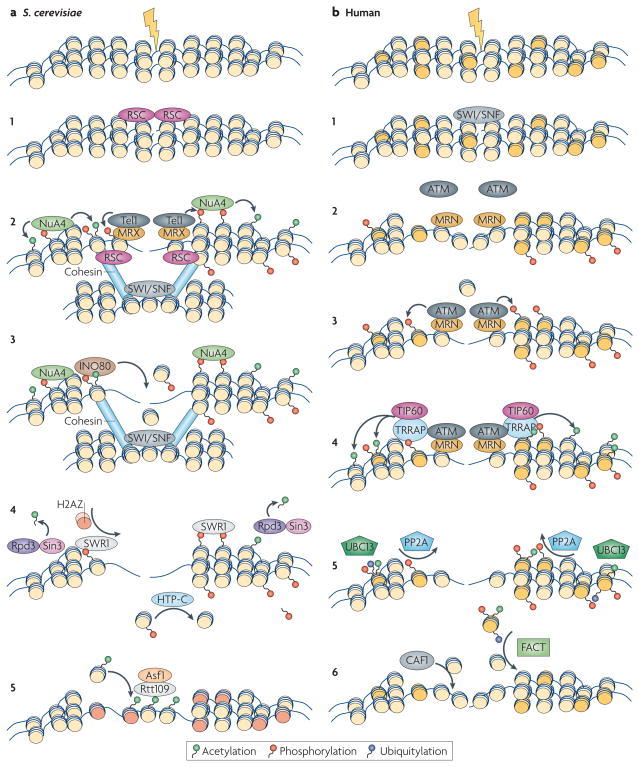
DNA repair occurs in the context of highly structured chromatin (FIG. 3). Emerging evidence suggests that the ability of repair factors to detect DNA lesions and to be retained efficiently at breaks is determined by histone modifications around the DSBs and involves chromatin-remodelling events37.
Figure 3. Chromatin events during DNA repair.
a | Saccharomyces cerevisiae. a1,2 | Chromatin remodelling makes the chromatin more accessible to the MRX (Mre11–Rad50–Xrs2) sensor complex and the transducer kinases, which phosphorylate H2A. Subsequently, the NuA4 histone acetyltransferase (HAT) complex is recruited by phospho-H2A and induces H2A and H4 acetylation. The SWI/SNF complex remodels the donor template, and phospho-H2A together with the RSC remodelling complex attracts cohesin, which facilitates sister chromatin pairing. a3 | The INO80 complex is recruited through its affinity for phospho-H2A and evicts nucleosomes, allowing resection. a4 | Several steps of chromatin restoration occur after double-stranded DNA break (DSB) repair. The Rpd3–Sin3 complex removes acetylation marks from histone H4. SWR1 removes phospho-H2A and substitutes it with the histone variant H2AZ (orange). The HTP-C phosphatase complex removes the phospho-group from the evicted H2A. a5 | The Asf1–Rtt109 chromatin assembly complex restores nucleosome structure around the break. b | Human. b1,2 | Recruitment of chromatin remodelling activities, including the SWI/SNF complex, increases the accessibility of the DSB to the sensor complex MRN (MRE11–RAD50–NBS1). b3 | Recruitment of MRN results in eviction of histone H2B (light yellow), recruitment of ATM and phosphorylation of H2AX (dark yellow). b4 | TRRAP facilitates the recruitment of the TIP60 HAT complex, which results in the acetylation of H2AX and H4. b5 | After repair, acetylated γ-H2AX is ubiquitylated via UBC13, facilitating the eviction of γ-H2AX, and the PP2A phosphatase complex dephosphorylates γ-H2AX. b6 | The chromatin-assembly factor CAF1 promotes the incorporation of a new H3–H4 dimer, and the FACT complex exchanges the variant histone dimer γ-H2AX–H2B with new dimer H2A–H2B.
Covalent chromatin modifications in DNA repair
DNA repair involves an extensive set of histone modifications (BOX 2). The most prominent DNA-damage-induced histone modification is the phosphorylation of the C-terminal tail of mammalian H2AX (H2A in yeast and H2Av in Drosophila melanogaster), referred to as γ-H2AX38,39 (FIG. 3). H2AX constitutes 2–25% of total H2A, depending on cell type, and H2AX can be phosphorylated by three transducer kinases — ATM, DNAPK and ATR. Mammalian cells that are deficient in H2AX show sensitivity to ionizing irradiation, elevated genomic instability and defects in sister-chromatin recombination and class-switch recombination40–44. The primary function of γ-H2AX seems to be a platform to attract and retain repair proteins. Although some factors such as MDC1 recognize γ-H2AX directly11,45, others, such as NBS1 and 53BP1, initially sense breaks through an H2AX-independent mechanism. But their efficient accumulation and retention in DSB chromatin ultimately requires γ-H2AX26,46. Consistent with a key role of H2AX in DDR, H2AX−/− mouse embryonic fibroblasts are defective in triggering the G2–M checkpoint after low doses of irradiation, and H2A is also important in checkpoint function in budding yeast47,48. γ-H2AX also acts as a recognition signal for several chromatin remodelling and structural proteins, including INO80, NuA4, Swr1 and cohesin in S. cerevisiae49–53. Ubiquitylation of H2AX is also emerging as a key step in foci formation and DDR signalling16–21 (BOX 2).
Box 2. Histone modifications during DNA repair.
Histone acetylation
Mutations in the N-terminal tail of yeast and mammalian histones H4 and H3 result in cell-cycle arrest, genomic instability and hypersensitivity to DNA-damaging agents107–110. Several histone acetyl transferases (HATs) and histone deacetylases (HDACs) interact with double-stranded DNA breaks (DSBs) and covalently modify H3 and H4 histone tails upon DNA damage, including the HAT TIP60 (which acetylates residues K5, K8, K12 and K16 of histone H4 and facilitates homologous recombination repair (HRR))111,112 and the HAT MOF113 (which acetylates H4K16). In yeast, recruitment of Esa1, the HAT subunit of the NuA4 complex, results in acetylation of residues K5, K8, K12 and K16 of H4 (REF. 107). Moreover, the HATs HAT1 and GCN5 interact with DSBs and contribute to H3 and H4 modifications after DNA damage66,109,110. H3K56 acetylation is mediated by the yeast HAT Rtt109 and occurs on newly synthesized histone H3, which is incorporated into chromatin during S phase and is important for DDR signalling114,115 and histone reassembly after DNA repair75.
Histone methylation
H3K79 methylation by DOT1 in both yeast and mammalian cells is not caused by damage, but its increased accessibility at DSBs seems to be important for the recruitment of 53BP1 (Rad9 in Saccharomyces cerevisiae)24,116,117. 53BP1 has an affinity for methylated histone H4 at residue K20 that is mediated by the histone methyltransferase PR-SET7 (REF. 118), and PR-SET7 appears to be crucial for the maintenance of genome integrity and S-phase progression119,120. Both phosphorylation of the fission yeast H2A and methylation of H4K20 by the methyltransferase Set9 are necessary for the initial recruitment of Crb2 (REFS 121,122).
Histone phosphorylation
In addition to phosphorylation of H2AX (see main text), histone H2B can be phosphorylated at residue S14 upon γ-irradiation in a γ-H2AX-dependent manner123. In S. cerevisiae, H2B is phosphorylated at residue S10 as a result of apoptotic stimuli124.
Histone ubiquitylation
The ubiquitin ligase RNF8 is recruited to DSBs by MDC1 and monoubiquitylates H2AX and H2A. RNF8 then serves as a binding site for the E2-conjugating enzyme UBC13, which catalyses K63-linked polyubiquitylation of histones H2A and γ-H2AX. Ubiquitylation of H2AX and H2A is also crucial for the recruitment and the retention of the downstream factor 53BP1 (REFS 16–21).
Several other histone modifications, including acetylation, methylation and phosphorylation of most core histones, have been linked to various aspects of DNA repair by facilitating access to the chromatin fibre, recruiting repair factors and chromatin-remodelling components, and propagating cell-cycle checkpoint signals (BOX 2).
Chromatin remodelling in DNA repair
In response to DNA damage, chromatin undergoes rapid local and global decondensation54,55, a process that has been proposed to facilitate genome surveillance by enhancing access of DDR proteins to sites of damage (FIG. 3). Repair-associated chromatin remodelling is carried out by chromatin- remodelling complexes, which use ATP hydrolysis to remove histones or alter their conformation.
While chromatin remodelling is probably important at multiple points in the repair process, it seems to be particularly involved in the late stages of repair. A yeast remodelling complex, RSC, binds early after damage and is thought to facilitate the break detection by mobilizing nucleosomes around the break56,57, whereas most chromatin remodellers bind later. In yeast, recruitment of the INO80 complex occurs late and is strictly dependent on phosphorylation of H2A49,51,53. INO80 is thought to remove nucleosomes around the DSB to facilitate resection, ssDNA formation and recruitment of Rad51 (REFS 49,51,53,58). The yeast SWR1 remodelling complex, another member of the INO80 family, has a similar mode of recruitment to that of the INO80 complex, but a distinct role. Although it is recruited to DSBs by phosphorylated H2A, SWR1 also has high affinity for H2AZ and can specifically replace H2A–H2B with H2AZ–H2B59,60. Interestingly, SWR1 seems to antagonize INO80, which contributes to escape from cell-cycle checkpoint arrest61. Whereas SWR1 functions in error-free NHEJ, INO80 acts predominantly in Mec1-dependent checkpoint activation62. In mammalian cells, the SWI/SNF complex seems to have a similar function as the RSC complex during the early stages of DSB repair. Inactivation of SWI/SNF results in reduced phosphorylation of H2AX and in efficient repair, suggesting that the remodelling activity of SWI/SNF is needed in the early steps of DSB repair to promote the phosphorylation of H2AX63.
Chromatin reconstitution after DNA repair
Repair of DSBs not only involves the restoration of genetic information by faithful replacement of the correct nucleotides, it also entails faithful re-establishment of the original chromatin organization around the break (FIG. 3). This serves to turn off the activated checkpoint signalling and to allow normal chromatin function. Two mechanisms are responsible for post-repair chromatin reconstitution: recruitment of chromatin-modifying enzymes with the opposite activity of those acting during repair, and the eviction and replacement of modified histones.
Reversal of repair-associated histone modifications is primarily carried out by HDACs. Hst4 in fission yeast (a homologue of the HDAC Sir2) and the Rpd3, Sir2 and Hst1 HDACs in budding yeast are essential for viability following DSB repair, and they are speculated to function in post-repair restoration of the acetylation status of chromatin64–66. Similarly, the HTP-C (histone H2A phosphatase) complex in S. cerevisiae67 and the PP2A and PP4 phosphatase complexes in mammalian cells reverse the phosphorylation of γ-H2AX68,69. Interestingly, the PP4 phosphatase specifically dephosphorylates ATR-mediated γ-H2AX that is generated during DNA replication69, pointing to the existence of distinct mechanisms of chromatin restorations following different types of DSBs.
An emerging idea for restoring normal chromatin structure after DNA repair is the replacement of core histones. In budding yeast, the remodelling complex SWR1, in cooperation with the NuA4 histone acetyl transferases (HAT) complex, deposits the histone-variant dimer H2AZ–H2B by replacing the pre-existing phospho- H2A–H2B at sites of damage56,59,61,70. In D. melanogaster and mammals, the TIP60 complex is responsible for acetylation of γ-H2AX and its eviction from the sites of damage50,71,72. Recently, the FACT complex, a multi- component complex that was identified based on its ability to facilitate transcription in chromatin, was also suggested to catalyse the exchange of γ-H2AX with H2A in nucleosomes at DSB chromatin73. But histone exchange is not limited to the H2AX–H2B dimer. In budding yeast, whole nucleosomes are depleted from a region that spans at least 2 kb on either side of a DSB58. In mammalian cells, a new H3–H4 dimer is incorporated during repair of ssDNA breaks and DSBs, and this exchange depends largely on the chromatin-assembly factor 1 (CAF1)74. Other general chromatin-assembly factors also participate in post-repair chromatin reassembly. In yeast, the histone chaperone anti-silencing function 1 (Asf1) together with Rtt109-dependent H3 acetylation is important for nucleosome reassembly at regions around DSBs following repair75–77. Furthermore, there is evidence that ASF1 works synergistically with CAF1 in nucleosome formation during NER in mammalian cells78.
Not all epigenetic changes that are induced by DNA damage are reversible79,80. Breaks induced at a CpG island-containing promoter result in DNA methylation and occasional heritable gene silencing79. Moreover, oxidative stress and DNA damage leads to a global redistribution of the chromatin modifier SIRT1, and it has been speculated that this might lead to permanent changes to the chromatin structure at sites of repair and to stable transcriptional deregulation, which is often observed in tumours80.
Higher-order chromatin structure and DNA repair
The importance of chromatin structure in DDR and DNA repair raises the question of whether breaks in different chromatin environments, particularly heterochromatin or euchromatin, are recognized and repaired with similar efficiency and using the same repair machinery and pathways. A hint of the importance of chromatin context in repair efficiency comes from the recent observation that DSBs in heterochromatic regions are characterized by slower repair kinetics than those in euchromatic regions81. The simplest explanation for the reduced repair efficiency in heterochromatin is that the repair machinery has less access to the highly compact hetero-chromatin regions. Supporting this model, the elimination of KAP1, a transcriptional co-repressor associated with heterochromatin, or heterochromatin protein 1 (HP1) — both architectural components of hetero-chromatin — alleviate the repair delay81. Importantly, the release of KAP1 is dependent on ATM, for which it is a direct substrate, suggesting that the removal of architectural chromatin proteins that are inhibitory to the repair process is yet another function of ATM55,81.
Removal of inhibitory architectural proteins is not limited to heterochromatin regions. Upon DNA damage by irradiation, the heterochromatin protein HP1β becomes more loosely bound in both euchromatin and heterochromatin and dissociates from sites of damage82. Its dispersion is dependent on its phosphorylation at residue T51 by casein kinase 2 and is also important for proper DDR signalling82. Along the same lines, loss of the nucleosome-binding protein HMGN1 affects optimum activation of DDR upon γ-irradiation83. HMGN1 mediates the irradiation-induced global acetylation of histone H3 on residue K14, and this modification is crucial for ATM autophosphorylation and activation. In the absence of breaks, HMGN1 decreases the affinity of ATM to chromatin, possibly preventing inappropriate and ectopic DDR activation83.
The impact of chromatin compaction in genome surveillance is also evident in embryonic stem cells (ESCs) from transgenic mice with reduced amounts of the linker histone H1, and thus less compacted chromatin. These cells exhibit high resistance to DNA-damaging agents as a result of an enhanced cellular response to DSBs84. Breaks that are induced in this environment seem to be more promptly sensed by DDR factors, suggesting their increased accessibility to the lesion. Moreover, the strength of the DDR signal that is generated at each break site is enhanced, suggesting that DDR is amplified in the context of open chromatin84. Similar results were obtained in cells that were treated with the deacetylase inhibitor TSA, which leads to global chromatin decondensation84. Consistent with these findings, heterochromatin is refractory to efficient γ-H2AX spreading from breaks that are induced in adjacent euchromatic regions85. These results demonstrate that chromatin structure influences the repair process and that these proteins may help fine-tune the cellular response to DDR. More importantly, observations on ESCs suggest that differential responsiveness to DNA damage might be of significant physiological relevance in genome maintenance. Each ESC must maintain its genome in a pristine state so as not to pass on damage to its offspring lineages. Interestingly, consistent with heightened response and increased signal strength in decondensed chromatin, ESCs have globally decompacted chromatin86, providing a possible mechanism for maintaining intact genomes in these universal progenitor cells.
Repair in the context of nuclear architecture
Recent observations suggest that the non-random organization of the nucleus affects where repair occurs, how it is organized in the nuclear space and how chromosomal translocations form in vivo.
Nuclear organization of DNA repair
A key feature of the nucleus of eukaryotic cells is the high degree of compartmentalization5. The mammalian nucleus contains many functionally distinct subnuclear bodies, which are thought to segregate distinct nuclear functions and thereby increase efficiency5. And, the major nuclear processes of transcription and replication occur in spatially limited regions of the nucleus where transcription and replication factors are concentrated5. The global compartmentalization of the nucleus raises the questions of whether repair can occur anywhere in the nucleus, whether repair efficiency is affected by the location of the lesion and whether dedicated repair centres exist that are optimized to increase repair efficiency.
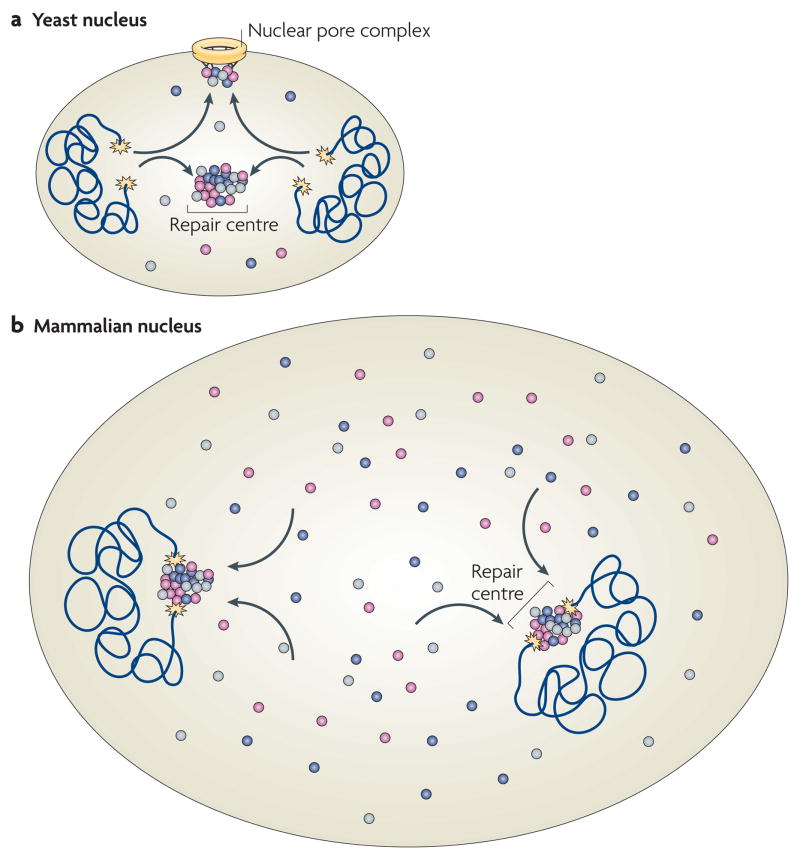
Observations in yeast suggest that distinct, dedicated DNA-repair centres exist as preferential sites of repair (FIG. 4a). Upon induction of multiple DSBs by irradiation, typically only a few DNA-repair foci form in which all lesions appear to be repaired87–89. More tellingly, although irradiation at a low dose leads to the formation of 2–4 repair foci, subsequent irradiation with higher doses to induce more lesions does not increase the number of foci, suggesting that the newly formed lesions are recruited to pre-existing repair centres in which multiple lesions are repaired87–89. When two independent DNA lesions are followed in yeast using fluorescent tagging near the break sites, they generally co-localize in foci that contain the HRR factor Rad52, further suggesting they are recruited to shared DNA-repair centres88. It has been suggested that centralized repair centres serve as a scaffold to facilitate the capture and alignment of a broken chromosome end with its template for HRR, although it could be argued that bringing multiple unrepaired broken chromosomes into spatial proximity might be dangerous as it increases the likelihood of illegitimate end-joining among chromosomes. Further evidence for spatially restricted repair in yeast comes from the observation that persistently unrepaired DSBs migrate from their internal nuclear positions to the nuclear periphery, where they associate with the pore, which might facilitate and coordinate repair90.
Figure 4. DNA repair in the context of nuclear architecture.
a | In yeast, double-stranded DNA breaks (DSBs; yellow stars) associate with repair centres, which serve multiple DSBs. Persistent breaks associate with the nuclear pore complex. b | In mammalian cells, DSBs are largely immobile and repair factors associate on them from a diffusible pool. Multiple DSBs in the same nucleus do not appear to coalesce into repair centres. The differences in the spatial organization of DSB repair are probably due to the smaller size of the yeast nucleus, which allows for multiple breaks to associate with each other due to the diffusional motion of chromatin. Distances in mammalian cells are too large to allow for efficient pairing of multiple DSBs on the basis of diffusional chromatin mobility. Not drawn to scale.
Although these observations suggest preferential localization of DNA repair in yeast, the situation seems different in mammalian cells. Broken chromosome ends visualized by fluorescent tags are essentially immobile in the nuclear space, preventing them from migrating to a preformed repair site or to the nuclear periphery91 (FIG. 4b). Furthermore, in contrast to the situation in yeast, multiple DSBs on several chromosomes do not coalesce on shared repair centres in mammalian cells91. These observations on targeted single DSBs are supported by earlier findings demonstrating that irradiation-induced globally damaged regions undergo no or only limited motion in the nucleus, ruling out their coalescence into shared repair sites54,92,93. Positional immobility might, however, not apply to all types of mammalian DNA damage. The mobility of defective, deprotected telomeres, which are repaired by NHEJ, has recently been found to be increased compared to intact telomeres94. Higher mobility of these defective telomeres, as well as joining of distant sites on the same chromosome during V(D)J recombination95, is dependent on 53BP1 and correlates with increased repair efficiency, although it is unclear whether, or how, 53BP1 contributes directly to chromatin motion or whether it exerts its effect by other means.
The distinct organization of DNA repair between yeast and mammalian nuclei is probably explained by the different nature of nuclear architecture in the two organisms. The yeast nucleus largely lacks the complex internal organization of the mammalian nucleus and the organization of chromatin in yeast appears to be more plastic5. More importantly, the yeast nucleus is roughly one- tenth of the diameter and thus one-thousandth of the volume of the mammalian nucleus, although the ability of chromatin to move in the nucleus is similar in both organisms. In both cases, chromatin undergoes limited, locally constrained diffusional motion with a radius of ~1 μm (REF. 96). Although in yeast this degree of motion is sufficient for a damaged chromatin region to rapidly search the entire nuclear volume for a pre-existing DNA-repair site, in mammalian cells it only allows sampling of a small subfraction of the nucleus and the search would therefore probably be unsuccessful unless many pre-formed repair foci existed. The same applies to the migration of a damaged locus to the nuclear pore. Although this can occur rapidly in yeast, the limited ability of chromatin to move over long distances in mammalian cells may prevent the efficient recruitment of an unrepaired site to the nuclear pore96. It thus seems that differential nuclear organization in lower and higher eukaryotes has led to the evolution of distinct ways of spatially organizing DNA repair.
Spatial genome organization and chromosome translocations
One of the most fundamental properties of the mammalian cell nucleus is the non-random arrangement of the genome in the nuclear space97,98. The non-randomness of spatial genome organization has recently emerged as a contributor to determining the outcome of chromosome translocations99. Striking evidence for a role of spatial genome organization in the formation of chromosome translocation came from correlative studies on the location of frequent translocation partners99. Murine lymphoma cells are characterized by the presence of various combinations of translocations between chromosomes 12, 14 and 15 (REF. 100). When the positions of these chromosomes were mapped in the inter-phase nucleus of normal lymphocytes, they were found to form highly non-random clusters, bringing them into close spatial proximity, and presumably favouring illegitimate rejoining101. Similar observations were reported for several other translocations including chromosomes 5 and 6 in mouse hepatomas102 and BCR–ABL t(9;22) and PML–RAR t(15;17) in humans103. A role for spatial proximity in determining translocation partners is even more strongly supported by observations in Burkitt’s lymphoma, in which MYC translocates with different frequencies with the IGH, IGκ or IGλ locus. Mapping of the position of these loci reveals that the distance of these loci from MYC corresponds well with their translocation frequency104. Furthermore, detailed visualization studies have demonstrated that there is a correlation between translocation frequency of chromosomes and the degree by which their genetic material physically intermingles105.
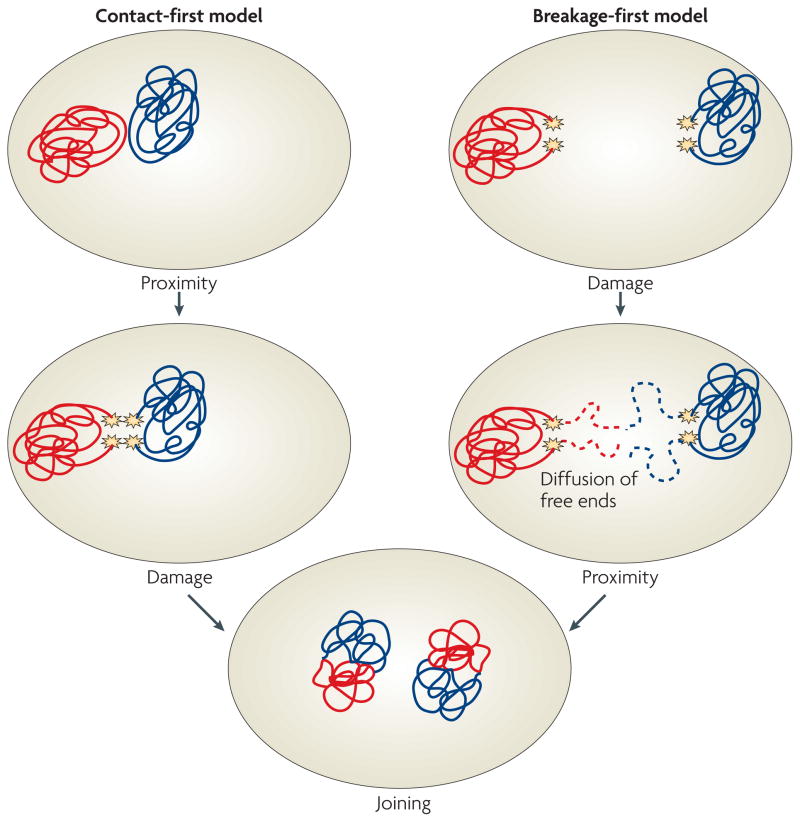
These observations point to a model in which translocations preferentially occur between chromosomes that are in close spatial proximity in the interphase nucleus due to their non-random arrangement. This scenario is generally referred to as the contact-first model (FIG. 5). However, an alternative possibility exists in which, as in yeast, DSBs occur first and the broken ends diffuse through the nucleus until they find other unrepaired DSBs with which they form a translocation (FIG. 5). This breakage-first model, although not strictly ruled out, is unlikely for chromosomal translocations involving DSBs in light of the limited mobility of DSBs in the mammalian nucleus91.
Figure 5. Models of chromosome translocations.
Two models for chromosome translocation formation have been proposed. In the contact-first model (left), translocations occur among chromosomes, which are in spatial proximity to each other due to the non-random organization of the genome in the cell nucleus. Upon concurrent damage of neighbouring chromosomes, the broken chromosome ends are illegitimately joined to form a translocation. In the breakage-first model (right), double-stranded DNA breaks (DSBs) occur first. Upon DNA damage on multiple, distant chromosomes, the broken chromosome ends roam the nuclear space by diffusion. They undergo illegitimate joining when they encounter another DSB. Recent evidence demonstrating the proximity of frequent translocation partners and the immobility of DSBs favour the contact-first model.
The contact-first model is attractive because it explains the well-established preferential occurrence of translocations in tumours from different tissues106. In mice, for example, chromosomes 5 and 6 frequently translocate in hepatoma cells, but not in lymphoma cells, which are often characterized by t(12;15). The arrangement patterns of chromosomes are tissue specific and chromosome 5–6 pairs are often seen in hepatocytes but not in lymphocytes, whereas 12–15 pairs are frequent102. The tissue-specific arrangement of chromosomes thus mirrors their propensity to undergo translocations in a tissue-specific manner, and it seems likely that the tissue-specific proximity of particular chromosomes favours their translocation.
The fact that in a contact-first model the formation of translocations is limited to neighbouring chromosomes and that broken chromosome ends cannot freely diffuse through the nucleus is probably also an important genome-protection mechanism. The inability of broken chromosome ends to roam the nucleus prevents their interaction, and illegitimate joining, with randomly occurring lesions that are present at all times in the nucleus even of unchallenged cells. An important corollary from this scenario is that cellular factors hold broken chromosome ends in place — and in this way limit their ability to move in the nucleus — are important for the protection of genome integrity. Proof-of-principle for the existence of such factors is provided by the finding that knockdown of the DNA-end-binding protein Ku80 leads to a loss of the positional immobility of broken chromosome ends and enables them to roam the nuclear space91.
Conclusions
DNA repair and maintenance of genome stability is fundamentally a cell-biological problem. These processes obviously occur in the context of the highly organized cell nucleus and in the higher-order structure of the chromatin fibre. The accumulation of repair components at sites of DNA damage to form repair foci occurs by recruitment from a freely diffusing pool of factors in the nucleoplasm, and the assembly of the repair machinery requires complex spatial and temporal coordination of repair factor interactions, both with each other and with chromatin. In addition to the repair machinery, the nature of the chromatin in which a DNA lesion occurs and chromatin remodelling events during and after the repair process affect the progress of the repair reaction. Finally, the organization of the repair site in the context of the nuclear space and the mobility of chromatin in the nucleus are emerging as key determinants in repair efficiency and in the maintenance of genome stability.
DNA repair has long been studied using reductionist approaches. The state of knowledge regarding the molecular basis of DNA repair is now developed to a level that we can begin to place it in the context of an intact cell and of higher-order chromatin. Recent efforts using cell-biological approaches and the development of experimental systems to follow the repair process in individual cells in vivo is revealing an entirely new layer of complexity, and presumably regulation, based on spatial and temporal control of repair factors. The next logical and exciting steps are to expand these nascent efforts so that we can fully characterize repair in vivo and link cell-biological properties of DSBs to what we know about the molecular basis of repair. But the most promising avenue of investigation relates to the profound physiological roles of DNA repair. DNA damage is central to major biological processes, such as cancer, maintenance of stem cell function and ageing. We can now begin to combine the molecular knowledge of repair with cell-biological approaches to address the key questions of why different cell types respond differently to insult, how the location of repair is related to its efficiency and, most importantly, how the organization of repair in the intact nucleus contributes to preventing loss of genome stability during physiological and pathological processes.
Acknowledgments
We thank V. Roukos for critically reading the manuscript. Misteli’s laboratory is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute and Center for Cancer Research. Soutoglou’s laboratory is supported by the National Centre for Scientific Research and Institute of Genetics and Molecular and Cellular Biology.
Glossary
- D-loop
A DNA structure in which the two strands of a double-stranded DNA are separated by a third strand. It is created after strand invasion during homologous recombination
- Holliday junction
A mobile junction between four strands of DNA, with homology between pairs of neighbouring DNA molecules
- Histone variant
A core histone other than the classical histones (H2A, H2B, H3 or H4)
- Chromatin-remodelling complex
A protein complex that alters the higher-order structure of the chromatin fibre by changing its compaction or repositioning nucleosomes, often using ATP hydrolysis
- Clamp loader
A protein that loads the clamp complex onto the DNA
- Clamp complex
A protein complex that wraps around DNA and facilitates the processivity of DNA replication
- Nucleotide-excision repair
The repair mechanism that removes damaged single nucleotides, mostly in response to ultraviolet damage
- Histone deacetylase
An enzymatic activity that removes an acetyl group from a histone tail
- Histone acetyl transferase
An enzymatic activity that adds an acetyl group to a histone tail
- Heterochromatin
Compact, closed chromatin that generally contains silenced genome regions
- Euchromatin
Decondensed, open chromatin that generally contains transcriptionally active genome regions
- Histone methyltransferase
An enzymatic activity that adds a methyl group to a histone tail
- V(D)J recombination
A recombination reaction that assembles genes encoding diverse T-cell receptor and immunoglobulin molecules from variable (V), diversity (D) and joining (J) gene segments. V(D)J recombination is necessary for the recognition of diverse foreign antigens
Footnotes
DATABASES
UniProtKB: http://ca.expasy.org/sprot
53BP1 | ATR | BRCA1 | CHK1 | CHK2 | HMGN1 | Hst1 | Hst4 | Ku70 | Ku80 | MDC1 | Mec1 | Mec3 | MRE11 | NBS1 | Rad17 | Rad24 | RAD50 | RAD51 | RAD52 | Rpd3 | Rtt109 | Sir2
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
Tom Misteli, Email: mistelit@mail.nih.gov.
Evi Soutoglou, Email: evisou@igbmc.fr.
References
- 1.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 2.Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nature Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 3.Kanaar R, Wyman C, Rothstein R. Quality control of DNA break metabolism: in the ‘end’, it’s a good thing. EMBO J. 2008;27:581–588. doi: 10.1038/emboj.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair. 2004;3:997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nature Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 7.Paull TT, Lee JH. The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle. 2005;4:737–740. doi: 10.4161/cc.4.6.1715. [DOI] [PubMed] [Google Scholar]
- 8.Petrini JH, Stracker TH. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- 9.Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:E110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11–Rad50–Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 11.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melander F, et al. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spycher C, et al. Constitutive phosphorylation of MDC1 physically links the MRE11–RAD50–NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci USA. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 19.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 20.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. A comprehensive analysis of the temporal sequence of recruitment events in S. cerevisiae. [DOI] [PubMed] [Google Scholar]
- 23.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 24.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. Introduces the concept of activation of the DDR in the absence of DNA breaks in S. cerevisiae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soutoglou E. DNA lesions and DNA damage response: Even long lasting relationships need a “break”. Cell Cycle. 2008;7:3653–3658. doi: 10.4161/cc.7.23.7178. [DOI] [PubMed] [Google Scholar]
- 26.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. Introduces the concept of activation of the DDR in the absence of DNA breaks in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukas C, Bartek J, Lukas J. Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma. 2005;114:146–154. doi: 10.1007/s00412-005-0011-y. [DOI] [PubMed] [Google Scholar]
- 28.Bekker-Jensen S, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Essers J, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukas C, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol Cell. 2008;30:486–497. doi: 10.1016/j.molcel.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Politi A, et al. Mathematical modeling of nucleotide excision repair reveals efficiency of sequential assembly strategies. Mol Cell. 2005;19:679–690. doi: 10.1016/j.molcel.2005.06.036. Reports the development of a mathematical model for the assembly process of the DNA-repair machinery in vivo. [DOI] [PubMed] [Google Scholar]
- 33.Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. Shows that a persistent DDR in the absence of breaks leads to cellular senescence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 35.Mazumdar M, et al. Tumor formation via loss of a molecular motor protein. Curr Biol. 2006;16:1559–1564. doi: 10.1016/j.cub.2006.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bencokova Z, et al. ATM activation and signalling under hypoxic conditions. Mol Cell Biol. 2008;29:526–537. doi: 10.1128/MCB.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Downs JA, Nussenzweig MC, Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 38.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 40.Bassing CH, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 41.Celeste A, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. Demonstrates the functional importance of H2AX phosphorylation in genome maintenance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie A, et al. Control of sister chromatid recombination by histone H2AX. Mol Cell. 2004;16:1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 46.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 47.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez-Capetillo O, et al. DNA damage-induced G2–M checkpoint activation by histone H2AX and 53BP1. Nature Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- 49.Downs JA, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 51.Morrison AJ, et al. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 52.Unal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 53.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 54.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 56.Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev. 2007;17:126–131. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobor MS, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 61.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park JH, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haldar D, Kamakaka RT. Schizosaccharomyces pombe Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryot Cell. 2008;7:800–813. doi: 10.1128/EC.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jazayeri A, McAinsh AD, Jackson SP. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci USA. 2004;101:1644–1649. doi: 10.1073/pnas.0304797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keogh MC, et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 68.Chowdhury D, et al. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 69.Chowdhury D, et al. A PP4-phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krogan NJ, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ikura T, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heo K, et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 74.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 75.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mousson F, Ochsenbein F, Mann C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma. 2007;116:79–93. doi: 10.1007/s00412-006-0087-z. [DOI] [PubMed] [Google Scholar]
- 77.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 78.Mello JA, et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goodarzi AA, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. Provides evidence for a role of higher-order chromatin structure in DDR activation. [DOI] [PubMed] [Google Scholar]
- 82.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 83.Kim YC, et al. The activation of ATM depends on chromatin interactions occurring prior to DNA damage induction. Nature Cell Biol. 2008;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murga M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. Provides evidence for an inhibitory effect of chromatin compaction on DNA-damage signalling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Efroni S, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lisby M, Antunez de Mayolo A, Mortensen UH, Rothstein R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle. 2003;2:479–483. [PubMed] [Google Scholar]
- 88.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 89.Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr Opin Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 90.Nagai S, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Soutoglou E, et al. Positional stability of single double-strand breaks in mammalian cells. Nature Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. Reports the development and use of an experimental system to visualize DSBs in living mammalian cells and demonstrates that mammalian DSBs are immobile in the cell nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aten JA, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 93.Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- 94.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. Shows that 53BP1 promotes chromatin mobility and end joining of deprotected telomeres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soutoglou E, Misteli T. Mobility and immobility of chromatin in transcription and genome stability. Curr Opin Genet Dev. 2007;17:435–442. doi: 10.1016/j.gde.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 98.Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- 99.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liyanage M, et al. Abnormal rearrangement within the α/β T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]
- 101.Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol. 2002;12:1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- 102.Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. The nuclear topography of ABL, BCR, PML, and RARα genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93:1197–1207. [PubMed] [Google Scholar]
- 104.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nature Genet. 2003;34:287–291. doi: 10.1038/ng1177. Demonstrates a correlation between the spatial proximity of genome regions and their likelihood of translocating, pointing to a significant role of non-random genome organization in determining translocation outcome. [DOI] [PubMed] [Google Scholar]
- 105.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. Demonstrates physical intermingling of the chromatin fibres of neighbouring chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nature Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 107.Bird AW, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 108.Megee PC, Morgan BA, Smith MM. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 109.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qin S, Parthun MR. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol Cell Biol. 2006;26:3649–3658. doi: 10.1128/MCB.26.9.3649-3658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Murr R, et al. Histone acetylation by Trrap–Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 112.Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- 113.Gupta A, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 116.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 117.Wysocki R, et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Houston SI, et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jorgensen S, et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 123.Fernandez-Capetillo O, Allis CD, Nussenzweig A. Phosphorylation of histone H2B at DNA double-strand breaks. J Exp Med. 2004;199:1671–1677. doi: 10.1084/jem.20032247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ahn SH, Henderson KA, Keeney S, Allis CD. H2B (Ser10) phosphorylation is induced during apoptosis and meiosis in S. cerevisiae. Cell Cycle. 2005;4:780–783. doi: 10.4161/cc.4.6.1745. [DOI] [PubMed] [Google Scholar]